Abstract
The activated amino acid response (AAR) and unfolded protein response (UPR) stress signaling pathways converge at the phosphorylation of translation initiation factor eIF2α. This eIF2α modification suppresses global protein synthesis but enhances translation of selected mRNAs such as that for activating transcription factor 4 (ATF4). An ATF4 target gene, SNAT2 (system A sodium-dependent neutral amino acid transporter 2), contains a C/EBP-ATF site that binds ATF4 and triggers increased transcription during the AAR. However, the present studies show that despite increased ATF4 binding to the SNAT2 gene during UPR activation in HepG2 human hepatoma cells, transcription activity was not enhanced. Hyperacetylation of histone H3 and recruitment of the general transcription factors at the HepG2 SNAT2 promoter occurred in response to the AAR but not the UPR. In contrast, the UPR did enhance transcription from a plasmid-based reporter gene driven by a SNAT2 genomic fragment containing the C/EBP-ATF site. Simultaneous activation of the AAR and the UPR pathways revealed that the UPR actually suppressed the increased SNAT2 transcription by the AAR pathway, demonstrating that the UPR pathway generates a repressive signal that acts downstream of ATF4 binding.
A wide variety of stress signals activate one or more of a set of eukaryotic initiation factor 2α (eIF2α)2 kinases (1). Phosphorylation of the translational initiation factor eIF2α at serine 51 by these kinases provokes a suppression of global protein synthesis and a paradoxical increase in the translation of selected mRNAs containing short upstream opening reading frames, including that of activating transcription factor 4 (ATF4) (2, 3). One of the eIF2α kinases is double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK), which is activated by ER stress conditions, such as perturbation of calcium homeostasis, glucose deprivation, or other causes of misfolded protein accumulation in the ER lumen. Experimentally, the drugs tunicamycin (Tm), an inhibitor of N-glycosylation, or thapsigargin (Tg), an ER Ca2+-ATPase blocker, can also be used to induce the UPR (4). PERK is one component of an adaptive response known as the unfolded protein response (UPR), which is comprised of three signal transduction pathways mediated by the ER membrane-resident stress sensors PERK, IRE1, and ATF6 (4–8). In contrast to the UPR, amino acid deprivation leads to an increase in uncharged tRNA, which binds to and activates the eIF2α kinase GCN2 (9, 10). This cascade of events is called the amino acid response (AAR) pathway.
Activation of either PERK or GCN2 leads to the induction of specific target genes, as a result of increased ATF4 synthesis (1). Amino acid deficiency in yeast results in an increase in GCN4 (the yeast counterpart to ATF4) translation affecting the transcription of hundreds of genes, which has led Hinnebusch and co-workers (11, 12) to characterize it as a “master regulator.” Array analysis in yeast and mouse revealed overlapping targets for GCN4 and ATF4, including genes involved in amino acid transport and metabolism (13). ATF4 is a member of the ATF subfamily of the basic leucine zipper (bZIP) factor superfamily (14). ATF members are known to heterodimerize within the ATF group as well as with other bZIP transcription factors, including members of the C/EBP family. However, ATF4 also interacts with non-bZIP proteins, including general transcription factors, RPB3 subunit of RNA polymerase II, ribosomal S6 kinase 2 (RSK2), and runt-related transcription factor 2 (RUNX2) (15–17). The interactions of ATF4 with its partners appear to be both stress-specific and tissue-specific, and its up-regulation represents a major component of the adaptive AAR and UPR stress responses. ATF4-dependent activation of transcription is mediated through binding to a C/EBP-ATF composite genomic element that is made up of half-sites for the C/EBP and ATF families, respectively (18).
Among the target genes up-regulated by ATF4 is SNAT2 (system A sodium-dependent neutral amino acid transporter 2). Both the expression of SNAT2 gene and its transport activity are up-regulated during amino acid deprivation, hypertonic stress, or hormonal stimulation (19–21). SNAT2 activity in the liver is induced by glucagon, and its role in supplying alanine and other gluconeogenic amino acids is likely to contribute to the excessive glucose biosynthesis in insulin-dependent diabetes (22). In addition, system A transport is elevated during the cell cycle and is constitutively high in nearly all transformed cells and tissues (23). Its adaptive regulation by substrate supply and hormones, as well as its increased expression in transformed cells and its role in diabetes, makes SNAT2 a potentially attractive therapeutic target.
Another ATF4-regulated gene is ASNS (asparagine synthetase). The two cis-acting elements, nutrient sensing response elements 1 and 2 (NSRE-1 and NSRE-2), in the proximal promoter of human ASNS mediate the transcriptional activation of the gene by either the AAR or the UPR pathway (24, 25). The NSRE-1 sequence is a C/EBP-ATF composite site that binds ATF4 following activation of either the AAR or the UPR (24, 26, 27). In contrast, the ATF4-responsive enhancer element in the SNAT2 gene is composed of a single 9-bp intronic sequence (5′-TGATGCAAT-3′) that is also a C/EBP-ATF composite site but differs in sequence by 2 bp from the ASNS NSRE-1 (5′-TGATGAAAC-3′) (28). Although ATF4 binding to the SNAT2 C/EBP-ATF site has been documented during AAR activation (29), whether or not there is ATF4 binding to SNAT2 during UPR activation has not been investigated. It is interesting to note that despite the increased ATF4 synthesis known to occur during the UPR and the presence of an ATF4-responsive C/EBP-ATF composite site within the SNAT2 gene, the cellular SNAT2 mRNA content and transport activity are not induced in response to UPR activation (30).
This study was designed to explore the differences in the mechanisms for transcriptional control of the SNAT2 gene during UPR and AAR activation. Three questions were addressed. 1) Does ATF4 bind to the SNAT2 C/EBP-ATF composite site during the UPR? 2) Is ATF4 binding to the C/EBP-ATF site the determinant event that induces SNAT2 gene transcription? 3) Are other components of the general transcriptional machinery assembled on the SNAT2 gene during the UPR? The experiments revealed that SNAT2 transcriptional activity remains at the basal level in the presence of ER stress despite increased synthesis of ATF4 and its subsequent enhanced binding to the SNAT2 C/EBP-ATF composite site. Chromatin immunoprecipitation (ChIP) analysis revealed no increase in histone H3 acetylation or general transcription factor (GTF) recruitment to the SNAT2 promoter following activation of the UPR pathway. Simultaneous activation of both pathways indicated that the UPR generates a suppressive signal that blocks the AAR-induced SNAT2 transcription activity downstream of ATF4 binding.
MATERIALS AND METHODS
Antibodies—The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA): ATF3, sc-188; ATF4 (CREB-2), sc-200; C/EBPβ, sc-150, sc-7962; RNA pol II, sc-899; TFIID (TBP), sc-204; TFIIB, sc-274; TFIIE-α, sc-237; and normal rabbit IgG, sc-2027. An ATF4 rabbit antiserum produced by Cocalico Biologicals (Reamstown, PA) was used for ATF4 protein immunoblotting. Antibodies against acetylated histone H3 (number 06-599, recognizes acetylated Lys-9 and Lys-14) and acetylated histone H4 (number 06-866, recognizes acetylated H4 at Lys 5, -8, -12, and -16) were purchased from Upstate Biotechnology, Inc. (Millipore).
Cell Culture—HepG2 human hepatoma cells and BNL-CL2 immortalized mouse fetal hepatocytes were cultured in modified Eagle's minimal essential medium (MEM; pH 7.4) (Mediatech, Herndon, VA) at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. The 293 human kidney cells, human fibroblasts, human MDA breast cancer cells, and mouse embryonic fibroblasts were cultured in Dulbecco's MEM. Both media were supplemented with 1× nonessential amino acids, 2 mm glutamine, 100 mg/ml streptomycin sulfate, 100 units/ml penicillin G, 0.25 mg/ml amphotericin B, and 10% (v/v) fetal bovine serum. Cells were replenished with fresh medium and serum 12 h before all experiments, to ensure that little or no nutrient deprivation occurred prior to initiating drug treatments. To induce the UPR pathway, cells were incubated for a specific time in medium lacking glucose or containing either 300 nm thapsigargin or 5 μg/ml tunicamycin. To activate the AAR, cells were incubated in medium either lacking the amino acid histidine (Invitrogen) or in complete medium supplemented with 1 or 2 mm l-histidinol (HisOH). HisOH blocks charging of histidine onto the corresponding tRNA (31) and thus mimics histidine deprivation thereby triggering activation of the AAR cascade (32, 33). We have documented that HisOH treatment mirrors histidine-deficient medium with regard to induction of transcription (34, 35). For incubations using histidine deprivation, the medium was supplemented with 10% dialyzed fetal bovine serum (Sigma).
Immunoblot Analysis—After incubation in MEM only, MEM + HisOH, or MEM + Tg for 0–12 h, total cell extracts were prepared for immunoblot analysis. Protein content was quantified by a Lowry assay, and 30 μg of protein were separated on a pre-cast Criterion Tris/HCl polyacrylamide gel (Bio-Rad). After electrotransfer to a Bio-Rad nitrocellulose membrane, the membrane was stained with Fast Green to check for equal loading and then incubated with 5% blocking solution (5% (w/v) Carnation nonfat dry milk and Tris-buffered saline/Tween (30 mm Tris base, pH 7.6, 200 mm NaCl, and 0.1% Tween 20)) for 1 h at room temperature with mixing. Immunoblotting for ATF4 was performed using a rabbit serum containing a polyclonal antibody against the ATF4 at a dilution of 1:5000 in 5% dry milk blocking solution for 5 h at room temperature with mixing. The membrane was washed (one time for 5 min and two times for 10 min) in TBS-T solution (30 mm Tris base, pH 7.6, 200 mm NaCl, and 0.1% Tween 20) on a shaker and then incubated with peroxidase-conjugated goat anti-rabbit secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD) at a 1:10,000 dilution in 5% dry milk blocking solution for 1 h at room temperature with mixing. The membrane was then washed (1 × 5 min, 3 × 10 min) in TBS-T solution. The bound secondary antibody was detected using an enhanced chemiluminescence kit (Amersham Biosciences) and exposing the membrane to Biomax® MR film (Eastman Kodak Co.). To provide a demonstration of equal loading beyond the Fast Green staining, some membranes were re-probed with a 1:5000 dilution of an antibody specific for β-actin (Sigma).
Transcriptional Activity Determination—Total RNA was isolated using the Qiagen RNeasy kit (Qiagen), including DNase I treatment before the final elution to eliminate any DNA contamination. To measure the transcriptional activity, qRT-RCR was used to amplify a product spanning the human SNAT2 exon 4 and intron 4 junction, the mouse SNAT2 exon 13 and intron 13 junction, the ASNS intron 12 and exon 13 junction, and the BiP/GRP78 exon 2 and intron 2 junction to measure the short lived unspliced transcript heterogeneous nuclear RNA (hnRNA). This procedure for measuring transcriptional activity is based on that described by Lipson and Baserga (36), except that we analyzed hnRNA levels by quantitative real time PCR (qRT-PCR) using the DNA Engine Opticon 2 system (Bio-Rad) and SYBR Green. Reactions without RT were performed as a negative control to rule out amplification from any residual genomic DNA. These tests were always negative. The transcription activity primers for amplification were as follows: for human SNAT2, sense 5′-GCAGTGGAATCCTTGGGCTTTC-3′ and antisense 5′-CCCTGCATGGCAGACTCACTACTTA-3′; for mouse SNAT2, sense 5′-GTCACCCTCACGGTCCCAGTAGTTA-3′ and antisense 5′-GCATACCCATAGCTGTCGCAGAAGT-3′; for ASNS, sense 5′-CCTGCCATTTTAAGCCATTTTGC-3′ and antisense 5′-TGGGCTGCATTTGCCATCATT-3′; and those for BiP/GRP78, sense 5′-AGGACATCAAGTTCTTGCCGTTCA-3′ and antisense 5′-CACCACCCACCCGTTCTCTAACT-3′. The reaction mixtures were incubated at 48 °C for 30 min followed by 95 °C for 10 min to activate the Taq polymerase and amplification of 35 cycles of 95 °C for 15 s and 63 °C (SNAT2), 58 °C (ASNS), or 60 °C (BiP) for 60 s. After PCR, melting curves were acquired by stepwise increase of the temperature from 55 to 95 °C to ensure that a single product was amplified in the reaction. The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a negative control for the drug-induced stress and as an indicator of the variation for the quantitative real time PCR analysis. The primers used to measure relative mRNA levels for GAPDH were as follows: sense primer 5′-TTGGTATCGTGGAAGGACTC-3′ and antisense primer, 5′-ACAGTCTTCTGGGTGGCAGT-3′. The SNAT2, ASNS, and BiP/GRP78 hnRNA values were determined relative to an RNA standard curve. The RNA was also analyzed for GAPDH mRNA content, which was unchanged by the AAR or UPR pathways, as a control to demonstrate that equal amounts of total RNA were used for hnRNA measurement. The PCRs were performed in duplicate for each sample, and samples were collected from at least three independent experiments. Values are expressed as means ± S.E.
ChIP—ChIP analysis was performed according to our previously published protocol (27). The reaction mixtures were incubated at 95 °C for 10 min, followed by amplification at 95 °C for 15 s and either 60 °C (SNAT2 C/EBP-ATF enhancer primers), 62 °C (SNAT2 promoter primers), or 61.4 °C (ASNS promoter primers) for 60 s for 35 cycles. The SNAT2 promoter primers were as follows: sense primer 5′-GCCGCCTTAGAACGCCTTTC-3′ and antisense primer 5′-TCCGCCGTGTCAAGGGAA-3′. The human SNAT2 C/EBP-ATF enhancer primers were as follows: sense 5′-GGGAAGACGAGTTGGGAACATTTG-3′ and antisense 5′-CCCTCCTATGTCCGGAAAGAAAAC-3′ and the mouse SNAT2 C/EBP-ATF site primers were as follows: sense 5′-ATCGGGTCTTGTGCCTCGAAA-3′ and antisense 5′-ATACCGAGGGGCGATTGATTGT-3′. The SNAT2 primers for exon 10 were as follows: sense 5′-CAGGTACAAGAGCTGTTGGCTGTGT-3 and antisense 5′-GTGTCCTGTGGAAGCTGCTTTGA-3′. The ASNS proximal promoter primers, where the C/EBP-ATF enhancer site is located (24), were as follows: sense 5′-TGGTTGGTCCTCGCAGGCAT-3′ and antisense 5′-CGCTTATACCGACCTGGCTCCT-3′.
Transient Transfections—HepG2 human hepatoma cells were seeded in 24-well plates at a density of 1.2 × 105 cells/well, supplied with complete MEM, and grown for 24 h. Transfection was performed with 1 μg of Firefly luciferase reporter driven by the SNAT2 genomic fragment of nt -512 to +770 that contains both the proximal promoter and the intronic C/EBP-ATF enhancer site (28, 29). To generate a C/EBP-ATF mutant sequence, a plasmid containing the human SNAT2 sequence from nt -512/+770 was used as a template, and mutagenesis was performed with the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The identity and fidelity of the constructs were verified by sequencing. The wild type sequence 5′-TATTGCATCA-3′ was mutated to 5′-TAATGCCCCA-3′, and the mutagenic primers used have been described previously (28). Transfection of 1.2 × 105 cells was accomplished using Superfect reagent at a ratio of 1:6 μg of DNA/μl reagent, according to the manufacturer's protocol (Qiagen, Valencia, CA). Each well also received 10 ng of pRL-SV40 plasmid (Renilla luciferase) to serve as a control for transfection efficiency. The amount of co-transfected transcription factor expression plasmid was 100 ng/well, and the total amount of transfected DNA was kept constant among experimental groups by the addition of empty pcDNA3.1 plasmid. After 3 h, cells were rinsed once with phosphate-buffered saline (PBS) and given fresh MEM. At 16 h post-transfection, the medium was removed; the cells were rinsed once with PBS and incubated for 10 h in 1 ml/well of either complete MEM or MEM lacking histidine (MEM-His) to induce the AAR or MEM lacking glucose or containing 300 nm Tg or 5 μg/ml Tm to induce the UPR, each supplemented with 10% dialyzed fetal bovine serum. After the completion of treatment, the cells were rinsed with PBS, lysed with 100 μl of 1× Passive lysis buffer (Promega), and then subjected to one freeze-thaw cycle to ensure complete disruption of the membranes. Firefly and Renilla luciferase activities were measured using the dual luciferase reporter assay system (Promega). Replicates of six transfections were performed for each experimental condition, and all experiments were repeated with separate batches of cells to ensure reproducibility of results.
Short Interfering RNA (siRNA) Transfection—The human ATF3, XBP1, ATF6α, and ATF6β (CREBL1) siGENOME SMARTpool (Dharmacon M-008663-01-0005, M-009552-02-0005; M-009917-00-0005; M-008805-00-0005), siControl nontargeting siRNA (D-001210-02), and DharmaFECT 4 transfection reagent were purchased from Dharmacon, Inc. (Lafayette, CO). HepG2 cells were seeded in 6-well plates at a density of 4 × 105 cells per well in MEM and grown for 16 h. Transfection was performed according to Dharmacon's instructions using 3 μl of DharmaFECT-4 and 100 nm per well final siRNA concentration. HepG2 cells were treated with transfection reagent for 24 h and then rinsed with PBS, given fresh MEM, and cultured for another 24 h. The medium was then removed and replaced with control MEM or MEM +Tg. At 8 h, protein extracts and total RNA were isolated and analyzed by immunoblotting (described above) or RT-PCR, respectively. To monitor mRNA, the primers were as follows: for ATF6α (sense 5′-GGAACAGGATTCCAGGAGAATGAACCCTAGTG-3′; antisense5′-GATGTGTCCTGTGCCTCTTTAGCAGAAAATCC-3′), for ATF6β (CREBL1) (sense 5′-CTGAAGCGGCAGCAGCGAATGATCAAG-3′; antisense 5′-CGAGCCTCCAGTCCCTGCAGATACTCTTTC-3′), and for XBP1 (sense 5′-CAGAGTAGCAGCTCAGACTGCCAGAGATCG-3′; antisense 5′-GCTGTTCCAGCTCACTCATTCGAGCC-3′).
RESULTS
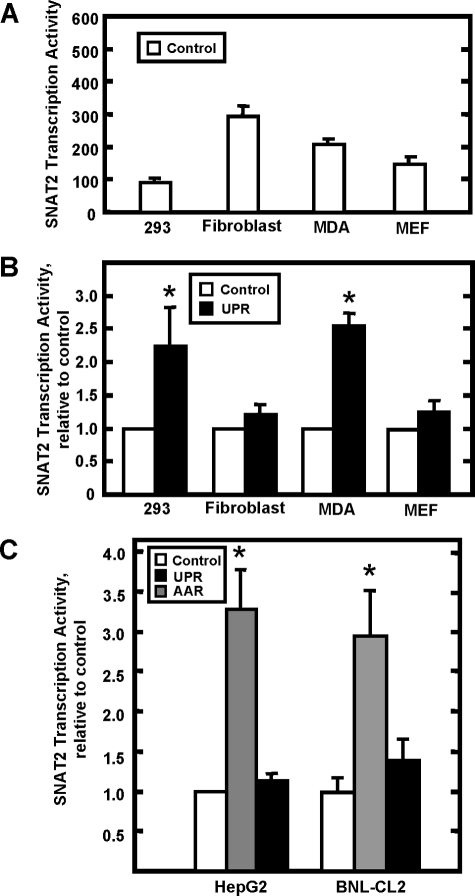
SNAT2 Transcriptional Activity Is Induced during AAR Activation but Not during UPR Activation—Despite the prediction that ER stress will activate C/EBP-ATF-containing genes via ATF4 action, previous studies have reported that the steady state mRNA content for SNAT2 is increased after amino acid deprivation but not after treatment of HepG2 human hepatoma cells to induce ER stress (30). To extend those observations, we monitored the effect of Tg treatment on SNAT2 transcription activity in several cell types (Fig. 1). The basal rate of SNAT2 transcription between cell types was quite variable (Fig. 1A). Interestingly, the UPR insensitivity of the SNAT2 gene was cell-specific (Fig. 1, B and C). Human kidney cells (293) and breast cancer cells (MDA) both exhibited increased SNAT2 transcription activity, whereas normal human fibroblasts, mouse embryonic fibroblasts (MEF), and mouse fetal liver cells (BNL-CL2) did not respond to ER stress. For the HepG2 human hepatoma cells and the mouse BNL-CL2 cells, the SNAT2 responsiveness to amino acid limitation was tested, and in both cell types transcription was induced as expected (Fig. 1C). These observations are consistent with the proposal that the UPR plays different roles in different cell types (1). To further investigate the mechanism of the UPR insensitivity, the time course for SNAT2 transcription activity during UPR or AAR activation was measured in HepG2 hepatoma cells (Fig. 2, top panel). The results revealed that the activity of the SNAT2 gene during AAR activation by HisOH started to increase at 2 h, peaked at 8 h with a 4-fold increase, and then declined at the 12-h time point. In contrast, after activation of the UPR by Tg, the SNAT2 transcriptional activity transiently fluctuated upward at 4 h but by 8 h had returned to the basal level observed in the control MEM condition. The MEM values at 2 and 4 h also rose modestly (Fig. 2, top panel). As a positive control for the UPR, BiP/GRP78, a known UPR-activated gene (37, 38), was monitored. In contrast to previous reports that have relied on the steady state mRNA content to assess BiP/GRP78 expression, we measured the actual transcription activity of the gene, which was enhanced at all time points at 2 h and beyond, with a maximum of nearly 8-fold at 8 h (Fig. 2, bottom panel). This result is in clear contrast to the short, transient rise observed for SNAT2 during the UPR. As reported previously (39), BiP/GRP78 expression was not responsive to amino acid deprivation.
FIGURE 1.
SNAT2 transcription activity in different cell lines during UPR or AAR activation. A shows the transcription activity of the SNAT2 gene in several human and mouse cell lines in the “control” or basal state (cultured in MEM). The transcription activity was assayed by measuring the SNAT2 hnRNA using primers spanning the human exon 4-intron 4 junction and for mouse, spanning the exon 13 to intron 13 junction. B shows the fold change, relative to the control value shown in A, in SNAT2 transcription activity after Tg treatment for 8 h to induce the UPR. Values are expressed as means ± S.E. C shows the SNAT2 transcriptional activity for two cell lines, HepG2 human hepatoma cells and BNL-CL2 mouse immortalized fetal hepatocytes, that do not activate the SNAT2 gene in response to the UPR but do respond to AAR. The asterisk denotes a value significantly different from the control at p < 0.05. The labels are as follows: HepG2, human hepatoma cells; 293, human kidney; fibroblast, immortalized normal human fibroblast; MDA, MDA-MB-231 human breast cancer; MEF, mouse embryonic fibroblast; BNL-CL2, mouse fetal hepatocytes.
FIGURE 2.
Transcription activity of the SNAT2 and BiP genes in HepG2 human hepatoma cells during activation of the UPR or AAR pathways. HepG2 cells were incubated for 0–12 h in MEM (Control), MEM + HisOH (AAR), or MEM + Tg (UPR). At the time points indicated, total RNA was isolated and analyzed by qRT-PCR. The transcription activity was assayed by measuring the SNAT2 hnRNA using primers spanning the exon 4-intron 4 junction, and the BiP primers spanned the exon 2-intron 2 junction. The results are expressed as fold change relative to the control value at time zero (t = 0). The PCRs were performed in duplicate for each sample, and samples were collected from three independent experiments. Values are expressed as means ± S.E. Where not shown, the error bars are within the symbol. The asterisk denotes a value significantly different from the control at p < 0.05.
Protein abundance of C/EBP-ATF-binding proteins is elevated by both amino acid deprivation and ER stress. A temporal sequence of events occurs in response to amino acid limitation during which target gene activation, induced by ATF4 binding, is followed by a subsequent period of transcriptional suppression. The decline in transcription parallels an increased de novo synthesis of ATF3 and C/EBPβ and their subsequent recruitment to the target gene (27, 40). Changes in the protein levels of these three transcription factors were assayed after either AAR or UPR activation in HepG2 cells (Fig. 3). An increase in the ATF4 protein level during either the AAR or UPR occurred within 1 h, and the maximum expression level at 4–12 h was similar for both stress conditions. Likewise, both stress pathways also induced the expression of ATF3 as well as the C/EBPβ-LAP* and C/EBPβ-LAP isoforms beginning at 4 h and continuing through the 12-h time period tested (Fig. 3). The truncated C/EBPβ liver-enriched inhibitory protein isoform was also induced, but the level of induction was lower than that for LAP* or LAP (Fig. 3).
FIGURE 3.
Immunoblot analysis of ATF4, ATF3, and C/EBPβ protein expression during amino acid deprivation or ER stress. Whole cell lysates of HepG2 cells incubated for 0–12 h in MEM (Control), MEM + HisOH (AAR), or MEM + Tg (UPR) were collected and subjected to immunoblot analysis. After transfer to a nitrocellulose membrane, the blot was probed with antibodies against ATF4, ATF3, C/EBPβ (showing the LAP*, LAP, and LIP (liver-enriched inhibitory protein) isoforms), or actin. The blots shown are representative of several experiments performed to ensure qualitative reproducibility.
Protein Binding at the SNAT2 C/EBP-ATF Enhancer Site— ChIP analysis of ATF4, ATF3, and C/EBPβ binding to the human SNAT2 C/EBP-ATF site revealed an increase in ATF4 binding within 2 h that reached a maximum between 4 and 8 h that was approximately equal in magnitude regardless of whether the AAR or the UPR stress signaling pathway had been activated (Fig. 4B). ATF4 association with the human SNAT2 gene approximately paralleled the AAR-induced transcription activity illustrated in Fig. 2. Consistent with the proposal that C/EBPβ expression is itself regulated by ATF4 (41) and that C/EBPβ action at an AARE temporally follows that of ATF4 (27), the ChIP analysis revealed a minimal increase in C/EBPβ binding at 2 h and a significant increase at 4 and 8 h (Fig. 4B). However, C/EBPβ binding to the SNAT2 gene exhibited a similar time course in response to activation of either the AAR or the UPR pathway. In contrast, the time course of ATF3 recruitment to the human SNAT2 gene was slightly different in response to the two stress-activated pathways. Tg treatment led to an increased association of ATF3 with the SNAT2 C/EBP-ATF site at 4 and 8 h, but the level of ATF3 binding was not increased during the initial 4 h after amino acid limitation, and the degree of association was still less than the Tg value at 8 h (Fig. 4B). Given that the SNAT2 gene in the mouse BNL-CL2 cells also showed an insensitivity to ER stress, the ATF4 recruitment to the SNAT2 C/EBP-ATF site in those cells was monitored (Fig. 4C). The results showed that, like HepG2 cells, ATF4 binding was enhanced following Tg treatment despite no increase in SNAT2 transcription activity (Fig. 1C). This result in the immortalized fetal cell line demonstrates that the lack of an induction following UPR activation is not unique to the transformed HepG2 cells.
FIGURE 4.
ATF4, ATF3, and C/EBPβ proteins bind to the SNAT2 C/EBP-ATF composite site following activation of either the AAR or the UPR pathway. A, scheme of human SNAT2 gene showing the qRT-PCR primer positions for ChIP analysis. B, ChIP analysis was performed on HepG2 cells incubated for 2, 4, or 8 h in MEM (Control), MEM + HisOH (AAR), or MEM + Tg (UPR). C, ChIP analysis was performed on mouse BNL-CL2 immortalized fetal hepatocytes incubated for 8 h in MEM (Control), MEM + HisOH (AAR), or MEM + Tg (UPR). The antibodies used in the assay were against ATF4, C/EBPβ, or ATF3. As a negative control, rabbit anti-chicken IgG was tested. and the values were always less than 5% of those with the transcription factor antibodies (for example, see C). Primers specific for the SNAT2 intronic C/EBP-ATF region were used for amplification during qRT-PCR. The data are presented as the fold change relative the MEM values (B) or the ratio to the input DNA (C). Each time point was calculated from analysis in duplicate for samples from at least three independent experiments and represents the mean ± S.E.
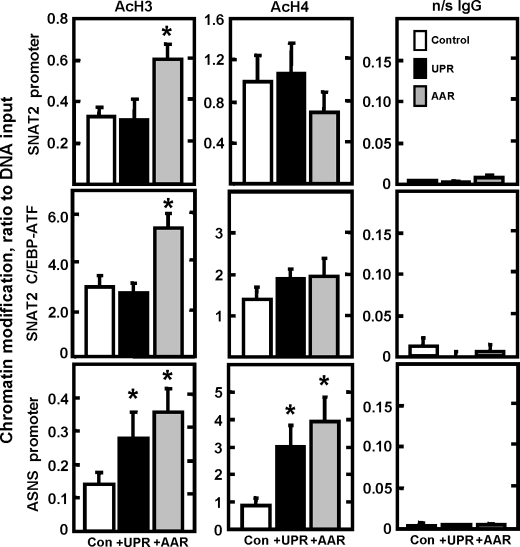
Neither Histone H3 Hyperacetylation Nor Assembly of the Transcription Preinitiation Complex Occurs at the SNAT2 Gene in Response to the UPR—Increased transcription from the SNAT2 gene following amino acid limitation is associated with increased histone acetylation and increased recruitment of the general transcription machinery (29). To determine whether ATF4 binding was sufficient to induce changes in chromatin structure at the SNAT2 gene, histone H3 and H4 acetylation at the SNAT2 promoter and the intronic C/EBP-ATF site was analyzed at 8 h following activation of the AAR or UPR pathways (Fig. 5). Increased H3 acetylation was observed at both the promoter and the C/EBP-ATF site following AAR activation, but relative to the MEM control values, there was little or no change in Tg-treated cells. Investigation of the histone H4 acetylation status of the SNAT2 gene revealed no significant difference upon activation of either the AAR or UPR pathway (Fig. 5). As a positive control, analysis of the chromatin status of the ASNS gene illustrated a significant increase in H3 and H4 acetylation during both UPR and AAR activation, consistent with previously published work (27). The ChIP assay was also employed to explore the recruitment to the SNAT2 promoter of several GTFs associated with the preinitiation complex (Fig. 6). At 1 h after activation of the AAR pathway, the association of RNA pol II, TBP, TFIIB, and TFIIE with the SNAT2 promoter remained at basal levels, whereas by 8 h the recruitment for each of these factors increased by 2–3-fold. In marked contrast, upon activation of the UPR, there was no enhanced recruitment of these GTFs to the promoter, even at 8 h. As a positive control, the binding of ATF4 at the SNAT2 C/EBP-ATF site was shown to increase following induction of both pathways (Fig. 6). The background value for the ChIP analysis was measured using a nonspecific IgG. Furthermore, to illustrate the degree of nonspecific binding for each of the GTF antibodies tested and their specificity for the promoter, primers for a downstream sequence within the SNAT2 gene (exon 10) were used (Fig. 6).
FIGURE 5.
ER stress does not provoke an increase in acetylation of histone H3 at the SNAT2 promoter or at the C/EBP-ATF site. ChIP analysis was performed on HepG2 cells incubated for 8 h in MEM (Con, control), MEM + HisOH (AAR), or MEM + Tg (UPR). Antibodies against acetylated histone H3, acetylated H4, or nonspecific IgG (n/s IgG) were used in the assay. Primers specific for the SNAT2 promoter or the SNAT2 C/EBP-ATF region, as shown in Fig. 4A, and the ASNS promoter regions were used for amplification during qRT-PCR. The data are presented as the ratio to input DNA. The values for each time point are calculated from duplicate assays for at least three independent experiments and are represented as the mean ± S.E. The asterisks denote a value that is statistically different (p < 0.05) from the MEM control.
FIGURE 6.
The preinitiation complex is not assembled on the SNAT2 promoter during UPR activation. ChIP analysis was performed on HepG2 cells incubated for 1 or 8 h in MEM (white bars), MEM + Tg (UPR, black bars), or MEM + HisOH (AAR, gray bars). Antibodies against RNA pol II, TBP, TFIIB, TFIIE, ATF4 and nonspecific IgG (n/s IgG) were used. Primers specific for the SNAT2 promoter (RNA pol II, TBP, TFIIB, and TFIIE) or the C/EBP-ATF (ATF4), as shown in Fig. 4A, were used for amplification during qRT-PCR. As an additional negative control for each antibody, PCR was performed with primers corresponding to the SNAT2 exon 10, a downstream region of the gene (bottom panel). The data are presented as the ratio to input DNA, and the values for each time point are calculated from duplicate assays for at least three independent experiments. The data shown represent the mean ± S.E., and an asterisk denotes values that are statistically different (p < 0.05) from the MEM control.
UPR-dependent Activation of Transcription from a Plasmid-based SNAT2-driven Reporter Gene—The lack of SNAT2-associated histone modification and recruitment of the GTFs in response to the UPR suggested that changes in chromatin structure may be a critical factor in triggering enhanced transcription via the C/EBP-ATF composite sequence. The less organized chromatin structure of a plasmid may allow for increased SNAT2 transcription during UPR. To further test this possibility, the transcriptional response of a Firefly luciferase reporter gene, driven by a SNAT2 genomic fragment containing the promoter and the C/EBP-ATF site (nt -512/+770), was monitored during either AAR or UPR activation (Fig. 7). Consistent with the known function of the SNAT2 C/EBP-ATF sequence as an amino acid-responsive enhancer, activation of the AAR resulted in a 25-fold induction of luciferase activity. Unexpectedly, in contrast to the lack of a response by the endogenous SNAT2 gene, UPR activation by medium lacking glucose and containing Tm or containing Tg led to an induction of SNAT2-driven transcription (Fig. 7). The contribution of the C/EBP-ATF site was investigated by transfection of a SNAT2/luciferase reporter construct containing mutations within the C/EBP-ATF sequence. Surprisingly, mutation of the C/EBP-ATF site, previously thought only to function as an AARE, abolished the transcriptional response to both amino acid limitation and ER stress.
FIGURE 7.
ER stress induces SNAT2 transcription from a reporter plasmid through the C/EBP-ATF composite site. HepG2 cells were transfected with a Firefly luciferase reporter gene driven by a SNAT2 genomic fragment (nt -512/+770), containing both the promoter and the wild type or mutated C/EBP-ATF enhancer sequence (nt +709/+717). The cells were co-transfected with an SV40-driven Renilla luciferase as a transfection control. Induction of the AAR pathway was achieved by incubation for 10 h in histidine-free MEM, whereas the UPR was activated by a 10-h incubation in MEM lacking glucose (-Glc), MEM + 300 nm thapsigargin (Tg), or MEM + 5 μg/ml tunicamycin (Tm). The data are the averages ± S.D. for 4–6 individual assays, and all differences marked with an asterisk are statistically significant at p < 0.01.
Role of ATF3 in the Regulation of SNAT2 Transcription—The data of Fig. 3 showed that the recruitment of ATF3 to the SNAT2 C/EBP-ATF site was different following activation of the AAR and the UPR. Particularly at 4 h, ATF3 binding was enhanced by the UPR but not by the AAR. Given the proposal that ATF3 serves as a repressor of ATF4 action at C/EBP-ATF sites (18, 27, 42), this difference in ATF3 binding at 4 h may explain the lack of Tg-induced transcription subsequent to this time period (Fig. 2). To test the hypothesis that a UPR-driven recruitment of ATF3 is responsible for the SNAT2 insensitivity to this pathway, HepG2 cells were transfected with an siRNA against ATF3 and then tested for SNAT2 transcription activity (Fig. 8A). Immunoblot analysis of ATF3 protein content after siRNA treatment documented an effective inhibition of expression under both basal and induced conditions (Fig. 8B). In the absence of ATF3 protein expression, activation of the UPR pathway still did not result in an increase in SNAT2 transcription (Fig. 8A). Similar results were obtained when ATF3 wild type and deficient mouse embryonic fibroblasts were used to monitor SNAT2 expression, i.e. SNAT2 transcription was not activated by ER stress in either cell population (data not shown). Thus, using two independent systems the results indicate that ATF3 is not a necessary component of the SNAT2 UPR insensitivity. The trend toward higher SNAT2 transcription activity in the siATF3-treated cells is consistent with the proposal that ATF3 is a repressor of this and other C/EBP-ATF-regulated genes (27, 42).
FIGURE 8.
Knockdown of ATF3 does not reverse the insensitivity of the SNAT2 gene to the UPR. HepG2 cells were transfected with either a control siRNA (siControl) or an siRNA against ATF3, as described under “Materials and Methods.” The cells were then incubated for 8 h in either MEM (Con, Control), MEM + HisOH (AAR), or MEM + Tg (UPR) prior to isolation of RNA for analysis of SNAT2 transcription activity (A). The PCRs were performed in duplicate for each sample, and samples were collected from three independent experiments. Values are expressed as means ± S.E. Isolation of whole cell extracts was performed for immunoblot analysis of ATF3 protein content and for actin as a loading control (B). The immunoblot results shown are representative of multiple experiments.
Activation of the UPR Pathway Blocks the Activation of the SNAT2 Gene by Amino Acid Limitation—The lack of activation of SNAT2 transcription by the UPR could result from either the lack of generating a positive secondary signal to the gene that complements ATF4 action or from production of a negative signal to repress the gene despite ATF4 binding. To distinguish between these two possibilities, HepG2 cells were treated to induce the AAR (MEM + HisOH or MEM lacking histidine) independently or in combination with Tg (Fig. 9). As expected, the AAR alone activated SNAT2 transcription, whereas the UPR did not. However, activating both pathways simultaneously resulted in almost a complete blockade of the AAR induction (Fig. 9A, left panel). To ensure that this effect of Tg was not a direct interaction with the HisOH itself, histidine-free MEM was used instead of HisOH to trigger the AAR pathway. Cells were subjected to histidine deprivation without or with simultaneous Tg treatment, and once again, the increased SNAT2 transcription resulting from activation of the AAR pathway was blocked by simultaneous activation of the UPR pathway (Fig. 9A, right panel). To demonstrate that the response was specific for the SNAT2 gene, the same samples were used to monitor the transcription activity from the ASNS gene (Fig. 9B). ASNS contains an ATF4-responsive C/EBP-ATF element (NSRE-1), which, in conjunction with a second element (NSRE-2), mediates activation of ASNS transcription in response to either the AAR or the UPR pathway (24). Consistent with its responsiveness to the UPR, the induction of ASNS transcription by the AAR was not blocked by simultaneous treatment with Tg, regardless of whether HisOH (Fig. 9B, left panel) or histidine-free medium (Fig. 9B, right panel) was used to trigger the AAR. As an aside, it should be noted that the simultaneous activation of the AAR and UPR pathways produced no change, or a slight decline, in transcription activity of ASNS relative to the AAR pathway alone (Fig. 9B). This observation is consistent with the proposal that both of these pathways activate this gene through the common set of genomic elements, NSRE-1 and NSRE-2 (24). The antagonism of the AAR pathway by Tg for the SNAT2 gene suggests that the lack of increased transcription following the UPR pathway arises from a UPR-dependent repressive activity, rather than simply a lack of an activating signal. Immunoblot analysis for ATF4 protein content did not reveal any difference between activation of the AAR alone or in combination with the UPR, suggesting that the action of the UPR was downstream of ATF4 translational control (Fig. 9C). Furthermore, ChIP analysis demonstrated that ATF4 binding at the SNAT2 C/EBP-ATF site occurred during concomitant activation of AAR and UPR pathways (Fig. 9D).
FIGURE 9.
Activation of the UPR pathway antagonizes the induction of the SNAT2 gene by the AAR pathway. HepG2 cells were incubated in control MEM or in the indicated conditions to activate either separately or concomitantly the AAR and UPR. To obtain the data in A (SNAT2) and B (ASNS), the AAR pathway was activated by two different treatments, either MEM containing HisOH (left-hand side) or MEM lacking histidine (right-hand side). At 0 and 8 h, total RNA was isolated and analyzed by qRT-PCR. The transcription activity was assayed by measuring the SNAT2 (A) and ASNS (B) hnRNA using primers spanning the exon 4-intron 4 and the intron 12-exon 13 junctions, respectively. The PCRs were performed in duplicate for each sample, and samples were collected from three independent experiments. Values are expressed as means ± S.E. C, illustrates an immunoblot analysis of ATF4 protein abundance in whole cell lysates prepared from HepG2 cells incubated in the indicated condition for 0–12 h (AAR = MEM-His; UPR = MEM + Tg). After the transfer, the blot was probed with an ATF4 antibody followed by an actin antibody, as described under “Materials and Methods.” D, ChIP analysis of ATF4 binding to the SNAT2 C/EBP-ATF site was performed on HepG2 cells treated for 8 h with the indicated AAR or UPR pathway activators. Samples were collected from three independent experiments. Values are expressed as means ± S.E. A, asterisk denotes that the value for the simultaneous treatment is significantly different (p < 0.05) from that for the HisOH or -His treatment alone. B and D, asterisk denotes a value statistically different (p < 0.05) from the MEM control.
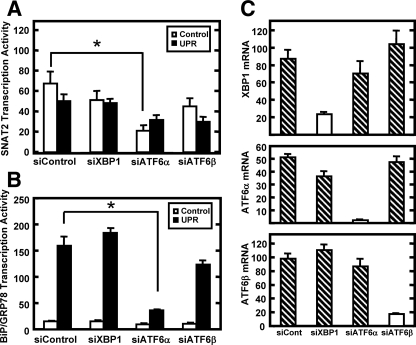
Knockdown of the XBP1 and ATF6 Expression Does Not Result in Activation of the SNAT2 Gene by ER Stress—Although an ERSE or UPRE element has not been described for the SNAT2 gene, the role of UPR effectors XBP1, ATF6α, and ATF6β was investigated to rule out the possibility that these transcription factors may contribute to the repression of SNAT2 transcription during ER stress. Thuerauf et al. (43) used an siRNA knockdown strategy against ATF6α or ATF6β in HeLa cells to test their role in transcription regulation of the ER stress target gene BiP. It was shown in that study that ATF6α acts as a transcriptional activator, whereas ATF6β functions as a transcriptional repressor of BiP/GRP78, although other investigators question whether or not ATF6β serves in this capacity (44). Yamamoto et al. (45) have demonstrated in IRE1α-/- MEFs that transcriptional up-regulation of BiP during ER stress is not affected by the absence of the Ire1-XBP1 arm.
To investigate whether or not the XBP1 and ATF6 arms of the UPR contribute to SNAT2 repression by ER stress, HepG2 cells were transfected with siRNA sequences against XBP1, ATF6α, or ATF6β (Fig. 10). The mRNA analysis for these factors demonstrated that the siRNA transfection was effective in lowering their expression (Fig. 10C), and as a control it was demonstrated that ATF6α knockdown did result in a suppression of UPR-dependent activation of BiP/GRP78 transcription activity (Fig. 10B). The results show that suppression of XBP1, ATF6α, or ATF6β expression did not alter the UPR-dependent repression of SNAT2 transcription. Collectively, these data indicate that the UPR effectors ATF6α, ATF6β, and XBP1 do not mediate the repressive effect of the UPR on SNAT2 transcription.
FIGURE 10.
Knockdown of the expression of XBP1, ATF6α, or ATF6β does not increase SNAT2 transcription activity during UPR activation. HepG2 cells were transfected with either a control siRNA (siControl) or an siRNA specific for XBP1, ATF6α, or ATF6β for 24 h followed by incubation in complete MEM for 24 h. The cells were then incubated for 8 h in MEM or MEM + Tg. Total RNA was isolated and transcription activity of SNAT2 (A), BiP/GRP78 (B), and steady state mRNA (C) of XBP1, ATF6α, and ATF6β were assayed. The qPCRs were performed in duplicate for each sample, and samples were collected from three independent experiments. Values are expressed as means ± S.E. An asterisk indicates a statistically significant difference (p < 0.05) relative to the respective siControl value.
DISCUSSION
The data obtained for HepG2 human hepatoma cells in this study led to the following novel observations, some of which are presented graphically in Fig. 11. 1) The responsiveness of the SNAT2 gene to activation by the UPR pathway is cell-specific. 2) In HepG2 human hepatoma cells and mouse BNL-CL2 fetal hepatocytes SNAT2 transcriptional activity remains near basal levels despite a readily detected increase in ATF4 recruitment and binding to the C/EBP-ATF site within the SNAT2 gene that equals that observed during the AAR. 3) The lack of increased SNAT2 transcription by the UPR results from an active repression of the gene rather than merely an absence of an activating signal. 4) The mechanism of this repression is unknown, but it appears to occur at a step downstream of ATF4 binding; it is not triggered by the ATF6 or XBP1 arms of the UPR, and it can override the activation by the AAR pathway. 5) The results demonstrate that the SNAT2 C/EBP-ATF composite site, previously considered to function exclusively as an AARE, can mediate a UPR-initiated induction of transcription outside the context of intact chromatin (i.e. a plasmid) in HepG2 hepatoma cells.
FIGURE 11.
Working model for the differential regulation of the SNAT2 gene in HepG2 cells during activation of the AAR or UPR pathways. A, SNAT2-driven transcription is induced by either AAR or UPR from a plasmid-based luciferase reporter that contains the SNAT2 promoter and C/EBP-ATF site. In contrast, SNAT2 transcription from the endogenous gene is induced only by the AAR, but not by the UPR pathway. B, AAR pathway triggers assembly of the ATF4 enhanceosome, increased H3 acetylation, and preinitiation complex (PIC) formation, events that lead to enhanced SNAT2 transcription activity. Conversely, despite the assembly of the ATF4 enhanceosome, the UPR represses SNAT2 transcription.
With regard to the genomic elements that mediate transcriptional activation by the AAR and UPR stress pathways, there are three classes of genes as follows: 1) those genes that are only induced by one or the other pathway via a specific genomic element (e.g. BiP/GRP78, containing an endoplasmic reticulum stress element (ERSE) but insensitive to the AAR); 2) those genes, such as CHOP, that are induced by both pathways but that contain two different genomic elements that exhibit distinct AARE and ERSE activities; and 3) the ASNS gene, which thus far is a unique case in that it is activated by both pathways through a common bipartite site consisting of NSRE-1, a C/EBP-ATF composite site, and NSRE-2 (24, 27, 46). Consistent with these distinctions, promoter deletion analysis by Jousse et al. (47) demonstrated that amino acid depletion and ER stress act via independent elements to activate the CHOP gene. However, there are also data indicating that the ERSE (nt -93 to -75) and AARE (C/EBP-ATF sequence at nt -310 to -302) sites both contribute to the CHOP induction in response to ER stress (48–50). In this regard, the results of Yoshida et al. (51) showed that a CHOP promoter construct (nt -870/+17) containing wild type ERSE-I and ERSE-II exhibited a 3-fold increase in transcription in response to UPR activation. However, even after mutating both ERSE sites, the construct yielded about a 2-fold increase, albeit the absolute values of luciferase activity were lower (see Fig. 4) (51). These data can be interpreted to suggest that the CHOP C/EBP-ATF site at nt -310/-302 may still allow this promoter fragment to respond to the UPR. The results of Yoshida et al. (51) for CHOP would be analogous to and consistent with the present results for UPR activation of SNAT2 in the context of a plasmid.
ATF4 has been shown to activate the transcription from many stress-sensitive genes that contain a C/EBP-ATF composite site (52). The C/EBP-ATF composite site within the intron of the SNAT2 gene functions as an AARE, mediating ATF4-dependent activation of the gene following amino acid deprivation (29). However, given that ER stress also induces ATF4 synthesis, the question of whether or not ATF4 bound to the SNAT2 C/EBP-ATF composite site during ER stress remained unanswered. The answer to this outstanding issue is mechanistically important because it is known that SNAT2 mRNA content and transport activity are not increased proportionally to the increase in ATF4 protein following ER stress in HepG2 cells (30). The present ChIP data demonstrate clearly that ATF4 protein, increased in abundance following UPR activation, is bound to the SNAT2 C/EBP-ATF site at a level equal to or greater than that observed after activation of the AAR pathway in both HepG2 human hepatoma cells and mouse BNL-CL2-immortalized fetal hepatocytes.
Induction of SNAT2 transcription during amino acid deprivation is controlled by the sequential binding and interplay of ATF4, ATF3, and C/EBPβ at the SNAT2 C/EBP-ATF site (29). Previous results had illustrated that the binding of these three factors at an AARE parallels their de novo synthesis (27), and this programmed sequence of factor synthesis and recruitment occurs in a qualitatively similar manner for each of six different C/EBP-ATF containing genes tested (40). ATF4 binding is at least a component of the transcriptional trigger because the time course of ATF4 recruitment closely parallels histone acetylation, GTF recruitment, including pol II, and the actual transcription activity (27). Conversely, increased ATF3 and C/EBPβ binding to the C/EBP-ATF site occurs later (after 6–8 h) and corresponds with the repression phase of transcription (27, 40). Remarkably, the ChIP data in this study show that the binding of all three of these factors at the SNAT2 C/EBP-ATF site occurs during ER stress, indicating that the subsequent recruitment of ATF3 and C/EBPβ may be triggered by bound ATF4, but it is not dependent on histone acetylation, assembly of the general transcriptional machinery, or on enhanced transcription from the gene. With one exception, the temporal and quantitative binding of ATF4, ATF3, and C/EBPβ following UPR activation is nearly identical to that for the AAR activation. Increased ATF3 binding during ER stress occurred at an earlier time (4 h) than during amino acid limitation (8 h), and UPR-associated ATF3 binding was followed by a complete repression of a transient rise in SNAT2 transcription. However, knockdown of ATF3 with siRNA did not reverse the UPR repression of the SNAT2 gene, indicating that ATF3 is not a critical mediator of that action.
The absence of sustained induced transcription activity from the SNAT2 gene following activation of the UPR pathway could have been the consequence of two different mechanisms. It was possible that increased transcription requires not only ATF4 binding to the C/EBP-ATF composite site but also a second activating signal. If this hypothesis were true, it would be that amino acid deprivation initiates this secondary signal, whereas ER stress does not. Conversely, it was possible that the UPR pathway triggers a repressive signal for the SNAT2 gene, which overrides the ATF4 binding and prevents an induction of transcription activity. Support for the latter proposal came from results demonstrating that the induction of SNAT2 transcription by amino acid deprivation was blocked by the concurrent activation of the UPR pathway. The nature of this signal remains to be elucidated, but as discussed below, the evidence presented here links the signal to chromatin structure. Furthermore, knockdown of the ATF6 and XBP1 arms of the UPR did not relieve the SNAT2 repression suggesting that neither of these transcription factors mediates the repression, either directly or indirectly.
To further explore the mechanistic basis for the lack of SNAT2 activation during ER stress, histone acetylation and recruitment of general transcription factors (GTFs) was investigated. Our ChIP data revealed that there is an increase in histone H3 acetylation at the SNAT2 promoter and the C/EBP-ATF region following AAR activation but not following UPR activation. Likewise, although the GTF proteins that make up the preinitiation complex are present on the SNAT2 promoter following amino acid limitation, they are not recruited following UPR activation. These data also document that histone modification and assembly of the preinitiation complex at the SNAT2 promoter is an event that occurs subsequent to ATF4 binding. ATF4 has been shown to interact directly with several GTFs, including TBP, TFIIB, and TFIIF (15) and the RPB3 subunit of RNA polymerase II (17). Indeed, co-expression of RPB3 with ATF4 enhances its transcriptional activation capabilities (17), so it is possible that ATF4 plays a direct role in the GTF recruitment. Our results suggest that the recruitment of chromatin-modifying complexes containing histone acetyltransferase activity may not occur. Why the bound ATF4 does not trigger histone modification and GTF recruitment to the SNAT2 gene when the initial stimulus was ER stress is not clear.
Another important observation of this study was that, in contrast to the endogenous SNAT2 gene, transcription from a SNAT2-driven luciferase reporter was induced upon UPR activation. The lack of higher order chromatin structure of a plasmid may confer the UPR sensitivity to the SNAT2 gene. Furthermore, mutations of the SNAT2 C/EBP-ATF composite site abolished the transcriptional induction by several different UPR activators, confirming that the response was dependent on the integrity of this intronic enhancer element. In addition to potential differences in chromatin structure, it is possible that there is a recruitment of a repressor/co-repressor to the SNAT2 gene in response to the UPR that does not occur following activation of the AAR. Another explanation for selective regulation of the SNAT2 gene may be a relocalization within the nucleus. For example, the recruitment of the SNAT2 promoter to a transcription factory during AAR activation, but not in the presence of ER stress, could also explain the difference in response between the two pathways. An example of such a relocation mechanism and of the association of a gene with a transcription factory is described for the murine β-globin locus during erythroid maturation (53, 54). If such a mechanism exists for SNAT2, UPR activation may block this relocalization. Regardless of the mechanism, these results suggest a link between SNAT2 activation by ATF4 and chromatin structure. Thus, the specificity of the SNAT2 C/EBP-ATF composite site to distinguish between the AAR and UPR signals depends on the context of the chromatin environment. It is interesting to speculate that ATF4 is a pioneer factor that binds to the C/EBP-ATF site in a nucleating event without the prerequisite of extensive chromatin modification, but then a second, chromatin-associated event becomes critical in triggering transcriptional activation. The SNAT2 gene provides an interesting model to investigate these and other mechanisms associated with transcriptional control in response to cellular stress and the associated signaling pathways.
Acknowledgments
We thank Dr. Roxana M. Coman and members of the Kilberg laboratory for technical advice and helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants DK-59315 and DK-52064 (NIDDK) (to M. S. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: eIF2α, eukaryotic initiation factor 2α; ATF4, activating transcription factor 4; AAR, amino acid response; AARE, amino acid-response element; ASNS, asparagine synthetase; BiP/GRP78, immunoglobulin heavy chain-binding protein/glucose-regulated protein 78; C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; CHOP, C/EBP homology protein; ERSE, endoplasmic reticulum stress element; GAPDH, glyceraldehyde-3-phosphate dehydrogenase gene; GCN2, general control nonderepressible; HisOH, histidinol; LAP, liver-enriched activating protein; NSRE, nutrient sensing response element; PERK, double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase; qRT, quantitative real time; RT, reverse transcriptase; SNAT2, system A sodium-dependent neutral amino acid transporter 2; TBP, TATA-binding protein; TFII, general transcription factor; Tg, thapsigargin; UPR, unfolded protein response; XBP1, X-box binding protein 1; MEM, minimal essential medium; nt, nucleotide; siRNA, short interfering RNA; pol, polymerase; GTF, general transcription factor; hnRNA, heterogeneous nuclear RNA; bZIP, basic leucine zipper; ER, endoplasmic reticulum; CREB, cAMP-response-element-binding protein.
References
- 1.Wek, R. C., and Cavener, D. R. (2007) Antioxid. Redox. Signal. 9 2357-2371 [DOI] [PubMed] [Google Scholar]
- 2.Vattem, K. M., and Wek, R. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11269-11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu, P. D., Harding, H. P., and Ron, D. (2004) J. Cell Biol. 167 27-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harding, H. P., Zhang, Y., Bertolotti, A., Zeng, H., and Ron, D. (2000) Mol. Cell 5 897-904 [DOI] [PubMed] [Google Scholar]
- 5.Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., and Ron, D. (2000) Mol. Cell 6 1099-1108 [DOI] [PubMed] [Google Scholar]
- 6.Schroder, M., and Kaufman, R. J. (2005) Annu. Rev. Biochem. 74 739-789 [DOI] [PubMed] [Google Scholar]
- 7.Ron, D. (2002) J. Clin. Investig. 110 1383-1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutkowski, D. T., and Kaufman, R. J. (2004) Trends Cell Biol. 14 20-28 [DOI] [PubMed] [Google Scholar]
- 9.Sood, R., Porter, A. C., Olsen, D. A., Cavener, D. R., and Wek, R. C. (2000) Genetics 154 787-801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, P., McGrath, B. C., Reinert, J., Olsen, D. S., Lei, L., Gill, S., Wek, S. A., Vattem, K. M., Wek, R. C., Kimball, S. R., Jefferson, L. S., and Cavener, D. R. (2002) Mol. Cell. Biol. 22 6681-6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natarajan, K., Meyer, M. R., Jackson, B. M., Slade, D., Roberts, C., Hinnebusch, A. G., and Marton, M. J. (2001) Mol. Cell. Biol. 21 4347-4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinnebusch, A. G., and Natarajan, K. (2002) Eukaryot. Cell 1 22-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harding, H. P., Zhang, Y., Zeng, H., Novoa, I., Lu, P. D., Calfon, M., Sadri, N., Yun, C., Popko, B., Paules, R., Stojdl, D. F., Bell, J. C., Hettmann, T., Leiden, J. M., and Ron, D. (2003) Mol. Cell 11 619-633 [DOI] [PubMed] [Google Scholar]
- 14.Ameri, K., and Harris, A. L. (2008) Int. J. Biochem. Cell Biol. 40 14-21 [DOI] [PubMed] [Google Scholar]
- 15.Liang, G., and Hai, T. (1997) J. Biol. Chem. 272 24088-24095 [DOI] [PubMed] [Google Scholar]
- 16.Yang, X., Matsuda, K., Bialek, P., Jacquot, S., Masuoka, H. C., Schinke, T., Li, L., Brancorsini, S., Sassone-Corsi, P., Townes, T. M., Hanauer, A., and Karsenty, G. (2004) Cell 117 387-398 [DOI] [PubMed] [Google Scholar]
- 17.De Angelis, R., Lezzi, S., Bruno, T., Corbi, N., Di Padova, M., Floridi, A., Fanciulli, M., and Passananti, C. (2003) FEBS Lett. 547 15-19 [DOI] [PubMed] [Google Scholar]
- 18.Fawcett, T. W., Martindale, J. L., Guyton, K. Z., Hai, T., and Holbrook, N. J. (1999) Biochem. J. 339 135-141 [PMC free article] [PubMed] [Google Scholar]
- 19.Kilberg, M. S., Stevens, B. R., and Novak, D. (1993) Annu. Rev. Nutr. 13 137-165 [DOI] [PubMed] [Google Scholar]
- 20.McGivan, J. D., and Pastor-Anglada, M. (1994) Biochem. J. 299 321-334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackenzie, B., and Erickson, J. D. (2004) Pfluegers Arch. 447 784-795 [DOI] [PubMed] [Google Scholar]
- 22.Barber, E. F., Handlogten, M. E., Vida, T. A., and Kilberg, M. S. (1982) J. Biol. Chem. 257 14960-14967 [PubMed] [Google Scholar]
- 23.Saier, M. H., Jr., Daniels, G. A., Boerner, P., and Lin, J. (1988) J. Membr. Biol. 104 1-20 [DOI] [PubMed] [Google Scholar]
- 24.Barbosa-Tessmann, I. P., Chen, C., Zhong, C., Siu, F., Schuster, S. M., Nick, H. S., and Kilberg, M. S. (2000) J. Biol. Chem. 275 26976-26985 [DOI] [PubMed] [Google Scholar]
- 25.Kilberg, M. S., and Barbosa-Tessmann, I. P. (2002) J. Nutr. 132 1801-1804 [DOI] [PubMed] [Google Scholar]
- 26.Siu, F., Bain, P. J., LeBlanc-Chaffin, R., Chen, H., and Kilberg, M. S. (2002) J. Biol. Chem. 277 24120-24127 [DOI] [PubMed] [Google Scholar]
- 27.Chen, H., Pan, Y. X., Dudenhausen, E. E., and Kilberg, M. S. (2004) J. Biol. Chem. 279 50829-50839 [DOI] [PubMed] [Google Scholar]
- 28.Palii, S. S., Chen, H., and Kilberg, M. S. (2004) J. Biol. Chem. 279 3463-3471 [DOI] [PubMed] [Google Scholar]
- 29.Palii, S. S., Thiaville, M. M., Pan, Y. X., Zhong, C., and Kilberg, M. S. (2006) Biochem. J. 395 517-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bain, P. J., LeBlanc-Chaffin, R., Chen, H., Palii, S. S., Leach, K. M., and Kilberg, M. S. (2002) J. Nutr. 132 3023-3029 [DOI] [PubMed] [Google Scholar]
- 31.Hansen, B. S., Vaughan, M. H., and Wang, L.-J. (1972) J. Biol. Chem. 247 3854-3857 [PubMed] [Google Scholar]
- 32.den Boer, M. L., Pieters, R., Kazemier, K. M., Rottier, M. M. A., Zwaan, C. M., Kaspers, G. J. L., Janka-Schaub, G., Henze, G., Creutzig, U., Scheper, R. J., and Veerman, A. J. P. (1998) Blood 91 2092-2098 [PubMed] [Google Scholar]
- 33.van Huizen, R., Martindale, J. L., Gorospe, M., and Holbrook, N. J. (2003) J. Biol. Chem. 278 15558-15564 [DOI] [PubMed] [Google Scholar]
- 34.Hutson, R. G., and Kilberg, M. S. (1994) Biochem. J. 303 745-750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiaville, M. M., Dudenhausen, E. E., Zhong, C., Pan, Y. X., and Kilberg, M. S. (2008) Biochem. J. 410 473-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipson, K. E., and Baserga, R. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 9774-9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida, H., Haze, K., Yanagi, H., Yura, T., and Mori, K. (1998) J. Biol. Chem. 273 33741-33749 [DOI] [PubMed] [Google Scholar]
- 38.Roy, B., and Lee, A. S. (1999) Nucleic Acids Res. 27 1437-1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbosa-Tessmann, I. P., Pineda, V. L., Nick, H. S., Schuster, S. M., and Kilberg, M. S. (1999) Biochem. J. 339 151-158 [PMC free article] [PubMed] [Google Scholar]
- 40.Pan, Y. X., Chen, H., Thiaville, M. M., and Kilberg, M. S. (2007) Biochem. J. 401 299-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen, C., Dudenhausen, E., Chen, H., Pan, Y. X., Gjymishka, A., and Kilberg, M. S. (2005) Biochem. J. 391 649-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan, Y.-X., Chen, H., Siu, F., and Kilberg, M. S. (2003) J. Biol. Chem. 278 38402-38412 [DOI] [PubMed] [Google Scholar]
- 43.Thuerauf, D. J., Marcinko, M., Belmont, P. J., and Glembotski, C. C. (2007) J. Biol. Chem. 282 22865-22878 [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto, K., Sato, T., Matsui, T., Sato, M., Okada, T., Yoshida, H., Harada, A., and Mori, K. (2007) Dev. Cell 13 365-376 [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, K., Yoshida, H., Kokame, K., Kaufman, R. J., and Mori, K. (2004) J. Biochem. (Tokyo) 136 343-350 [DOI] [PubMed] [Google Scholar]
- 46.Zhong, C., Chen, C., and Kilberg, M. S. (2003) Biochem. J. 372 603-609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jousse, C., Bruhat, A., Harding, H. P., Ferrara, M., Ron, D., and Fafournoux, P. (1999) FEBS Lett. 448 211-216 [DOI] [PubMed] [Google Scholar]
- 48.Ma, Y., Brewer, J. W., Diehl, J. A., and Hendershot, L. M. (2002) J. Mol. Biol. 318 1351-1365 [DOI] [PubMed] [Google Scholar]
- 49.Ma, Y., and Hendershot, L. M. (2004) J. Biol. Chem. 279 13792-13799 [DOI] [PubMed] [Google Scholar]
- 50.Pirot, P., Ortis, F., Cnop, M., Ma, Y., Hendershot, L. M., Eizirik, D. L., and Cardozo, A. K. (2007) Diabetes 56 1069-1077 [DOI] [PubMed] [Google Scholar]
- 51.Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2000) Mol. Cell. Biol. 20 6755-6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kilberg, M. S., Pan, Y. X., Chen, H., and Leung-Pineda, V. (2005) Annu. Rev. Nutr. 25 59-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ragoczy, T., Bender, M. A., Telling, A., Byron, R., and Groudine, M. (2006) Genes Dev. 20 1447-1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraser, P., and Engel, J. D. (2006) Genes Dev. 20 1379-1383 [DOI] [PubMed] [Google Scholar]