Abstract
MreB is a bacterial ortholog of actin that affects cell shape, polarity and chromosome segregation. Although a significant body of work has explored its cellular functions, we know very little about the biochemical behavior of MreB. We have cloned, overexpressed in E. coli, and purified untagged MreB1 from Thermotoga maritima. We have characterized the conditions that regulate its monomer-to-polymer assembly reaction, the critical concentrations of that reaction, the manner in which MreB uses nucleotides, its stability, and the structure of the assembled polymer. MreB requires a bound purine nucleotide for polymerization and rapidly hydrolyzes it following assembly. MreB assembly contains two distinct components, one that does not require divalent cations and one that does, which may comprise the nucleation and elongation phases of assembly, respectively. MreB assembly is strongly favored by increasing temperature or protein concentration but inhibited differentially by high concentrations of monovalent salts. Polymerization rate increases and bulk critical concentration decreases with increasing temperature, but in contrast to previous reports, MreB is capable of polymerizing across a broad range of temperatures. MreB polymers are shorter, stiffer, and scatter more light than eukaryotic actin filaments. Due to rapid ATP hydrolysis and phosphate release, we suggest that most assembled MreB in cells is in the ADP-bound state. Because of only moderate differences between the ATP and ADP critical concentrations, treadmilling may occur, but we do not predict dynamic instability in cells. Because of the relatively low cellular concentration of MreB and the observed structural properties of the polymer, a single MreB assembly may exist in cells.
The untested assumption that bacteria lack a cytoskeleton (1) was overturned by the discoveries of prokaryotic orthologs (2, 3) of tubulin (4), actin (5, 6), and even intermediate filament proteins (7). The tubulin ortholog FtsZ drives contraction of the septum during cytokinesis; the intermediate filament ortholog crescentin causes the distinctive, eponymous shape of Caulobacter crescentus; and the actin ortholog MreB drives the establishment or maintenance of cell shape in all asymmetrical eubacteria yet studied (8). Although the primary structure of MreB is poorly conserved with respect to eukaryotic actins, MreB shares a nearly superimposable tertiary structure with the eukaryotic actin monomer (6) and can assemble into polymers in vitro (6, 9, 10) and in cells (5, 11).
Although the cell biology of MreB has developed at a rapid pace (12), our understanding of MreB at the biochemical level is currently lacking. Low cellular abundance and difficulty in overexpression and purification have thus far precluded biochemical characterization of the molecule in native form (9, 10). Nonetheless, six decades of work on the eukaryotic actin cytoskeleton has taught us that the cell biology of the system can not be understood without a rigorous, quantitative understanding of the molecules and interactions that comprise it (13).
Thus, we have begun a program to biochemically define the assembly properties and intermolecular interactions of the bacterial actin cytoskeleton. Here we present a characterization of the conditions that govern the monomer-polymer reaction of MreB from the thermophile Thermotoga maritima. Our findings explain quantitatively the original observations of van den Ent, et al. (6). Several of our key findings contrast with those of Esue, et al., (9, 10), who observed robust assembly of MreB only at high temperatures. We provide a likely explanation for the differing results.
Experimental Procedures
Cloning
The T. maritima mreB1 gene was amplified from E. coli K12 genomic DNA (ATCC catalog # 10798D) by polymerase chain reaction using Pfu Ultra DNA polymerase (Stratagene) following the manufacturer’s instructions. Forward 5′ GGGAATTC CA’T ATG TTG AGA AAA GAC ATA GGA ATA GAT C 3′ and reverse 5′ ATAAGAAT G’TCGAC TCA CCC GGC ACC CTG AAG CTT C 3′ primers (IDT) were based on the T. maritima MreB1 gene (TM0588) from GenBank (accession number NC_000853). PCR products were digested with NdeI and XhoI (Promega) and ligated into the NdeI/XhoI site of pET23A(+) (Novagen). Cloning was carried out in BL21 (DE3) cells. The native MreB1 construct had the primary structure: MLRKDIGIDLGTANTLVFLRGKGIVVNEPSVIAIDSTTGEILKVGLEAKNMIGKTPATIKAI RPMRDGVIADYTVALVMLRYFINKAKGGMNLFKPRVVIGVPIGITDVERRAILDAGLEA GASKVFLIEEPMAAAIGSNLNVEEPSGNMVVDIGGGTTEVAVISLGSIVTWESIRIAGDE MDEAIVQYVRETYRVAIGERTAERVKIEIGNVFPSKENDELETTVSGIDLSTGLPRKLT LKGGEVREALRSVVVAIVESVRTTLEKTPPELVSDIIERGIFLTGGGSLLRGLDTLLQKET GISVIRSEEPLTAVAKGAGMVLDKVNILKKLQGAG.
Expression vectors were sequenced at the University of Wisconsin Biotechnology Center using ABI Big Dye and T7 forward and reverse primers.
Protein expression
Expression vectors were transformed into C43(DE3) cells (Avidis) (14). Cultures were grown at 37°C in LB broth supplemented with 50μg/ml ampicillin to an OD600 of 0.6 to 0.8. Protein expression was induced by adding IPTG to a final concentration of 0.375 mM for 3 hours. Cells were then collected by centrifugation and stored at -20°C until needed.
Protein extraction and Purification
12-15 g of frozen cell pellets were extracted twice in 8 ml per g of 1X CaG8 buffer (2mM Tris pH 8.0, 200μM ATP, 0.02% sodium azide, 0.1mM CaCl2, 0.5 mM DTT), homogenized, and sonicated with a probe sonicator. The extract was cleared by centrifugation at 4°C at 100,000 × g for 30 minutes in a 70 Ti rotor. Polymerizataion of MreB was induced immediately in supernatants by adding KCl to 20mM, MgCl2 to 1mM, EGTA to 1mM and MES buffer (pH 6.0) to 10mM. After mixing, the samples were allowed to polymerize overnight on ice at 4°C. The next morning, polymerized MreB was collected by centrifugation at 4°C at 100,000 × g for 30 min in a 70 Ti rotor. Pellets were combined and resuspended in 100mL of CaG8 buffer using a dounce homogenizer, then depolymerized at 4°C for 1-2 hours with gentle stirring. Depolymerized pellets were then loaded onto a 25mL DE52 DEAE column (Whatman), and step-eluted off by increasing KCl concentration in CaG8 buffer. 125-175 mM KCl fractions were treated one of two ways. In the first method, MreB was seen visibly to precipitate following overnight 4°C incubation of elution fractions. Fractions were pooled and centrifuged at 4°C at 20,000 × g in a JA-20 rotor for 20 minutes. Pellets were suspended in 10-20 ml of CaG8 buffer and depolymerized by dialysis against CaG8 buffer at 4°C for at least 72 hours. The dialysate was then centrifuged at 4°C at 100,000 × g for 30 min to remove oligomers and the supernatant was collected and quantified on by SDS-PAGE/Coomassie Blue densitometry in ImageJ using rabbit actin as a standard. Alternately, the 125-175 mM KCl fractions were pooled and diluted with CaG8 buffer to a KCl concentration of less than 50 mM. The sample was then concentrated by rebinding to DE52 and eluting in CaG8 buffer with a single step of 500 mM KCl. Fractions containing pure MreB were pooled and dialyzed against polymerization buffer (CaG8 supplemented with 10mM imidazole pH 7.0, 20mM KCl, 1mM MgCl2 and 1 mM EGTA) overnight at 4°C. The following morning, the dialysate was centrifuged at 4°C at 100,000 × g for 30 minutes. The pellet was homogenized in CaG8 then dialyzed against CaG8 overnight at 4°C. After 48-72 hrs, the dialysate was centrifuged at 4°C at 100,000 × g for 30 minutes, and the supernatant was loaded onto an S-300 gel filtration column (Amersham) equilibrated in CaG8 buffer and eluted in the same by gravity flow. Post-peak MreB fractions were pooled and concentrated using a Centriplus YM10 (Millipore) at 4°C following the manufacturer’s instructions and quantified as above. MreB prepared by the two methods behaved indistinguishably and was stable for up to four weeks stored on ice at 4°C.
Light Scattering
90° perpendicular light scattering experiments (15) were carried out in a PC1 spectrofluorometer (ISS) equipped with a temperature control jacket and under control of Vinci software (ISS) v1.4.9.5. Excitation and emission monochrometers were set at 400nm and slit pairs (typically 1 mm) were employed in both excitation and emission paths.
Polymerization timecourse
Unless stated otherwise, non-protein reaction components were mixed on ice in proportions such that the final reactions contained 200uM ATP, 10mM imidazole pH 7.0, 20 mM KCl, 1mM MgCl2 and 1mM EGTA (KMEI). Separately on ice, MreB in CaG8 storage buffer was mixed with 1/9th volume of 10X cation exchange buffer (1 mM MgCl2, 10 mM EGTA) and incubated on ice for one minute. Polymerization was initiated by combining the two samples and light scattering measurements were begun in a cuvette preequilibrated at 20°C after an approximately fifteen second experimental lag. Data was collected at 1 s intervals, analyzed in Excel and plotted using Kaleidagraph.
Nucleotide exchange
MreB protein in CaG8 storage buffer was mixed with 1/10th volume of Dowex resin 1X8-400Cl (Avacado Research) and incubated on ice for 10 min, vortexing every 2-3 min (16). The mixture was centrifuged at 4°C at 18,000 × g for 1 min. Supernatant was transferred to a separate tube and incubated on ice with the desired nucleotide at 200uM for 10 minutes. This procedure was carried out three times to ensure complete nucleotide exchange. For nucleotide concentration experiments, the final supernatant was supplemented instead with nucleotide at the stated concentration. Exchanged MreB was then immediately used as described for polymerization reactions.
Nucleotide dissociation
The nucleotide dissociation rate constant was determined by monitoring the decrease in fluorescence of εATP following dilution into ATP-containing buffer. 10 μM Mg-εATP MreB was prepared by Dowex treatment as above followed by Mg/EGTA exchange. Mg-εATP MreB was then diluted 10x into 2 mM Tris-HCl, pH 8.0 containing 1 mM ATP. εATP fluorescence was monitored with excitation at 360 nm and emission at 410 nm (17-19). The rate constant (k-1) for the dissociation of εATP from MreB was determined by fitting the time course to a single-exponential equation using Kaleidagraph software.
Critical concentration
MreB was polymerized as described above at varying protein concentrations. Reactions were equilibrated either for one hour at the experimental temperature or overnight at 4°C followed by one hour at the experimental tempertaure prior to measuring light scattering intensity as described above. Light scattering values were the average of one minute of acquired signal. Critical concentrations were determined by the intersection of two linear fits to the data points above and below the apparent inflection point in the graph as determined by visual inspection.
Sedimentation
MreB was polymerized as described and centrifuged either at 4 °C for 30 minutes at 100k × g in a Beckman tabletop ultracentrifuge or at room temperature for 10 minutes at 100k × g in a Beckman airfuge.
Phosphate assay
Phosphate production was determined with the EnzChek kit (Molecular Probes) employing a procedure modified from the manufacturer’s instructions because of significant absorbance of 360 nm light by polymerized MreB. Parallel reactions were carried out with and without the EnzChek enzyme and reactant components. Both reactions were read in real-time in a Beckmann DU640 spectrometer at 360 nm. The absorbance of MreB alone was subtracted from those of the complete reactions. A standard curve of Pi concentrations was employed as per the manufacturer’s instructions. Phosphate release curves were overlain onto the 400 nm light scattering curves of parallel polymerization reactions carried out in the ISS fluorimeter. Rabbit skeletal muscle actin was used as control.
Fluorescent labeling
The mreB1 gene was re-cloned as described above, using reverse primers modified to cause the mutations K331C, L332C, or Q333C. Each mutant was expressed and purified as for wild-type MreB above. Purified Cys-substituted MreB was dialyzed against DTT-free 1X CaG8 buffer for 4 hours followed by addition of a 6x molar excess of thiol-reactive dye and allowed to stir overnight at 4°C. The following morning, the labeling reactions were quenched by adding DTT to 1mM, the reactions were centrifuged at 4°C at 18,000 × g for 5 min, and protein was separated from free dye by gel filtration. Labeling efficiency (typically 60-100%) was calculated after measuring protein and fluorophore concentration of peak fractions by SDS-PAGE and spectrometry, respectively. MreB was labeled with tetramethylrhodamine-5-maleimide, Alexa Fluor 350 C5-maleimide, Lucifer Yellow Iodoacetamide, Alexa-Fluor 488 C5-maleimide, Alexa-Fluor 555 C2-maleimide, and N-(1-pyrene) iodoacetamide (all from Molecular Probes). All fluorescent derivatives of MreB were characterized for assembly properties by light scattering, pelleting and/or fluorescence microscopic assays.
Fluorescence resonance energy transfer
L332C MreB was labeled separately with either Alexa488 or Alexa555. Donor and acceptor molecules were mixed with black wild-type MreB at 3% and 15% of the total concentration, respectively. Polymerization reactions were carried out under standard conditions and polymerization was followed by excitation at 490 nm and measuring emission fluorescence intensity at 518 and 565 nm. Dual emission wavelengths were chosen to measure simultaneously the decrease in donor emission and increase in acceptor emission to maximize signal-to-noise. The 565/518 nm emission ratio was calculated and then normalized to represent the extent of polymerization.
Fluorescence microscopy
Alexa555-labeled L332C MreB and black native MreB were mixed and polymerized at room temperature for 1h in standard buffer conditions at 1 μM total concentration. Samples were applied directly to coverslips and viewed on an Olympus IX-71 epifluorescence microscope using a 60× 1.45 n.a. objective lens and rhodamine filter set (Chroma). Images were acquired using a Hamamatsu Orca 285 camera under control of Wasabi software. Image manipulation, limited to contrast and brightness enhancement, was carried out in Adobe Photoshop CS2. Polymer lengths were measured manually and analyzed using Excel and Kaleidagraph.
Circular dichroism
Ellipticity spectra were recorded between 200 and 255 nm following 5 min equilibration at increasing 5 °C steps in a Model 202SF Circular Dichroism Spectrophotometer (Aviv Biomedical). Melting profiles were determined by plotting 222 nm ellipticity versus temperature and fitting to a standard two-state model.
Results
We cloned and expressed MreB1 from T. maritima without purification tags and purified it to apparent homogeneity in the constant presence of ATP, using ion exchange and gel filtration chromatography and differential centrifugation. The purified protein eluted from gel filtration columns as a single peak consistent with a globular 36 kDa protein. MALDI-TOF confirmed the protein as the 36 kDa product of the TM_0588 gene, MreB1. SDS-PAGE demonstrated that the protein ran as a single band of 36 kDa and we determined by densitometry that the protein was greater than 95% pure. The protein was entirely soluble at 100k × g at 4 °C (Fig. 1). Upon addition of KMEI, MreB polymerized into structures that were insoluble at 100k × g. At 20 °C, MreB polymerized partially in the absence but fully in the presence of KMEI (see also discussion of critical concentration, Fig. 11, and Table 2).
Fig. 1.
Sedimentation of MreB. 4 μM MreB was incubated in CaG8 buffer (2 mM Tris-HCl pH 8.0, 0.1 mM CaCl2, 200 μM ATP, 0.5 mM DTT, 0.02% NaN3) with or without 1x KMEI (10mM imidazole, pH 7.0, 1mM MgCl2, 1 mM EGTA and 20 mM KCl) for either 1 h at 20 °C or overnight at 4 °C. Equivalent volumes of total (T) and 100k × g supernatants (S) and pellets (P) are shown by SDS-PAGE and Coomassie Blue.
Fig. 11.
Bulk critical concentration determination of MreB. MreB was polymerized overnight in A, 10mM imidazole, pH 7.0, 1mM MgCl2, 1 mM EGTA, 20 mM KCl and 200uM ATP or B, 2mM Tris pH 8.0, 0.1mM CaCl2, 200μM ATP and 0.5 mM DTT at 4 °C. Samples were equilibrated to 20 °C for 1 hr and light scattering intensity was measured. Linear fits to the data yield a critical concentration of 500 nM (A) and 1280 nM (B), respectively.
Table 2.
Bulk critical concentration of MreB Critical concentrations were determined as in Fig. 9 using MreB bound to ATP, ADP or GTP. Samples were equilibrated overnight at 4 °C under polymerizing conditions (+KMEI) or in MgG8 buffer (-KMEI) following 1 hr equilibration to the experimental temperature
| Nucleotide | Temp | + KMEI | - KMEI |
|---|---|---|---|
| ATP | 5 | 2100 | n/d |
| 20 | 500 | 1280 | |
| 40 | 260 | 290 | |
| 60 | 55 | n/d | |
| ADP | 20 | 1700 | 2900 |
| GTP | 20 | 660 | n/d |
MreB assembled into polymers that scattered light with approximately fifty-fold greater intensity than did soluble monomers (Fig. 2). The polymerization rate, as determined by the time required for half-maximal assembly, varied with protein concentration. The steady state light scattering intensity varied directly with protein concentration over the range of 1 to 12 μM. We next sought to characterize the requirements for MreB polymerization. MreB polymerizes over a wide range of pH conditions, but is most strongly favored at lower pH (Fig. 3). Divalent cations are required for rapid, extensive polymerization (Ca2+ Fig. 4, Mg2+ not shown). The effects of Ca2+ Mg2+ are quantitatively similar at both 20 °C and 37 °C; each cation achieved approximately 80% of its maximal stimulatory effect by 0.5 mM, and no further increases were observed between 1 mM and 5 mM.
Fig. 2.
Concentration-dependent polymerization of MreB. MreB in Ca-storage buffer was converted to Mg and polymerization was subsequently induced at 20 °C by dilution into buffer containing 10mM imidazole, pH 7.0, 1mM MgCl2, 1 mM EGTA, 20 mM KCl and 200uM ATP. Polymerization timecourse was monitored by 90 ° light scattering at 400 nm. [MreB]: 1 μM (open circles), 2 μM (closed circles), 3 μM (open diamonds), 4 μM (closed diamonds), 5 μM (open triangles), 6 μM (closed triangles), 8 μM (open squares), 10 μM (closed squares), 12 μM (open inverted triangles).
Fig. 3.
pH dependence of MreB polymerization. 8 μM MreB was polymerized at 20 °C in 0.1 mM EGTA, 1 mM MgCl2, 20 mM KCl, 200 μM ATP and 10 mM MES pH 5.5 (open circles), MES pH 6.0 (closed circles), imidazole pH 6.5 (open diamonds), imidazole pH 7.0 (closed diamonds), Tris pH 7.5 (open triangles), Tris pH 8.0 (closed triangles), Tris pH 8.5 (open squares), Tris pH 9.0 (closed squares) or Tris pH 9.5 (open inverted triangles). Polymerization timecourse was followed by 400 nM light scattering.
Fig. 4.
Divalent cation dependence of MreB polymerization. 8 μM MreB was polymerized at 20 °C in 10 mM imidazole pH 8.0 and 200 μM ATP containing CaCl2 at 0.1 (open circles), 0.2 (closed circles), 0.3 (open diamonds), 0.4 (closed diamonds), 0.5 (open triangles), 0.7 (closed triangles), 1.0 (open squares), 1.3 (closed squares), 1.6 (open inverted triangles), or 2 (closed inverted triangles) mM. Polymerization timecourse was followed by 400 nM light scattering.
T. maritima MreB1 has no native cysteine residues, so we introduced cysteines individually at positions 331, 332 and 333, corresponding approximately to the position of the native Cys374 of eukaryotic actins (6). Cysteine at each position efficiently accepted a variety of thiol-reactive fluorophores. In light scattering and pelleting assays, Cys332 MreB behaved indistinguishably from wild-type MreB and was selected for further use. Cys333 and Cys331 differed only mildly in their behavior from Cys332 and wild-type (not shown). Each of the three molecules accepted pyrene labeling, but none showed the characteristic increase in pyrene fluorescence exhibited by eukaryotic actin labeled with pyrene (20).
As a complementary approach to light scattering and pelleting, we followed MreB polymerization by fluorescence resonance energy transfer. We labeled Cys322 MreB with Alexa488 or Alexa555 and found (see Figure 13) that both assembled indistinguishably from wild-type. We doped Alexa488-and Alexa555-labeled Cys332 MreB into black, wild-type MreB and monitored fluorescence emission by both the donor and acceptor fluorophores in timecourse polymerization experiments. We observed efficient energy transfer only under polymerizing conditions and over a wide range of fluorophore ratios. The timecourse of FRET increase approximately followed that of the light scattering signal over a range of MreB concentrations (Fig. 5). After normalization of the two signals to reflect complete polymerization at the endpoints, FRET produced a somewhat lower intensity during the early stages of the reactions as compared to the light scattering signal. This quantitative difference likely results from nonlinearity of the light scattering signal with respect to the physical size of the assembled polymer (15).
Fig. 13.
Epifluorescence microscopy of MreB polymers. A, 1 μM MreB (40% Alexa555-labeled) was polymerized in 10mM imidazole, pH 7.0, 1mM MgCl2, 1 mM EGTA, 20 mM KCl and 200uM ATP for 1 h and imaged directly without dilution by epifluorescence microscopy. Scale bar = 10 μm. B, histogram of poymer lengths. 3.4 +/- 1.6 mm (mean +/- std. dev.) (n=100).
Fig. 5.
MreB polymerization measured by light scattering and fluorescence resonance energy transfer. MreB. Parallel polymerization experiments were carried out in 10mM imidazole, pH 7.0, 1mM MgCl2, 1 mM EGTA, 20 mM KCl and 200uM ATP, using 82% wild-type, unlabeled MreB doped with 3% Alexa488 L322C and 15% Alexa555 L322C MreB. Polymerization timecourse was followed by either 400 nm light scattering or by FRET intensity. 5 μM light scattering (open circles), 5 μM FRET (closed circles), 10 μM light scattering (open triangles), 10 μM FRET (closed triangles), 15 μM light scattering (open squares), 15 μM FRET (closed squares).
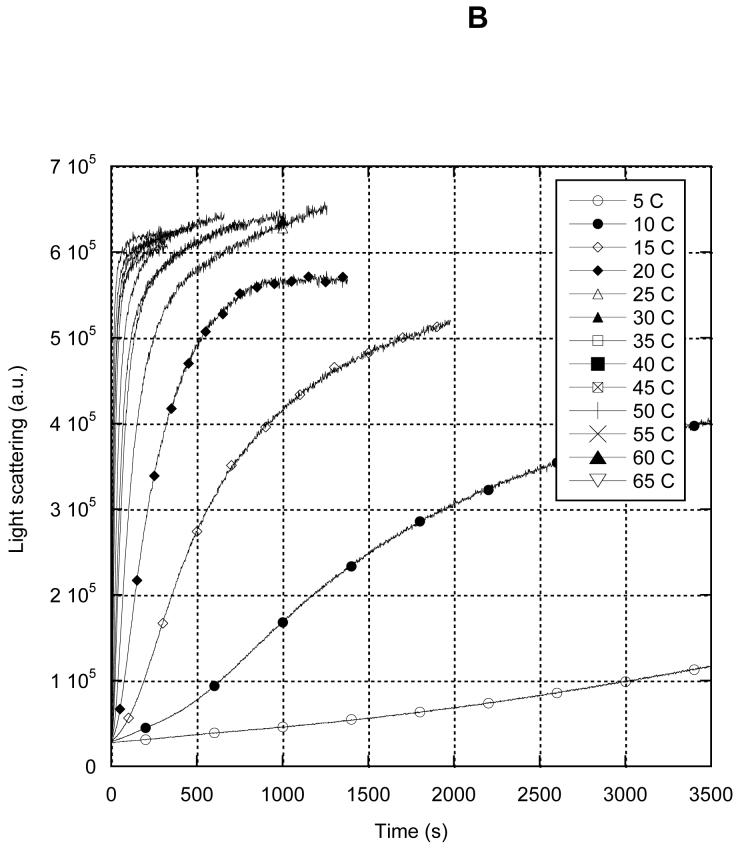
In the presence of divalent cations and ATP, MreB polymerized over a broad range of temperatures (Fig. 6). At 65 °C, polymerization was essentially complete within 1 minute (Fig. 6A), while at 5 °C, several hours were required. At temperatures as low as 15 °C, MreB fully assembled within one hour (Fig. 6B). To ensure that the thermophilic MreB retained a native fold at high temperatures, we determined its thermal denaturation profile by circular dichroism (Fig. 7). We found that in the presence of ATP and divalent cations in an imidazole buffer, MreB retained structure up to 70°C. However under higher salt, more acidic conditions without ATP or divalent cations, the protein melted at 53 °C. This observation may reflect upon previous studies of his-tagged MreB carried out at 65 °C under the latter conditions (9, 10).
Fig. 6.
Temperature-dependence of MreB polymerization. 8 μM MreB was polymerized in 10mM imidazole, pH 7.0, 1mM MgCl2, 1 mM EGTA, 20 mM KCl and 200uM ATP at different temperatures. Polymerization timecourse was followed by 400 nM light scattering. A, first 120 s of reactions. B, full 3500 s of reactions. 5 °C (open circles), 10 °C (closed circles), 15 °C (open diamonds), 20 °C (closed diamonds), 25 °C (open triangles), 30 °C (closed triangles), 35 °C (open squares), 40 °C (closed squares), 45 °C (crossed squares), 50 °C (+), 55 °C (X), 60 °C (closed triangles), 65 °C (open inverted triangles).
Fig. 7.
Thermal stability of MreB. Thermal denaturation of 2.2μM MreB was measured by circular dichroism at 222 nm. Temperature was increased in 5 °C steps with 5 min equibration. Buffer conditions: 100 mM Tris-HCl pH 7.0, 100 mM NaCl (squares) or 2 mM Tris-HCl, pH 8.0, 100 μM CaCl2, 3 μM ATP (triangles). Melting temperatures are 53 °C and 70 °C, respectively.
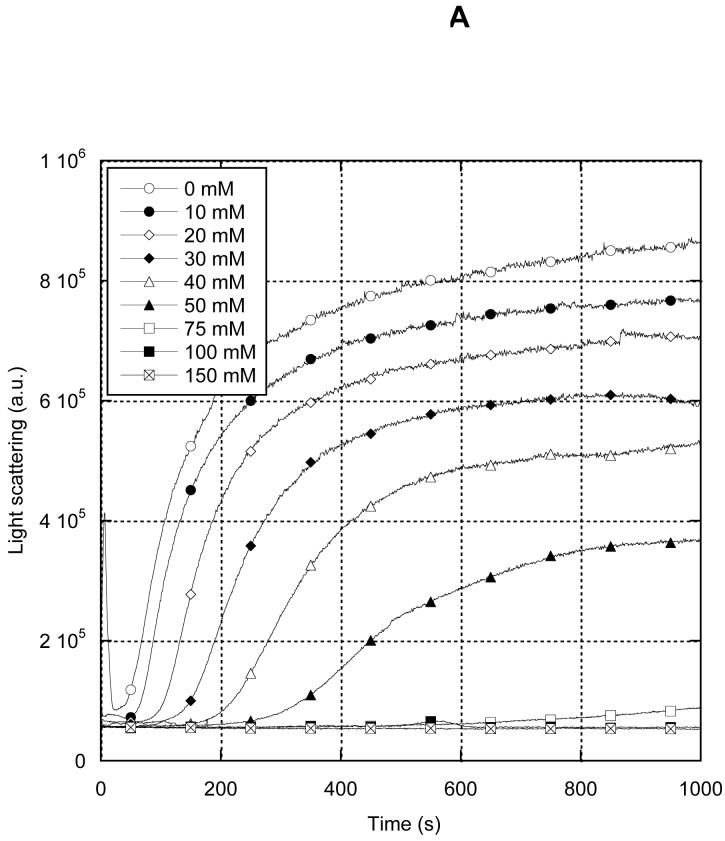
MreB polymerizes similarly in the presence of 1 mM either Ca2+ or Mg2+(Fig. 8A,B, respectively). In both conditions, KCl concentrations as low as 75-100 mM strongly inhibit polymerization and lengthen the lag phase of the reaction. In Mg2+, MreB exhibited a somewhat higher resistance to KCl inhibition and, unlike in Ca2+, assembled much more rapidly in 20 mM than in 0 KCl. In contrast, Van den Ent, et al., (6) observed MreB assembly at high NaCl concentrations. We therefore determined the effects of a variety of monovalent salts on MreB assembly (Table 1). We found that at both 20 °C and 37 °C, all salts tested were inhibitory, NaCl having the most potent effect, followed sequentially by NaOAc, KCl and K-glutamate. In all cases, increasing temperature modulated the inhibitory effects of increasing salt.
Fig. 8.
Salt-sensitivity of MreB polymerization. 8 μM MreB was polymerized in 10 mM imidazole, pH 7.0, 200 μM ATP, varying concentrations of KCl, and 1 mM CaCl2 (A) or MgCl2 (B). 0 mM KCl (open circles), 10 mM (closed circles), 20 mM (open diamonds), 30 mM (closed diamonds), 40 mM (open triangles), 50 mM (closed triangles), 75 mM (open squares), 100 mM (closed squares), 150 mM (crossed squares). 200 mM (+).
Table 1.
Inhibition of MreB assembly by monovalent salts The half-maximal inhibition of MreB assembly was determined by titration of monovalent salts into light scattering polymerization reactions under otherwise standard conditions, as in Figure 8B. Results were confirmed by pelleting assays performed in parallel
| Salt | 20 °C | 37 °C |
|---|---|---|
| NaCl | 30 mM | 70 mM |
| NaOAc | 60 mM | 110 mM |
| KCl | 90 mM | 120 mM |
| K-glut | 150 mM | 200 mM |
Because NaCl was most strongly inhibitory, we sought to explain the previous observation of MreB assembly in 100 mM NaCl. Under buffer conditions identical to those used (6), we found that the inhibitory effects of NaCl were highly dependent on protein concentration (not shown). Increasing the MreB concentration from 5 μM to 14 μM had the effect of increasing the half-maximal NaCl concentration from 50 mM to 200 mM at 20 °C. Therefore the salt inhibition effect can be overcome by increasing protein concentration. Because the cellular concentration of MreB is low (5) but the native temperature of T. maritima is high, we investigated the relative effects of temperature and salt. Under approximately physiological conditions of 65 °C, 1 mM Mg2+ and high salt, the assembly-promoting effect of temperature overcame the salt inhibition, resulting in robust polymerization (not shown).
We followed the ATPase activity of MreB by measuring inorganic phosphate production. MreB did not exhibit measurable ATPase activity under monomeric conditions, but following initiation of polymerization, phosphate release closely mirrored the timecourse of assembly (Fig. 9). Phosphate was produced stoichiometrically with MreB and unlike muscle actin (data not shown) (21), did not lag measurably behind polymerization within the temporal resolution of the present assay, suggesting that ATPase and phosphate release both occur within seconds of polymerization.
Fig. 9.
Phosphate release by MreB. MreB was polymerized in 10 mM imidazole, pH 7.0, 1 mM MgCl2, 20 mM KCl, 200 μM ATP at 20 °C. Polymerization timecourse (open symbols) was determined by 400 nm light scattering. Phosphate production (closed symbols) was determined in parallel experiments using the colorimetric Enz-chek assay. MreB concentration: 6 μM (circles) or 12 μM (triangles). Error bars represent standard deviation of n=3.
We determined the nucleotide release rate from MreB using ε-ATP (18). MreB exchanges nucleotides rapidly, with a release half-time of 19 seconds and a k of 0.036 s-1 (Fig. 10). We then determined the ability of MreB to polymerize when re-bound to a variety of purine nucleotides. MreB polymerizes fully when bound to stoichiometric concentrations of ATP, GTP or AMP-PNP but not at all when no nucleotide is present (data not shown).
Fig. 10.
Nucleotide release by MreB. Dissociation of RATP from Mg-MreB in the presence of 1 μM ATP was followed by excitation at 360 nm and emission at 410 nm. Fitting the normalized 410 nm fluorescence intensity to a single exponential curve-fit gave a k of 0.036 s-1 and T1/2 of 19 s.
We determined the bulk critical concentration for MreB polymerization by light scattering (Fig. 11) and FRET and sedimentation (not shown). In the presence of milimolar divalent cations at 20 °C, the critical concentration of ATP-MreB is 500 nM, roughly similar to eukaryotic actins (22) (Fig. 11A). GTP-MreB exhibited a slightly higher critical concentration (Table 2). ADP-MreB exhibited a critical concentration of 1700 nM, approximately three-fold higher than ATP. ATP-MreB critical concentration varied inversely with temperature, reaching 55 nM at 60 °C and 2100 nM at 5 °C.
We were surprised to find that MreB bound to either ATP or ADP in CaG8 buffer displayed a measurable critical concentration that also varied inversely with temperature and that the specific light scattering intensity as reflected by the slope of the curve was approximately five-to ten-fold less than in polymerization buffer (Fig 11B). These values explain both the incomplete pelleting of MreB in KMEI at 4 °C and the partial pelleting of MreB without KMEI at 20 °C in Fig. 1 and have significant implications for the nucleation mechanism (see discussion).
We compared the magnitude of light scattering by polymerized MreB to that of muscle actin. Although each molecule assembled with a roughly similar timecourse, polymerized MreB scattered light approximately 35 times more strongly than equal concentrations of muscle actin (Fig. 12). This observation is consistent with MreB polymers in solution having much greater bulk than individual actin filaments (23). Early timepoints in the polymerization reactions showed an approximately five-fold greater light scattering intensity by MreB than muscle actin. This may represent the presence of oligomers in the MreB solution initiated by warming.
Fig. 12.
Light scattering intensity of MreB and muscle actin filaments. MreB or rabbit skeletal muscle actin was polymerized at 20 °C in (MreB) 10mM imidazole, pH 7.0, 1mM MgCl2, 1 mM EGTA, 20 mM KCl and 200uM ATP or (actin) 10mM imidazole, pH 7.0, 1mM MgCl2, 1 mM EGTA, 50 mM KCl and 200uM ATP at different concentrations. Polymerization was followed by light scattering. Note logarithmic y-axis. Actin (open symbols) or MreB (closed symbols) at 2 μM (circles), 6 μM (triangles) and 12 μM (squares).
We visualized polymerized MreB covalently labeled with Alexa555 dye (Fig 13A). Unlike eukaryotic actin (24, 25), MreB polymers were readily apparent by standard epifluorescence microscopy at micromolar concentrations, consistent with large bundles of protofilaments. The polymers were uniformly rigid, but varied in fluorescence intensity both along their lengths and between polymers. Average polymer length was 3.3 +/- 1.6 μm (Fig 14B). We observed no evidence for flexibility, branching, severing, growth or other visible dynamics under steady state conditions, other than simple Brownian motion of polymers attached transiently to the coverslips. Electron microscopy confirmed the assembly of MreB into parallel bundles under these conditions (manuscript in preparation).
All light scattering experiments concerning pH, ionic, temperature, and protein concentration effects described above were verified by sedimentation assays (not shown).
Discussion
Here we present the first biochemical characterization of MreB without purification tags and under conditions that maintain a native fold and explain inconsistencies in the literature with respect to MreB assembly. We followed MreB assembly using five complementary assays: pelleting, light scattering, FRET, phosphate production and epifluorescence microscopy.
Two previous studies of T. maritima MreB were carried out at 65 °C in strong tris-based buffers that acidified at the experimental temperature, using his-tagged MreB prepared without ATP or divalent cation (9, 10). Because these studies yielded images of MreB polymers by electron microscopy, at least a fraction of the protein must have retained function. However, we have shown by circular dichroism that under these conditions, most MreB is irreversibly denatured. The strong light scattering observed by Esue, at al., only at high temperatures may largely reflect MreB denaturation or aggregation. Thus, the quantitative components of these studies are called into question. In contrast, we have shown that when purified and stored in the presence of Ca-ATP and maintained at neutral pH, un-tagged MreB is structurally stable; polymerizes at both low and high temperatures; and exhibits a non-zero critical concentration that varies with temperature and nucleotide state.
Consistent with the results of van den Ent (6), we find that MreB undergoes reversible polymerization when tightly bound to a purine nucleotide. Nucleotide binding, independent of species or the presence of a gamma phosphate, is an absolute requirement for assembly of T. maritima MreB. Other factors compete with one another in a quantitative fashion. Polymerization is strongly favored in the presence of millimolar concentrations of divalent cations, but is inhibited by high concentrations of monovalent salts. Polymerization is favored at low pH, high protein concentration and high temperature, but these conditions are not absolute requirements for assembly. Under ionic and temperature conditions that are physiological for T. maritima, MreB does polymerize. Like eukaryotic actins, MreB rapidly hydrolyzes ATP only subsequent to polymerization (21). However, unlike vertebrate actin, but like yeast actin (26), MreB releases inorganic phosphate within seconds of polymerization. As a result, most polymerized MreB in cells will be in the ADP-bound form that less strongly favors assembly. Phosphate production plateaus stoichiometrically with MreB, indicating that each protomer hydrolyzes a single ATP and does not undergo rapid polymerization dynamics subsequent to its initial incorporation into a polymer. If MreB is to turn over rapidly in cells, accessory factors must be involved.
The bulk critical concentration of MreB in the presence of millimolar divalent cations is approximately equivalent to that of eukaryotic actins when measured at the same temperatures (13). When bound to ADP rather than ATP, the critical concentration increases by approximately three-fold. Because of the modest difference between ATP and ADP values, MreB is unlikely to exhibit dynamic instability, as has been observed for the related, plasmid-encoded actin, ParM (27). We have not yet measured the polymerization reaction rates at the two ends of the polymer or discerned the individual critical concentrations at either end. If the values at the two ends differ, then monomers may treadmill directionally through polymers. Such in vivo dynamics are suspected in B. subtilis MreB (28). Because B. subtilis and T. maritima MreB share only limited homology, it will be crucial to characterize the biochemical properties of that molecule directly.
The absence of known rate constants also means that what we and others currently describe as “polymerization rate” reflects both the concentration of polymers in solution and the rate at which each individual polymer elongates. This shortcoming illustrates the need to quantitatively distinguish the two components of the assembly reaction dynamics.
MreB polymerizes much more rapidly at high temperatures, but is fully capable of assembling at temperatures as low as 5 °C. Critical concentration also varies inversely with temperature, such that polymerization is increasingly favored at high temperatures. Unlike muscle actins, which have evolved to maintain approximately equal critical concentrations at the physiological temperatures of species adapted to hot or cold environments (29), T. maritima MreB is likely polymerized to a much greater extent in vivo. We have not directly measured the enthalpic and entropic contributions to the free energy change of the MreB assembly reactions. Nonetheless, the strong temperature dependence of the critical concentration may indicate that T. maritima MreB represents a lower limit for the enthalpic contribution to the polymerization of all actins and that T. maritima MreB polymerization is almost entirely entropy-driven.
We were most surprised to find that MreB polymerizes in the absence of millimolar divalent cations when warmed above 4 °C. Under such conditions MreB scatters significantly less light, but nonetheless pellets at high g-forces and exhibits a distinct critical concentration much higher than that in the presence of divalent cations. We interpret these results as evidence that the polymerization reaction includes both divalent cation-dependent and -independent components. We propose that short oligomers form in a temperature-dependent manner in the absence of divalent cations, but that the presence of millimolar divalent cations drives MreB to assemble into the large ordered bundles seen by electron and fluorescence microscopy and that may exist in cells.
The polymers we observe by fluorescence microscopy are much brighter than equivalently labeled individual eukaryotic actin filaments and exhibit a rigidity that is inconsistent with flexible small-diameter polymers (30). Furthermore, the ability to clearly visualize the polymers through a background of several hundred nanomolar of fluorophores suggests a much larger polymer assembly more closely related to an actin filament bundle or a microtubule than to an individual filament. This is also consistent with previous observations of MreB sheets or bundles by electron microscopy (6, 9, 10) or by fluorescence microscopy of E. coli MreB expressed in S. pombe (11). Assembly of bundles appears to be a property intrinsic to MreB, although the degree of order within the bundles is not yet clear. Because the observed polymers exhibit no curvature over lengths equivalent to those of the cells in which they would reside, bending MreB polymers into the curved or helical structures observed in cells likely results in significant physical forces exerted against the cell wall that may directly affect cell shape. This differs from the small-radius curvature sometimes observed in single MreB protofilaments (9), a discrepancy which still requires resolution.
With the basic parameters that define polymerization established, the next set of questions remains: What is the nucleation mechanism for MreB? What is the structure of the MreB polymer? What are the dynamics of the assembly reaction at each end of the polymer? What is the specific mechanism of action of the drug A22? What molecules affect MreB polymerization? What molecules are affected by MreB? Furthermore, because MreB from extremophiles is highly divergent from those of more generally useful model organisms, it will be important to extend biochemical analyses to MreB from common laboratory bacterial species.
Acknowledgments
We are grateful to Melanie Dayton and Allison Montgomery for technical assistance; to John White and Bill Bement for critical reading of the manuscript; and especially to Josh Mayer for technical assistance, critical reading and many helpful discussions.
This work was supported by an American Heart Association Scientist Development Award to K.J.A. (award number 0430162N). Circular dichroism data were obtained at the University of Wisconsin - Madison Biophysics Instrumentation Facility, which was established with support from the University of Wisconsin-Madison and grants BIR-9512577 (NSF) and S10 RR13790 (NIH).
Abbreviations
- FRET
Fluorescence Resonance Energy Transfer
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- ATP
Adenosine triphosphate
- ADP
Adenosine diphosphate
- GTP
Guanosine triphosphate
- AMP-PNP
5′-adenylyl-beta,gamma-imidodiphosphate
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- EGTA
ethylene glycol tetraacetic acid
- ε-ATP
1,N6-etheno-ATP
- Pi
inorganic phosphate
- kDa
kilodalton
References
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J. Molecular Biology of the Cell - Second Edition. Garland; New York: 1989. [Google Scholar]
- 2.Fitch W. Distinguishing homologous from analogous proteins. Systematic Zoology. 1970;19:99–113. [PubMed] [Google Scholar]
- 3.Koonin EV. Orthologs, Paralogs, and Evolutionary Genomics. Annual Review of Genetics. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli Cell Division Protein FtsZ is a Guanine Nucleotide Binding Protein. PNAS. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones LJF, Carballido-Lopez R, Errington J. Control of Cell Shape in Bacteria: Helical, Actin-like Filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 6.van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 7.Ausmees N, Kuhn JR, Jacobs-Wagner C. The Bacterial Cytoskeleton: An Intermediate Filament-Like Function in Cell Shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 8.Moller-Jensen J, Lowe J. Increasing complexity of the bacterial cytoskeleton. Current Opinion in Cell Biology. 2005;17:75–81. doi: 10.1016/j.ceb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Esue O, Cordero M, Wirtz D, Tseng Y. The Assembly of MreB, a Prokaryotic Homolog of Actin. J. Biol. Chem. 2005;280:2628–2635. doi: 10.1074/jbc.M410298200. [DOI] [PubMed] [Google Scholar]
- 10.Esue O, Wirtz D, Tseng Y. GTPase Activity, Structure, and Mechanical Properties of Filaments Assembled from Bacterial Cytoskeleton Protein MreB. J. Bacteriol. 2006;188:968–976. doi: 10.1128/JB.188.3.968-976.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasan R, Mishra M, Murata-Hori M, Balasubramanian MK. Filament Formation of the Escherichia coli Actin-Related Protein, MreB, in Fission Yeast. Current Biology. 2007;17:266–272. doi: 10.1016/j.cub.2006.11.069. [DOI] [PubMed] [Google Scholar]
- 12.Carballido-Lopez R. The Bacterial Actin-Like Cytoskeleton. Microbiol. Mol. Biol. Rev. 2006;70:888–909. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard TD, Blanchoin L, Mullins RD. Molecular Mechanisms Controlling Actin Filament Dynamics in Nonmuscle Cells. Annu. Rev. Biophys. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 14.Miroux B, Walker JE. Over-production of Proteins inEscherichia coli: Mutant Hosts that Allow Synthesis of some Membrane Proteins and Globular Proteins at High Levels. Journal of Molecular Biology. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 15.Wegner A, Engel J. Kinetics of the cooperative association of actin to actin filaments. Biophys. Chem. 1975;3:215–225. doi: 10.1016/0301-4622(75)80013-5. [DOI] [PubMed] [Google Scholar]
- 16.De La Cruz E, Pollard TD. Nucleotide-free actin: Stabilization by sucrose and nucleotide binding kinetics. Biochemistry. 1995;34:5452–5461. doi: 10.1021/bi00016a016. [DOI] [PubMed] [Google Scholar]
- 17.Waechter F, Engel J. The kinetics of the exchange of G-actin-bound 1,N6-ethenoadenosine 5′-triphosphate with ATP as followed by fluorescence. Eur. J. Biochem. 1975;57:453–459. doi: 10.1111/j.1432-1033.1975.tb02320.x. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, Pollard TD. Profilin Binding to Poly-L-Proline and Actin Monomers along with Ability to Catalyze Actin Nucleotide Exchange Is Required for Viability of Fission Yeast. Mol. Biol. Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida E. Opposite effects of cofilin and profilin from porcine brain on rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry. 1985;24:1160–1164. doi: 10.1021/bi00326a015. [DOI] [PubMed] [Google Scholar]
- 20.Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl) iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur. J. Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- 21.Blanchoin L, Pollard TD. Hydrolysis of bound ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41:597–602. doi: 10.1021/bi011214b. [DOI] [PubMed] [Google Scholar]
- 22.Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang JX, Janmey PA. The Polyelectrolyte Nature of F-actin and the Mechanism of Actin Bundle Formation. J. Biol. Chem. 1996;271:8556–8563. doi: 10.1074/jbc.271.15.8556. [DOI] [PubMed] [Google Scholar]
- 24.Blanchoin L, Amann KJ, Higgs HN, Kaiser DA, Marchand J-B, Mullins RD, Pollard TD. Molecular Biology of the Cell; Abstracts. 2000. Role of ADF/cofilin, Arp2/3 complex, capping protein and profilin in the dynamics of branched actin filament networks. [Google Scholar]
- 25.Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. PNAS. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao X, Rubenstein PA. F-actin-like ATPase Activity in a Polymerization-defective Mutant Yeast Actin (V266G/L267G) J. Biol. Chem. 2001;276:25598–25604. doi: 10.1074/jbc.M011797200. [DOI] [PubMed] [Google Scholar]
- 27.Garner EC, Campbell CS, Mullins RD. Dynamic Instability in a DNA-Segregating Prokaryotic Actin Homolog. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- 28.Defeu Soufo HJ, Graumann PL. Dynamic localization and interaction with other Bacillus subtilis actin-like proteins are important for the function of MreB. Molecular Microbiology. 2006;62:1340–1356. doi: 10.1111/j.1365-2958.2006.05457.x. doi:10.1111/j.1365-2958.2006.05457.x. [DOI] [PubMed] [Google Scholar]
- 29.Swezey R, Somero G. Polymerization thermodynamics and structural stabilities of skeletal muscle actins from vertebrates adapted to different temperatures and hydrostatic pressures. Biochemistry. 1982;21:4496–4503. doi: 10.1021/bi00261a047. [DOI] [PubMed] [Google Scholar]
- 30.Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASp/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]