Abstract
In this study we have exploited the power of insertional mutagenesis to elucidate tumor progression pathways in mice carrying two oncogenes (MYC/Runx2) that collaborate to drive early lymphoma development. Neonatal infection of these mice with Moloney murine leukemia virus (MLV) resulted in accelerated tumor onset with associated increases in clonal complexity and lymphoid dissemination. Large-scale analysis of retroviral integration sites in these tumors revealed a profound bias towards a narrow range of target genes including Jdp2 (Jundm2), D cyclin and Pim family genes. Remarkably, direct PCR analysis of integration hot-spots revealed that every progressing tumor consisted of multiple clones harbouring hits at these loci, giving access to large numbers of independent insertion events and uncovering the contrasting mutagenic mechanisms operating at each target gene. Direct PCR analysis showed that high frequency targeting occurs only in the tumor environment in vivo and is specific for the progression gene set. These results indicate that early lymphomas in MYC/Runx2 mice remain dependent on exogenous growth signals and that progression can be achieved by constitutive activation of pathways converging on a cell cycle checkpoint that acts as the major rate-limiting step for lymphoma outgrowth.
Keywords: RUNX, MYC, insertional mutagenesis, lymphoma, JDP2
Introduction
Over-expression of the c-MYC gene is widespread in human cancer and was the first oncogenic lesion to be modelled in transgenic mice (1,2). These mouse strains have been used extensively to uncover oncogenes and tumor suppressor genes that can collaborate with Myc in tumorigenesis, which include the Runx gene family of transcription factors (3). The RUNX genes also play important roles in human cancer with evidence of both gain and loss of function in the context of different lineages and tumor types (reviewed in (4)). Indeed, RUNX1 is frequently involved in human leukemias where it is subject to a variety of chromosomal translocations causing gene fusions as well as gene amplification, deletion and inactivating point mutations (4,5). Of note for the present study, RUNX1 and MYC are among the most highly over-expressed genes in childhood acute lymphocytic leukemias (6).
We identified all three murine Runx genes as targets for insertional mutagenesis and over-expression in a MYC transgenic model in which this oncogene is directed to the T-cell compartment under the control of a CD2 expression cassette (3,7,8), suggesting that the Runx genes share a redundant oncogenic function in the context of deregulated Myc. To explore this aspect of Runx function, we have studied CD2-Runx2 transgenic mice that are prone to lymphoma development and display impaired thymocyte maturation with an accumulation of immature CD8 cells. Crossing with CD2-MYC mice leads to early tumor onset (9) and our recent studies of the underlying mechanism have indicated that ectopic Myc over-rides the Runx2-imposed proliferation block, while Runx2 expression confers a low apoptotic rate, apparently neutralising the propensity of Myc to induce apoptosis in tumor cells (10,11).
Despite the rapid onset of tumors in MYC/Runx2 transgenic mice, it appears that further events are required to complete oncogenic transformation. Rearranging gene analyses indicate that the tumors arise as outgrowths from an initially polyclonal population in the postnatal thymus (9,12). The identification and characterization of progression genes in MYC/Runx2 tumors is therefore of considerable interest for the further elucidation of this collaboration mechanism.
Retroviral insertional mutagenesis is a classical method of identifying genes relevant to cancer, and has been particularly effective in the study of haematopoietic malignancies (13). Based on the assumption that retroviral insertion is effectively random, the occurrence of a common insertion site in independent tumors is indicative of a selective process driving tumorigenesis and the proximity of a gene whose expression or function is affected by retroviral integration. The development of high throughput PCR methods and completion of human and murine genome sequences has led to a resurgence of interest in the use of retroviruses as genetic screening tools in cancer. Analysis of mice infected with strains of MLV or retrotransposons has revealed many genes with the potential to be targeted1. More refined developments of this approach include collaboration tagging, where the technique is used to detect co-operating genes in mice carrying a dominant oncogene or with a defect in a tumor suppressor gene (3,14-17) and complementation tagging, where mutagenesis is used to tag functional homologues of genes in mice deleted in one or more known targets (18). More recently, infection of mice with a defect in DNA repair has been employed to shift the target gene spectrum towards tumor suppressor loci (19). In this study we show that retroviral insertional mutagenesis can be used to elucidate the rate-limiting steps in tumor progression and identify the gene families and pathways that drive this process.
Materials and Methods
Transgenic mice and lymphomas
CD2-MYC/CD2-Runx2 transgenic mice (hereafter described as MYC/Runx2) on a C57Bl/6xCBA/Ca strain were generated as previously described (9). Newborn bi-transgenic mice were infected with 105 infectious units of Moloney MLV (15) within 24h birth. Genotypes of mice were identified by Southern blot hybridization analysis carried out on DNA extracted after tail biopsy. MYC/Runx2 animals revealed exclusively multicentric lymphomas from which high molecular weight DNA was isolated. All animal work was carried out in line with the UK Animals (Scientific Procedures) Act of 1986.
Cloning of proviral insertion sites
Proviral insertion sites were amplified using the splinkerette-based approach as previously described (18) with slight modifications (A.Uren, The Netherlands Cancer Institute, personal communication). Briefly 3μg of tumor DNA was digested with BstYI (New England Biolabs). Following inactivation of enzyme, 300ng of digested DNA was ligated to 0.12pmole of the splinkerette adaptor with 4U T4 DNA ligase (Roche) overnight at 16°C. To avoid subsequent amplification of the internal 3′ MoMLV fragment, the ligated mixture was digested with an excess of EcoRV followed by DNA purification using Qiagen columns. Proviral/genomic DNA junction fragments were isolated after two rounds of PCR amplification. 100ng of ligated DNA was used in the primary PCR containing 4U Pfu Turbo hotstart (Stratagene) and 200nM of each primer (Splink1 and LTR#5, sequences available on request). The hot-start PCR conditions were 3 min at 94°C (1 cycle), 15 sec at 94°C, 30 sec at 68°C, 5 min at 72°C (2 cycles), 15 sec at 94°C, 30 sec at 66°C, 5 min at 72°C (27 cycles) 5 min at 72°C (1 cycle). A nested PCR was carried out using 2μl of the primary PCR with 200nM of primer (Splink2 and LTR#1, primer sequences available on request) and 12.5μl Qiagen Multiplex PCR kit mix (Qiagen). The PCR conditions were 15min at 94°C (1 cycle), 15 sec at 94°C, 90 sec at 60°C, 3 min at 72°C (25 cycles), 5 min at 72°C (1 cycle). PCR products were visualized on 4% polyacrylamide gels and a ladder of fragments obtained ranging from ∼100bp to ∼2000bp. 1μl of the nested PCR reaction was shotgun cloned in the Topo TA vector (Invitrogen) and DNA from 24 transformants isolated and analysed by both EcoRI digestion followed by gel electrophoresis and DNA sequencing (BigDye terminator mix V2.0, Applied BioSystems). Gel electrophoresis allowed comparison of the cloned products with the nested PCR to ensure that all of the prominent bands in the PCR were represented in the shotgun cloning.
Sequence analysis
Homology searches of all of the sequences isolated were carried out using the publically available BLASTn in GenBank databases and mouse genome database, February 2006 draft assembly2 and identified annotated candidate genes located near each retroviral insertion site. We compared these sites with previously identified insertion sites in the Mouse Retroviral Tagged Cancer Gene Database (RTCGD)3.
Direct DNA PCR
Amplification was carried out on 1μg aliquots of genomic DNA with 50pmol of primer pairs in 2 X Reddy Mix (Abgene). MLV-LTR specific primers were LTR-S (5′-CCACCTGTAGGTTTGGCAAGC), LTR-AS (5′-CCAAACCTACAGGTGGGGTCTTTC or 5′- CTGTTCCATCTGTTCCTGACC). Gene specific primers were as follows: Jdp2 exon3 (JDP-R 5′-CATCTGGCTGCAGCGACTTTG), Pim1 exon 6 (PIM-F 5′-GGACAGCAATGACAACTCATTCC or 5′- GAAATCCGGAACCATCCATGG), Ccnd1 exon 1 (CCND-R 5′-CGCTGCCTCGCGCTGTACTG), Gfi-1 exon1 (GFI1-R 5′-ACATGCTCTTGCTAACAGCTGGC), Rorc exon 2 (RORC-F 5′-GCTGGCTGCAAAGAAGACCCA). Amplification conditions were 5 min at 94°C (1 cycle), 1 min at 94°C, 1 min at 60°C, 1min at 72°C (30 cycles), 5 min at 72°C (1 cycle) for all MLV/gene specific amplifications with the exception of PIM-F/LTR-AS PCR. Amplification conditions were 5 min at 94°C (1 cycle), 1 min at 94°C, 1 min at 55°C, 1min at 72°C (30 cycles), 5 min at 72°C (1 cycle). PCR products were separated on 1-2%TBE agarose gels and visualized by staining in EtBr.
RT-PCR
1μg of total RNA was reverse transcribed using ImProm-II Reverse Transcription System (Promega). Amplification was performed on aliquots of one-twentieth of the sample with 40pmole each of primer pair MLV-RU5 (5′-GCAGTTGCATCCGACTTGTGG) and JDP-R. Amplification was performed in 2x Reddy Mix (Abgene) at 95°C 30secs, 60°C 30secs, 72°C 30secs for 30 cycles.12.5μl aliquots were separated on 2% TBE agarose gels and visualized by staining in EtBr
DNA Hybridization analysis
High molecular weight DNA from mouse lymphomas and radiolabelled probes were prepared as previously described (9). MLV proviral sequences were detected using a probe derived from the U3 domain of the LTR (20). Pim1 probe (Pim1A) as previously described (19).
Results
Neonatal infection with Moloney MLV accelerates tumor onset and increases clonal complexity in MYC/Runx2 mice
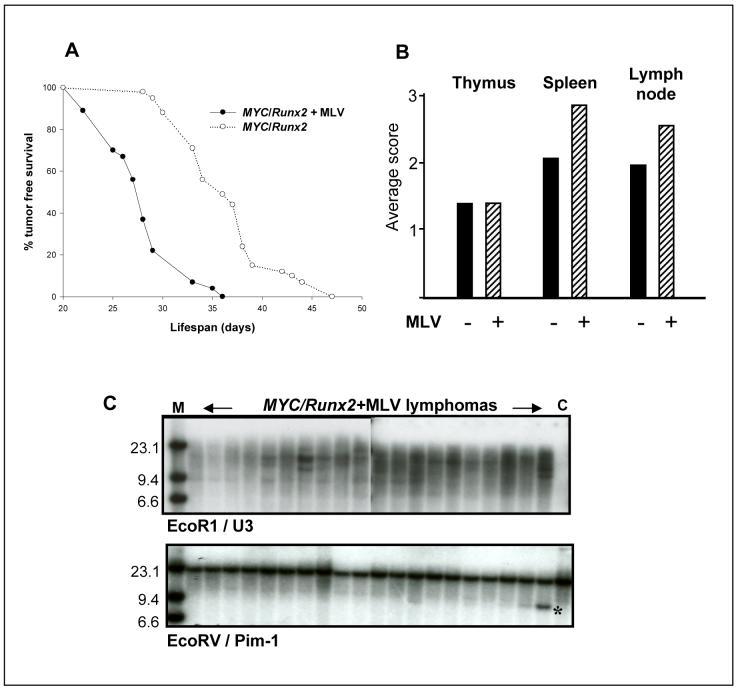
To test the ability of MLV to drive tumor progression, MYC/Runx2 mice were infected at birth and monitored for development of disease. Uninfected MYC/Runx2 mice develop lymphomas in the thymus and peripheral lymphoid tissues at an average age of 36 days (9,12). MLV caused a significant acceleration (P = < 0.0001) of tumor development, with infected animals developing multi-centric lymphomas by 28 days (Fig.1A). While tumor pathology was grossly similar, it was notable that the virus-accelerated tumors displayed greater involvement of extra-thymic lymphoid tissues (Fig.1B). Analysis of these lymphomas for clonal complexity by T-cell receptor gene rearrangement revealed a pattern of increased complexity in the infected tumors, suggesting that viral infection increases the number of expanding tumor cell clones and not merely the growth rate of transformed cells (not shown). As an independent marker of tumor cell clonality and a clue to the role of insertional mutagenesis in rapid tumor progression, the pattern of proviral integration junction fragments was assessed by hybridization with a U3 probe. This analysis showed that the lymphomas from MLV infected mice contained multiple viral copies with some evidence of emerging dominant clones as identified by unique junction fragments (Fig.1C). Screening for rearrangements at candidate target genes was carried out using common insertion site probes including Pim-1, Pim-2, c-Myb, Ahi-1, Bmi-1, Evi-5 and Gfi-1, most of which have been found as targets in virus-accelerated tumors of Runx2 mice (12). The results were negative, apart from Pim-1 where we noted a clonal and subclonal rearrangement (Fig.1C) and Pim-2 where a single tumor with a subclonal rearrangement was observed (not shown).
Figure 1.
Retroviral infection accelerates tumor onset and increases clonal complexity in MYC/Runx2 mice. A, Tumor free survival of MYC/Runx2 (o; n=41) and MYC/Runx2 infected with MLV (•; n=27) mice. B, Tumor involvement in thymus, spleen and lymph nodes was scored on a scale of 0-3 in MYC/Runx2 (filled columns; -MLV (n=28) and MYC/Runx2 infected (hatched columns; +MLV (n=19)) mice. C, Southern analysis of genomic DNA from MLV infected MYC/Runx2 mice and uninfected littermate control (C) DNA digested with EcoRI or EcoRV and probed with a viral specific U3-LTR (upper panel) and Pim1 3′UTR (lower panel) specific probes. *; rearranged Pim1 allele detected in a single MLV-infected MYC/Runx2 mouse, M; high molecular weight DNA markers
Direct PCR reveals multiple insertions at Pim-1 in all virus-accelerated MYC/Runx2 lymphomas
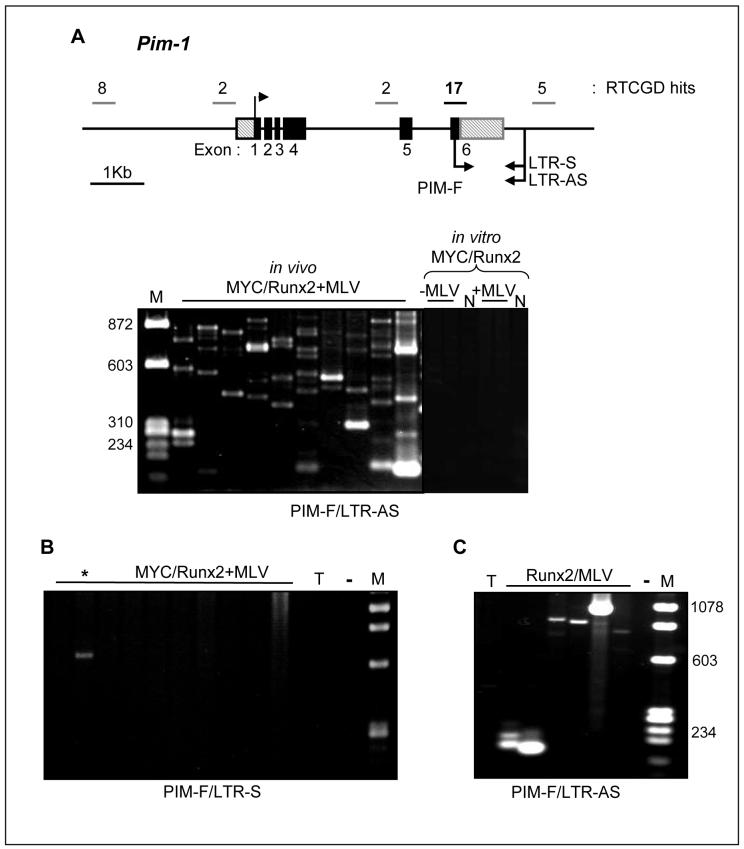
Detection of Pim-1 as a potential progression gene was consistent with the fact that an Eμ-Pim-1 transgene accelerates tumor onset in MYC/Runx2 mice (12). As MLV insertions that activate Pim-1 frequently cluster within the 3′UTR (15), we considered the possibility that the Southern blot analysis underestimated the prevalence of hits at Pim-1 due to the clonal complexity of the tumors and devised a direct PCR assay to detect insertions at this site. As shown in Figure 2A, this analysis revealed a remarkably high number of hits with every tumor containing multiple independent MLV insertions, lying in the same orientation as Pim-1, distributed throughout the 3′UTR. The authenticity of these insertions was confirmed by probe analysis and sequencing. In contrast, use of a LTR primer to detect proviruses in the antisense orientation at this site revealed only a single insertion from the tumor panel, confirming the bias in orientation of activating insertions at Pim-1 observed in previous studies (Fig.2B). The detection of multiple insertions in the MYC/Runx2 + MLV tumors contrasted with longer-latency tumors of Runx2 mice infected with MLV and harbouring insertions at Pim-1, which displayed a much simpler pattern, indicative of only one or a small number of cell clones with these insertions (Fig.2C). The band intensities in these cases were significantly higher and from dilution experiments we estimate that most of the oligoclonal insertions in the MYC/Runx2 tumors represent from 0.1-1% of the tumor cell mass, indicating that the cells carrying these insertions have undergone significant clonal expansion (not shown).
Figure 2.
Pim1 direct DNA PCR assay reveals multiple sense MLV integrations in 3′UTR. A, Upper panel, genomic organization of Pim1 gene with location and number of mapped MLV integrations indicated (RTCGD hits). 3′UTR integration hot spot is shown in bold. Upper arrow indicates transcription start site, lower arrows indicate direction of PCR primers, LTR sense (LTR-S), antisense (LTR-AS) and Pim1 exon 6 (PIM-F). Lower panel; PCR products amplified from a selection of MYC/Runx2+MLV lymphomas, MYC/Runx2 cell lines and control cells (N; NIH3T3) infected in vitro. B, lack of anti-sense MLV integrations in MYC/Runx2+MLV lymphomas. *; single PCR product amplified in one lymphoma. C, Single PCR products amplified in Runx2 infected mice. T; uninfected thymus DNA, -; negative control for PCR assay, M; low molecular weight DNA markers. Primer pairs used for individual assays are indicated.
High frequency gene targeting is a specific feature of the in vivo tumor environment
To investigate the possibility that the sensitive direct PCR assay was detecting an intrinsic bias of MLV integration rather than the result of growth selection of clones carrying insertions at Pim-1, we infected cells with Moloney MLV in vitro and grew these for 28 days to recapitulate the in vivo tumor latent period after neonatal infection. We infected NIH3T3 fibroblasts and a series of T-cell tumor cell lines established from the MYC/Runx2 background to model as closely as possible the genetic and transcriptional environment of the in vivo target cell. Direct PCR on DNA from these cells revealed no detectable hits at the Pim-1 3′UTR hot-spot in fibroblasts (Fig.2A). Southern blot analysis and hybridization with a U3 specific probe confirmed successful infection with MLV (data not shown). Faint bands seen on the stained gels were mostly non-specific and did not hybridize to a Pim-1 probe, although low-level insertions could be detected in one of the in vitro infected lymphoma lines (not shown).
Large-scale analysis of retroviral insertion sites reveals multiple hits at a narrow range of target genes including Jdp2, D cyclin and Pim genes
The discovery of Pim-1 as a frequent progression target encouraged us to conduct a wider analysis for other genes that might play an analogous role. For an unbiased screen of proviral insertion sites, we used a splinkerette based PCR approach (18). Shot-gun libraries of clones were generated from individual tumors and insert sizes were determined to ensure that all unique integrants were sequenced (see Methods). In total, 480 putative viral junction fragments were isolated, cloned and sequenced. Following elimination of non-informative clones and duplicate clones from the same tumor, 272 independent retroviral integration sites were identified. Homology (BLASTn) searches of the sequences isolated were carried out and candidate genes located near each tag identified. Genes targeted more than once in the dataset are shown in Table 1 (and Supplementary Table 1 for precise chromosomal coordinates). It is evident that the target loci detected in this screen are far from a random selection from the murine genome. Comparison with the RTCGD3 shows that 12 of the 18 genes targeted more than once correspond to known common insertion sites. Of the remaining 6 genes, 5 have annotated functions of potential relevance to cancer. Notably, genes that regulate survival and cell cycle progression are highly represented in the target set.
Table1.
Summary of genes represented more than once in a library of 272 insertion sites cloned from 20 lymphomas arising in MLV infected MYC/Runx2 transgenic mice
| Gene | # hits (tumors)1 | Known CIS2 | Product | Reported function relevant to cancer |
|---|---|---|---|---|
| Jdp2 | 16 (8) | Y | b-ZIP protein | Transformation, survival |
| Ccnd1 | 12 (9) | Y | Cyclin | Cell cycle, survival |
| Pim2 | 8 (6) | Y | Pim family protein kinase | Survival, proliferation |
| Rasgrp1 | 5 (4) | Y | Ras guanyl releasing protein | Proliferation, transformation |
| Pim1 | 4 (4) | Y | Pim family protein kinase | Survival, proliferation |
| Ccnd3 | 4 (4) | Y | Cyclin | Cell cycle, survival |
| Cstad | 4 (4) | Y | Mitochondrial protein | Mitochondrial biogenesis |
| Otx2 | 3 (3) | N | Homeodomain protein | Survival, tissue specification |
| Pik3r5 | 3 (2) | Y | PI3 kinase regulatory subunit | Survival, cell cycle |
| Tnfsf13b | 3 (2) | N | TNF family cytokine (Baff, BlyS) | Lymphocyte survival, cell cycle |
| Akt1 | 2 (2) | Y | Akt protein kinase | Survival, cell cycle |
| Bfsp2 | 2 (2) | N | Cytoskeletal protein | - |
| Ccrk | 2 (2) | N | Cell cycle related kinase (p42, PNQARLE) | Survival, cell cycle |
| Eif3s2 | 2 (2) | N | Translation initiation factor | Cell growth |
| Mad1l1 | 2 (2) | N | Mitotic spindle assembly checkpoint protein | Cell cycle |
| Rai17 | 2 (2) | Y | MIZ zinc finger domain | Localization to replication foci |
| Slc9a9 | 2 (2) | Y | Solute carrier | - |
| Ssbp3 | 2 (1) | Y | Single-stranded DNA binding | DNA replication |
Numbers in parenthesis represent number of tumors carrying gene specific insertions.
CIS, common insertion site
The most frequently targeted gene, Jdp2 (Jun dimerising protein 2; or Jundm2) has been observed previously as a common insertion site in a number of MLV-induced tumors from p27-deficient or wild-type mouse backgrounds (17,21) and encodes a b-ZIP protein that binds to Jun family proteins and represses AP-1 transcription (22). The D-cyclin genes Ccnd1 and Ccnd3 were also highly represented in the progression gene set with a total of 16 hits. The insertions corresponded to favored sites for gene activation, either upstream and in the reverse orientation or in the sense orientation within the 3′UTR. Two members of the Pim kinase family, Pim-1 and Pim-2, are also represented in the progression gene set with 4 and 8 hits respectively. As expected from the direct PCR analysis in Figure 2A, insertions cluster in the 3′UTR hotspot for Pim-1 and upstream of Pim-2 in the opposite orientation, the preferred mode for MLV activation each gene (13). While all of the frequently targeted gene set have been observed previously in wild-type mouse models3, the percentage hit rates recorded here are unprecedented.
Genes targeted 2 or 3 times in this study might conceivably have been observed by chance (23), but this seems unlikely in cases where the pattern and clustering of insertions are indicative of gene activation (Pik3r5, Akt1, BLyS, Otx2) or inactivation (Mad1l1) (Supplementary Table 1). Moreover, these rarer targets include known common insertion sites (Pik3r5, Akt1) and novel targets (Ccrk, BLyS, Mad1l1) with functional relevance to cancer (24-26).
Direct PCR confirms positive and specific selection of the progression gene set
As a further test for the specificity of the progression set, we devised direct PCR assays based on hot-spots in Ccnd1, which was frequently targeted in the MYC/Runx2+MLV tumors, and Gfi-1, which was not detected. The lack of hits at Gfi-1 was of interest as this is a potent collaborator with Myc that has also been observed as a target in virus-infected Runx2 mice (12). These assays reinforced the findings of the large-scale random cloning analysis, with frequent insertions detected at Ccnd1 (Fig. 3A), but none at the Gfi-1 cluster (Fig. 3B). We also generated a PCR assay for a locus that was targeted only once in the random cloning panel (Rorc). This assay detected a product only from the index tumor and showed no amplification products in the rest of the panel (Fig. 3C). Direct DNA PCR was carried out on MLV-induced tumors from strain matched non-transgenic control mice, revealing clonal insertions in only a subset of the tumors (Ccnd1 2/13, Pim-1 3/13, Jdp2 4/13, not shown), emphasising the bias towards these loci in the MYC/Runx2 tumors where 28/28 scored positive. Again, a contrast was evident with Gfi-1, where direct PCR assay revealed 2/13 insertions in the infected wild-type and 0/28 in the MYC/Runx2 tumors (not shown).
Figure 3.
Direct PCR confirms specificity of progression gene set. A, Integrations upstream of the Ccnd1 gene were assayed by direct DNA PCR. PCR products, amplified from a representative selection of MYC/Runx2+MLV lymphomas using sense (LTR-S) and anti-sense LTR (LTR-AS) primers in combination with a Ccnd1 specific exon 1 primer (CCND-R) are shown. B, Lack of MLV integrations observed in a commonly targeted region of the Gfi1 gene using LTR-AS and Gfi-1 exon 1 (GFI-R) primer pair. C, Rorc DNA PCR assay on a selection of MYC/Runx2+MLV lymphomas amplified a unique PCR product (*) in the index tumor only. PCR products visible across all lanes including uninfected thymus control (T) represent non-specific amplimers. -; negative control for PCR assay, M; low molecular weight DNA markers.
A major integration hot-spot in Jdp2 reveals selection for fusion transcripts and protein truncation
The most highly targeted gene in the large-scale analysis was Jdp2, where 80% (13/16) of the tags mapped to the 3′end of intron 2. A direct Jdp2 DNA PCR assay was devised to detect similar insertions (Fig. 4A). This analysis revealed multiple insertions in all of the MYC/Runx2 lymphomas and the authenticity of these products was again confirmed by blot hybridization analysis with a Jdp2-specific probe and by sequencing of selected clones. Integrations were detected in both the sense and antisense orientation with a slightly greater representation of antisense amplicons. Despite the very high hit rate in the tumors, we again found no evidence of insertions at this site in cells infected in vitro (Fig. 4A).
Figure 4.
Identification of the Jdp2 gene as a frequent target for MLV insertion in MYC/Runx2 lymphomas. A, Genomic organisation of Jdp2 gene with transcription start site (upper arrow) in exon 2 and PCR primers indicated. Direct DNA PCR (upper panel) using specific MLV LTR (LTR-S/LTR-AS) and Jdp2 exon 3 (JDP-R) primers amplifies multiple products from MYC/Runx2+MLV lymphomas in vivo. No integrations at Jdp2 were detected in in vitro infected T-cell tumor cell lines established from the MYC/Runx2 background and control fibroblasts (N). RT-PCR (lower panel) using a sense MLV primer (R-U5) and Jdp2 exon3 (JDP-R) primer amplifies multiple products from MYC/Runx2+MLV lymphomas in vivo. T; uninfected thymus, E; uninfected T-cell line EL4. B, Clustering of MLV integration sites in Jdp2 intron 2 and structure of fusion transcripts shown. Filled in arrows depict site and orientation of integration. sd;splice donor, sa;splice acceptor, P?; cryptic promoter element, M; methionine. C, Predicted amino acid sequence of truncated Jdp2 proteins derived from fusion transcripts. Amino acids in bold and underlined indicate histone binding domain located in exon 2. Only the first 9 amino acids of Jdp2 exon 3 are shown.
The presence of insertions in the sense orientation suggested that these might drive the expression of hybrid transcripts and this prediction was confirmed by RT-PCR analysis (Fig. 4A). We mapped a total of 83 independent insertion sites in intron 2 by either direct PCR or RT-PCR, and the locations of these are shown in Figure 4B. Direct sequence analysis of the RT-PCR products showed that the fusion transcripts derive from the viral 3′LTR, suggesting that they arose by read-through or de-repression of the 3′LTR promoter. An interesting difference between the sense and antisense insertions is that the latter do not occur close to the exon boundary (Fig. 4B). Scanning of the intron sequence for polII promoter sequences4 reveals a marginal prediction at the point shown by an arrow in Figure 4B, suggesting that the insertions in the antisense orientation may activate a cryptic promoter element.
The fusion transcripts contain in-frame ATGs derived from Jdp2 intron 2 close to the exon boundary or from further upstream due to splicing into exon 3 (Fig. 4B). Insertions in the opposite orientation would be predicted to drive the expression of similar products if the cryptic promoter element in intron 2 is activated. As shown in Figure 4C, a notable feature of the fusion proteins predicted from these insertions is that they lack an N-terminal domain that has been shown to be important for histone binding and inhibition of p300-mediated histone acetylation at target promoters (27). The products should therefore be defective in the AP1 repressive activity of the wild-type Jdp2 protein, but may be expected to have novel biological properties as they retain the b-ZIP domain through which Jdp2 heterodimerizes with c-Jun and related proteins (22).
Discussion
This study has shown that retroviral insertional mutagenesis can be used to drive tumor progression in vivo and to identify relevant target genes in tumors arising in highly tumor-prone mice. Large-scale analysis of integrated proviruses was able to detect a strong skew towards a subset of MLV targets that can be linked mechanistically to the initiating oncogenic programme. The ability of one of the frequent targets, Pim-1, to collaborate independently with Myc and Runx2 (12,28), and with the combination of both genes (12) confirms the relevance of the progression gene set detected in this study. Moreover, direct PCR of integration hot-spots validated the findings of random sampling and confirmed that the bias towards specific loci is a function of selection operating on cells in the in vivo tumor environment rather than an intrinsic integration bias of Moloney MLV. It seems likely that each clone arises from a single mutagenic hit, as the probability of hitting two loci simultaneously in a single round of infection is very low, and tumor onset is probably too fast to allow the generation and spread on MLV recombinants which could circumvent envelope-mediated interference (29).
The skew towards integrations at specific target genes observed here is even more profound than that seen previously in panels of retrovirus-induced tumors from wild-type mice or those carrying a single genetic lesion (17,18,30,31), presumably reflecting the limited repertoire of genes capable of efficient synergy with the MYC/Runx2 oncogenic programme. The predominance of hits at Ccnd1 is somewhat surprising in light of its low endogenous expression levels in lymphoid cells (32), and this observation lends further support to the argument that gene targeting by retroviruses in cancer is not simply a function of preferred integration into highly expressed genes (23).
An unanticipated advantage of the progression tagging approach we have described here is that the polyclonal nature of the tumors allows many insertions to be sampled from a relatively small series of tumors, providing superior insights into underlying mechanisms of gene deregulation. Thus, insertions within the Pim1-3′ UTR were entirely consistent with the established enhancer insertion mechanism (33), displaying a strong bias with regard to orientation but no evident selection for precise location within this domain. In contrast, insertions in Jdp2 intron 2 often appeared to select for disruption of the coding sequence and the expression of truncated proteins.
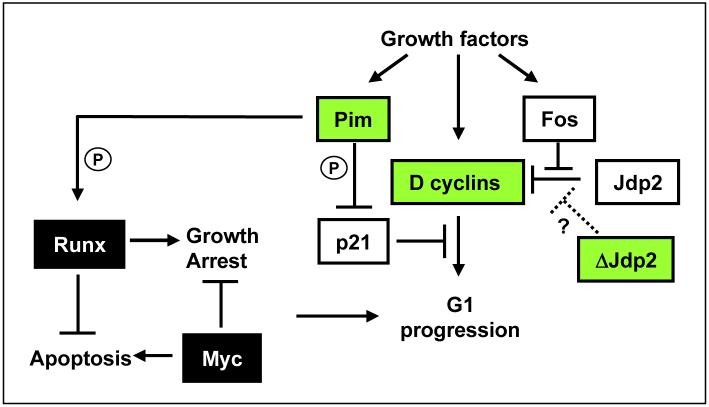
Figure 5 presents a model based on our current understanding of the oncogenic collaboration of MYC and Runx2 (4,11), incorporating the progression set as accessory factors interacting with the basic feedback loop. Our previous studies have indicated that Runx2 and MYC collaborate by neutralising each other’s fail-safe responses. The induction of growth arrest by ectopic Runx2 (10) is not fully understood but may conceivably involve cdk inhibitors such as p21Waf1 that can be induced or repressed by Runx (34) and can be counteracted by ectopic Myc (35). Runx2 appears to inhibit Myc-induced apoptosis in vivo although the underlying pathways remain to be identified (11). The Pim kinases have recently been shown to phosphorylate Runx proteins directly (36), but may also feed into G1 checkpoint controls through phosphorylation and inactivation of p21Waf1 (37). However, it should be noted that this model may represent only a subset of relevant Pim functions as these kinases have additional targets in cell cycle and apoptosis control (38).
Figure 5.
Proposed model based on our current understanding of the oncogenic collaboration of MYC and Runx2, incorporating the progression set as accessory factors interacting with the basic feedback loop.
The frequent targeting of D cyclins implies that G1 checkpoint controls are rate-limiting for the growth of MYC/Runx2 expressing cells. This observation is reminiscent of classical studies in fibroblasts where Myc was characterized as a competence factor capable of mediating G0/G1 transition, but unable to complete progression to S phase unless complemented by progression factors (39,40). It is conceivable that progression signals are provided in the MYC/Runx2 thymus by stromally presented growth factors or TCR ligation, allowing limited expansion of pre-neoplastic cells. In this model, tumor progression results from genetic or epigenetic changes mediating release from the requirement for external mitogenic signals. The progression gene set we have observed fits this model well, as the expression of both D-cyclins and Pim kinases is strongly dependent on mitogenic signals (38,41), and this process should be short-circuited by proviral gene activation. The model is also consistent with the biology of virus-accelerated tumors which display wider lymphoid dissemination (Fig. 1B). In further support of this model, some of the less frequently targeted common insertion sites we identified such as Akt1, Pi3kr5, BLyS would also be expected to feed into this checkpoint through induction of D cyclin expression (25,42).
The role of Jdp2 in cancer has been the subject of conflicting findings, but we favor a model in which the truncated proteins feed into the same D cyclin-responsive checkpoint through mimicry of growth factor-induced c-Fos activation. Jdp2 protein was discovered as a repressor of AP-1 signalling that is expressed in quiescent cells where it forms heterodimers with Jun family proteins and is displaced by c-Fos upon serum induction (22). Recent studies support its putative tumor suppressor role by showing that the full-length protein can suppress both Ras transformation of 3T3 fibroblasts and the tumorigenicity of prostate cancer xenografts (43). Notably, the truncated proteins that are predicted to be expressed as a result of retroviral insertions in Jdp2 intron 2 lack a domain that has been identified recently to be critical for histone binding and transcriptional repression (27). These observations suggest a scenario where the truncated Jdp2 proteins block repression of AP1-responsive promoters by homodimerizing with wild-type Jdp2 or by forming non-functional heterodimers with other Jun family members. Such an explanation leaves us with a puzzle with regard to those cases where MLV has inserted upstream of the gene and appears to be associated with enhanced transcription (17,21), although it should be noted that the integrity of the Jdp2 sequence has not been analysed in such cases. Functional analysis of full-length and truncated Jdp2 variants will be required to answer these questions.
Our findings show that insertional mutagenesis is a powerful genetic tool in the study of tumor progression. However, there is a darker side to these findings as the ability of viral insertions to drive tumor progression with unexpectedly high efficiency raises further concerns about the safety of retroviral vectors in human cancer treatment, where they have been used for the purposes of cell marking (44) and delivery of prodrug-activating enzymes (45). In light of these results, it would seem prudent to avoid the use of integrating vectors for such applications, or at least ensure that their powerful mutagenic potential has been neutralized.
Supplementary Material
Acknowledgements
Grant support: We are grateful to Cancer Research UK and the Leukemia Research Fund who provided major support for this work and to the UK Department of Health for support to LS. We thank Finn Pedersen for sharing unpublished data on Jdp2 and also thank Sharon Mackay and Margaret Bell for technical assistance and Ashleigh Manning who assisted with PCR assays.
Footnotes
References
- 1.Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–37. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 2.Adams JM, Harris AW, Pinkert LM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–8. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 3.Stewart M, Terry A, Hu M, et al. Proviral insertions induce the expression of bone-specific isoforms of PEBP2alphaA (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci U S A. 1997;94:8646–51. doi: 10.1073/pnas.94.16.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blyth K, Cameron ER, Neil JC. The Runx gene family: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–87. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 5.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukemia. Nat Rev Cancer. 2002;2:502–13. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 6.Niini T, Kanerva J, Vettenranta K, Saarinen-Pihkala UM, Knuutila S. AML1 gene amplification: a novel finding in childhood acute lymphoblastic leukemia. Haematologica. 2000;85:362–6. [PubMed] [Google Scholar]
- 7.Stewart M, Mackay N, Cameron ER, Neil JC. The common retroviral insertion locus Dsi1 maps 30kb upstream of the P1 promoter of the murine Runx3/Cbfa3/Aml2 gene. J Virol. 2002;76:4364–9. doi: 10.1128/JVI.76.9.4364-4369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wotton S, Stewart M, Blyth K, et al. Proviral insertion indicates a dominant oncogenic role for Runx1/AML1 in T-cell lymphoma. Cancer Res. 2002;62:7181–5. [PubMed] [Google Scholar]
- 9.Vaillant F, Blyth K, Terry A, et al. A full length Cbfa1 gene product perturbs T-cell development and promotes lymphomagenesis in synergy with MYC. Oncogene. 1999;18:7124–34. doi: 10.1038/sj.onc.1203202. [DOI] [PubMed] [Google Scholar]
- 10.Vaillant F, Blyth K, Andrew L, Neil JC, Cameron ER. Enforced expression of Runx2 perturbs T cell development at a stage coincident with beta selection. J Immunol. 2002;169:2866–74. doi: 10.4049/jimmunol.169.6.2866. [DOI] [PubMed] [Google Scholar]
- 11.Blyth K, Vaillant F, Mackay N, et al. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res. 2006;66:2195–201. doi: 10.1158/0008-5472.CAN-05-3558. [DOI] [PubMed] [Google Scholar]
- 12.Blyth K, Terry A, Mackay N, et al. Runx2: a novel oncogenic effector revealed by in vivo complementation and retroviral tagging. Oncogene. 2001;20:295–302. doi: 10.1038/sj.onc.1204090. [DOI] [PubMed] [Google Scholar]
- 13.Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–72. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 14.Adams JM, Cory S. Oncogene cooperation in leukaemogenesis. Cancer Surv. 1992;15:119–41. [PubMed] [Google Scholar]
- 15.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E-mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–52. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 16.Clurman BE, Hayward WS. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol Cell Biol. 1989;9:2657–64. doi: 10.1128/mcb.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang HC, Martins CP, Bronkhorst Y, et al. Identification of oncogenes collaborating with p27(Kip1) loss by insertional mutagenesis and high-throughput insertion site analysis. Proc Nat Acad Sci U S A. 2002;99:11293–8. doi: 10.1073/pnas.162356099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikkers H, Allen J, Knipscheer P, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32:153–9. doi: 10.1038/ng950. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J. 2006;25:3422–31. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuypers HT, Selten G, Quint W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37:141–50. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen MH, Sorensen AB, Morris DW, et al. Tumor model-specific proviral insertional mutagenesis of the Fos/Jdp2/Batf locus. Virology. 2005;337:353–64. doi: 10.1016/j.virol.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Aronheim A, Zandi E, Hennemann H, Elledge SJ, Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol Cell Biol. 1997;17:3094–102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu XL, Luke BT, Burgess SM. Redefining the common insertion site. Virology. 2006;344:292–5. doi: 10.1016/j.virol.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 24.Wohlbold L, Larochelle S, Liao JCF, et al. The cyclin-dependent kinase (CDK) family member PNQALRE/CCRK supports cell proliferation but has no intrinsic CDK-activating kinase (CAK) activity. Cell Cycle. 2006;5:546–54. doi: 10.4161/cc.5.5.2541. [DOI] [PubMed] [Google Scholar]
- 25.Huang XG, Di Liberto M, Cunningham AF, et al. Homeostatic cell-cycle control by BLyS: Induction of cell-cycle entry but not G(1)/S transition in opposition to p18(INK4c) and p27(Kip1) Proc Nat Acad Sci U S A. 2004;101:17789–94. doi: 10.1073/pnas.0406111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsukasaki K, Miller CW, Greenspun E, et al. Mutations in the mitotic check point gene, MAD1L1, in human cancers. Oncogene. 2001;20:3301–5. doi: 10.1038/sj.onc.1204421. [DOI] [PubMed] [Google Scholar]
- 27.Jin CY, Kato K, Chimura T, et al. Regulation of histone acetylation and nucleosome assembly by transcription factor JDP2. Nat Struct Mol Biol. 2006;13:331–8. doi: 10.1038/nsmb1063. [DOI] [PubMed] [Google Scholar]
- 28.Verbeek S, van Lohuizen M, van der Valk M, Domen J, Kraal G, Berns A. Mice bearing the E-mu-myc and E-mu-pim-1 transgenes develop pre-B cell leukemia prenatally. Mol Cell Biol. 1991;11:1176–9. doi: 10.1128/mcb.11.2.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baxter EW, Blyth K, Donehower LA, Cameron ER, Onions DE, Neil JC. Moloney murine leukemia virus induced lymphomas in p53 deficient mice: overlapping pathways in tumor development? J Virol. 1996;70:2095–100. doi: 10.1128/jvi.70.4.2095-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki T, Shen H, Akagi K, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–74. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 31.Lund AH, Turner G, Trubetskoy A, et al. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet. 2002;32:160–5. doi: 10.1038/ng956. [DOI] [PubMed] [Google Scholar]
- 32.Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 Transgene Impedes Lymphocyte Maturation and Collaborates in Lymphomagenesis with the Myc Gene. EMBO J. 1994;13:2124–30. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuypers HT, Selten G, Zjilstra M, de Goede R, Melief C, Berns A. Tumor progression in murine leukemia virus-induced T-cell lymphomas: monitoring clonal selections with viral and cellular probes. J Virol. 1986;60:230–1. doi: 10.1128/jvi.60.1.230-241.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lutterbach B, Westendorf JJ, Linggi B, Isaac S, Seto E, Hiebert SW. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J Biol Chem. 2000;275:651–6. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- 35.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–34. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 36.Aho TL, Sandholm J, Peltola KJ, Ito Y, Koskinen PJ. Pim-1 kinase phosphorylates RUNX family transcription factors and enhances their activity. BMC Cell Biol. 2006;7:21–9. doi: 10.1186/1471-2121-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang ZP, Bhattacharya N, Mixter PF, Wei WY, Sedivy J, Magnuson NS. Phosphorylation of the cell cycle inhibitor p21(Cip1/WAF1) by Pim-1 kinase. BBA Mol Cell Res. 2002;1593:45–55. doi: 10.1016/s0167-4889(02)00347-6. [DOI] [PubMed] [Google Scholar]
- 38.Bachmann M, Moroy T. The serine/threonine kinase pim-1. Int J Biochem Cell Biol. 2005;37:726–30. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Kaczmarek L, Hyland JK, Watt R, Rosenberg M, Baserga R. Microinjected C-Myc As A Competence Factor. Science. 1985;228:1313–5. doi: 10.1126/science.4001943. [DOI] [PubMed] [Google Scholar]
- 40.Jones SM, Kazlauskas A. Growth-factor-dependent mitogenesis requires two distinct phases of signalling. Nat Cell Biol. 2001;3:165–72. doi: 10.1038/35055073. [DOI] [PubMed] [Google Scholar]
- 41.Musgrove EA. Cyclins: Roles in mitogenic signaling and oncogenic transformation. Growth Factors. 2006;24:13–9. doi: 10.1080/08977190500361812. [DOI] [PubMed] [Google Scholar]
- 42.Gille H, Downward J. Multiple Ras effector pathways contribute to G(1) cell cycle progression. J Biol Chem. 1999;274:22033–40. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- 43.Heinrich R, Livne E, Ben Izhak O, Aronheim A. The c-Jun dimerization protein 2 inhibits cell transformation and acts as a tumor suppressor gene. J Biol Chem. 2004;279:5708–15. doi: 10.1074/jbc.M307608200. [DOI] [PubMed] [Google Scholar]
- 44.Brenner MK, Rill DR, Moen RC, et al. Gene-Marking to Trace Origin of Relapse After Autologous Bone-Marrow Transplantation. Lancet. 1993;341:85–6. doi: 10.1016/0140-6736(93)92560-g. [DOI] [PubMed] [Google Scholar]
- 45.Kan O, Kingsman S, Naylor S. Cytochrome P450-based cancer gene therapy: current status. Expert Opin Biol Ther. 2002;2:857–68. doi: 10.1517/14712598.2.8.857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.