Abstract
Vesicular secretion of macromolecules has recently been described in the basidiomycete Cryptococcus neoformans raising the question as to whether ascomycetes similarly utilize vesicles for transport. In the present study, we examine whether the clinically important ascomycete Histoplasma capsulatum produce vesicles and utilized these structures to secrete macromolecules. Transmission electron microscopy (TEM) show transcellular secretion of vesicles by yeast cells. Proteomic and lipidomic analyses of vesicles isolated from culture supernatants reveals a rich collection of macromolecules involved in diverse processes including metabolism, cell recycling, signaling, and virulence. The results demonstrate that H. capsulatum can utilize a trans-cell wall vesicular transport secretory mechanism to promote virulence. Additionally, TEM of supernatants collected from Candida albicans, Candida parapsilosis, Sporothrix schenckii, and Saccharomyces cerevisiae document that vesicles are similarly produced by additional ascomycetes. The vesicles from H. capsulatum react with immune serum from patients with histoplasmosis providing an association of the vesicular products with pathogenesis. The findings support the proposal that vesicular secretion is a general mechanism in fungi for the transport of macromolecules related to virulence and that this process could be a target for novel therapeutics.
Keywords: Histoplasma capsulatum, ascomycete, vesicles, transcellular transport
Introduction
Histoplasma capsulatum, a dimorphic fungus of the phylum Ascomycota, is a major human pathogen with a worldwide distribution (Kauffman, 2007). The fungus usually causes a mild, often asymptomatic respiratory illness, but infection may progress to life-threatening systemic disease, particularly in immunocompromised individuals, infants, or the elderly. H. capsulatum grows as a saprophytic mould in the environment but undergoes phase transition to a yeast form at mammalian physiological temperatures. Within macrophages, H. capsulatum modifies its microenvironment over a broad pH range, survives nutrient-starvation, resists reactive oxygen and nitrogen species, and survives exposure to degradative enzymes (Woods, 2002). In the yeast form, several important exoantigens have been described, including the H and M antigens, pluripotent glycoproteins that elicit both humoral and T-cell-mediated immune responses (Deepe and Gibbons, 2001b; Fisher and Woods, 2000; Zancope-Oliveira et al., 1999), and a virulence-related, phase specific protein (YPS3p), that is found at the cell wall (Bohse and Woods, 2007; Bohse and Woods, 2005). Yeast cells secrete a calcium-binding protein (CBP) that enables the fungus to grow in calcium-limiting conditions (Sebghati et al., 2000). Heat shock proteins are also produced at a high level, which is consistent with the thermally dimorphic nature of the organism (Burnie et al., 2006).
In contrast to prokaryotic organisms, secretory pathways in eukaryotic cells involve vesicular traffic of molecules to the plasma membrane (van Meer and Sprong, 2004; Ponnambalam and Baldwin, 2003). Fungal cells have complex cell walls and are therefore expected to require additional mechanisms to transfer periplasmic components from the plasma membrane to the extracellular space. The mechanisms by which macromolecules reach the extracellular environment and how they are transported through the cell wall, however, have not been rigorously explored in fungi. It has been recently described that the yeast-like pathogen Cryptococcus neoformans produces secretory vesicles that transport its major capsular polysaccharide to the extracellular space (Rodrigues et al., 2008; Rodrigues et al., 2007; Yoneda and Doering, 2006). The polysaccharide is synthesized intracellularly (Garcia-Rivera et al., 2004; Feldmesser et al., 2001) and packaged into lipid vesicles, which cross the cell wall and the capsule network by still unknown mechanisms to reach the extracellular environment. At the extracellular space, the polysaccharide is released and presumably used for capsule assembly (Rodrigues et al., 2007). Furthermore, bioactive fungal lipids, including glucosylceramides and sterols, are secreted by C. neoformans vesicles (Rodrigues et al., 2007). It remains unknown whether other pathogenic fungal species use the same mechanism to secrete extracellular molecules.
In the present study, we demonstrate that the parasitic yeast stage of H. capsulatum produces heterogeneous vesicles that are secreted extracellularly. A considerable variety of molecules, including phospholipids and proteins associated to stress responses, pathogenesis, cell wall architecture and virulence comprise the H. capsulatum vesicles. Furthermore, we analyzed whether additional ascomycetes, including Candida albicans, Candida parapsilosis, Sporothrix schenckii and Saccharomyces cerevisiae, produced vesicles. Finally, proteins extracted from H. capsulatum vesicles reacted with immune sera from patients with histoplasmosis suggesting that the vesicles are involved in host-pathogen interactions. These results show that vesicular secretion is a common mechanism of extracellular delivery in fungi.
Results
H. capsulatum produces extracellular vesicles
Extracellular vesicles were obtained from H. capsulatum yeast. Using our growth conditions, H. capsulatum is in exponential phase growth for the first 72–76 hours. At the time of collection, the yeast cells were >99% viable by propidium iodine staining, which makes the possibility of the vesicles arising from dead or dying cells exceedingly unlikely. TEM of the material recovered by ultracentrifugation of supernatants from H. capsulatum revealed the presence of bilayered, spherical vesicles (Fig. 1). Five hundred and eight vesicles were analyzed and were found to range in size from 10 to 350 nm (Fig. 2). The electron density of the vesicles varied considerably, suggesting distinct contents (Fig. 1). The protocol used for the isolation of H. capsulatum extracellular vesicles was based on that used for C. neoformans (Rodrigues et al., 2007), in which organelles from dead cells were not found. Similarly, organelles were not found in culture supernatants of heat-killed H. capsulatum yeast cells examined by TEM (data not shown). Notably, we identified vesicular structures in internal and outer regions of the cell wall, as well as in the extracellular environment (Fig. 3), which is in accordance with the proposal that vesicle secretion is an active mechanism in living H. capsulatum cells. Vesicles were identified in and adjacent to the cell walls of all yeast cells analyzed (n = 200) indicating that this is a pervasive process.
Fig.1.
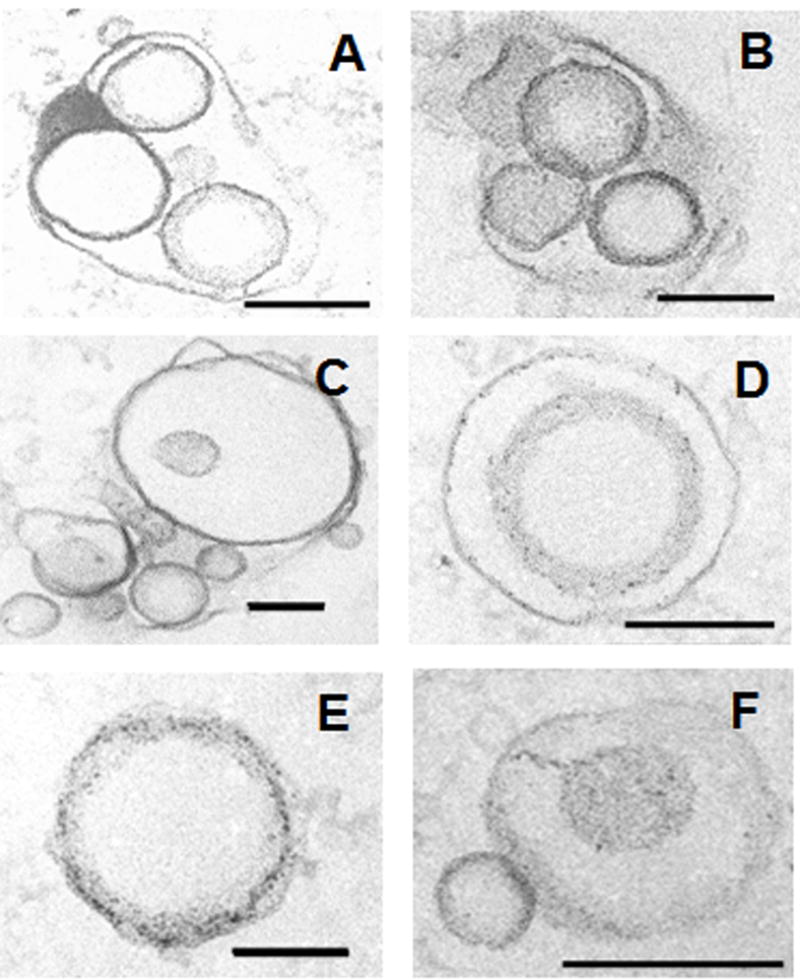
TEM of extracellular vesicles obtained by ultracentrifugation of culture supernatants from Histoplasma capsulatum showing bilayered membranes and different profiles of electron density. Bars, 100 nm (B, C and E) and 200 nm (A, D and F).
Fig.2.
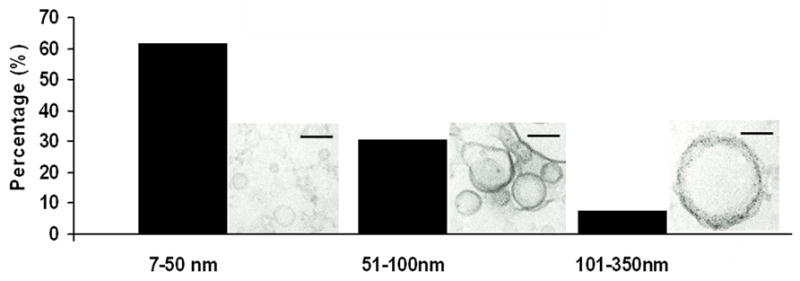
Size analysis of vesicles from H. capsulatum. Five hundred and eight vesicles were analyzed and the size ranged from 10 to 350 nm.
Fig.3.
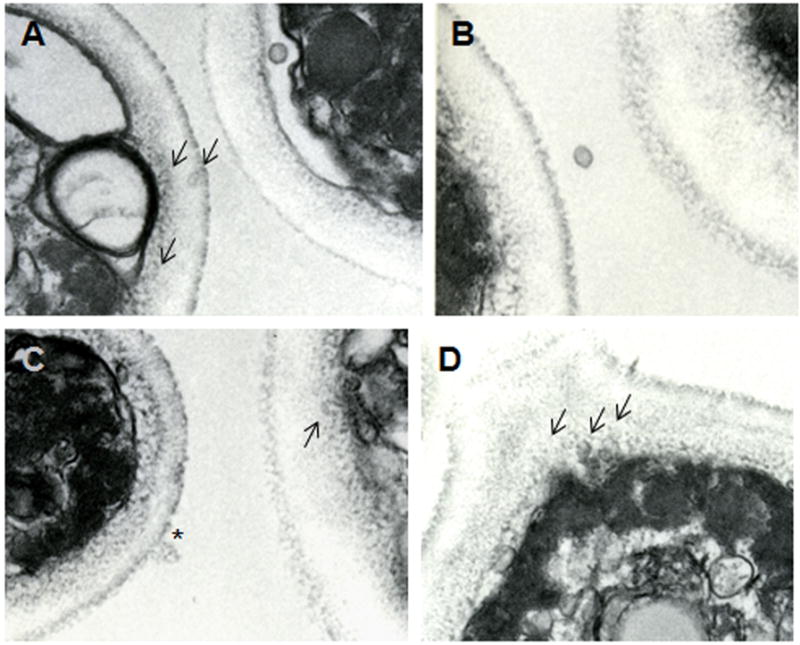
Vesiclular structures were observed in association with the cell wall (A, C and D) and the extracellular environment (B).
Membrane phospholipids are present in vesicular lipid extracts
Lipids were fractioned and analyzed by ESI-MS, in negative or positive-ion mode. The regions of the spectra in which molecular masses corresponding to phospholipids were expected are presented in Figure 4. The major peaks observed in both spectra were subjected to MS/MS analysis (Supplemental Figure 1), resulting in the identification of 17 different phospholipids (Table 1). In the negative-ion mode analysis, only phosphatidylethanolamine (PE) species were detected as major phospholipid species (Fig 4, Table 1). As shown in the Supplemental Figure 1A–E, diagnostic ions for PE were found at m/z 140.1 and 196.1, corresponding to ethanolamine phosphate (EtNP) and dehydrated glyceroethanolaminephosphate (GroEtNP/H2O), respectively. Fragment ions corresponding to the carboxylate ions of the acyl chains were also detected. On the other hand, the positive-ion mode analysis revealed mainly PE, phosphatidylserine (PS), and phosphatidylcholine (PC) as the major phospholipid species (Table 1, Supplemental Figure 1F-P). MS/MS spectra of PE species revealed diagnostic ions corresponding to the presence of cyclic ethanolaminephosphate (EtNPc) plus 2 Li+ adducts (m/z 152.0), and the neutral losses of ethanolamine (EtN) and ethanolaminephosphate (EtNP). MS/MS spectra of PC species were characterized by the presence of the diagnostic ion choline (Cho) at m/z 86.0, and neutral losses of trimethylamine (Me3N) and phosphocholine (ChoP). Finally, MS/MS spectra of PS species were characterized by the presence of dehydrated serinephosphate (SerP - H2O) and serinephosphate (SerP) ion species at m/z 168.0 and 186.0, respectively. The neutral loss of carboxyl group from Ser was also detected in most PS species (Supplemental Figure 1). For all phospholipid species analyzed in the positive-ion mode, the composition of acyl chains was determined by the neutral loss of these structures. In sum, PE and PC, followed by PS, were the most abundant phospholipids found in the MS analyses, consistent with the typical lipid distribution in pathogenic yeasts (Rattray et al., 1975).
Fig.4.
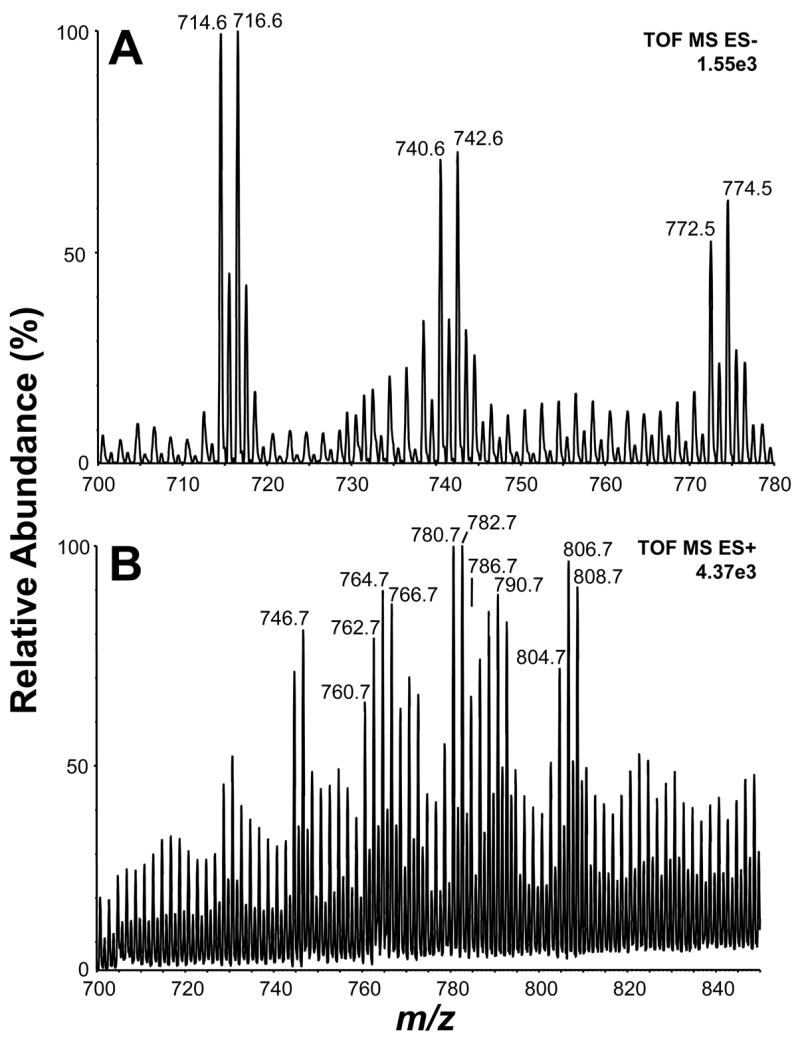
Lipid analysis by mass spectrometry of H. capsulatum vesicular components. Total phospholipids were fractionated by silica gel 60 chromatography and analyzed by ESI-MS, in negative- (A) or positive-ion (B) mode. The ion species corresponding to major phospholipids are indicated. These ions were subjected to MS-MS analysis, allowing the identification of 18 phospholipids (Table 1; Supplemental Figure 1). m/z, mass to charge ratio.
Table 1.
Lipid analysis and composition of the major phospholipids from H. capsulatum vesicles.
| MS Ion-Mode | Observed m/z, [ion species] | Proposed Phospholipid Compositiona | Predicted Mass (Da) | Observed Mass (Da) |
|---|---|---|---|---|
| Negative | 714.6, [M - H]− | C16:0/C18:2-PE | 715.5 | 715.6 |
| Negative | 716.6, [M - H]− | C16:0/C18:1-PE | 717.5 | 717.6 |
| Negative | 740.6, [M - H]− | C18:2/C18:1-PE | 741.5 | 741.6 |
| Negative | 742.6, [M - H]− | C18:1/C18:1-PE | 743.6 | 743.6 |
| Negative | 772.5, [M - H + Na+ + Cl−]+ | C16:0/C18:2-PE | 715.5 | 715.5 |
| Negative | 774.5, [M - H + Na+ + Cl−]+ | C16:0/C18:1-PE | 717.5 | 717.5 |
| Positive | 746.7, [M+ - H + Li+]+ | C16:0/C20:4-PE | 739.5 | 739.7 |
| Positive | 760.7, [M + H]+ | C16:0/C18:2-PS | 759.5 | 759.7 |
| Positive | 762.7, [M + H]+ | C16:0/C18:1-PS | 761.5 | 761.7 |
| Positive | 764.7, [M+ - H + Li+]+ | C16:0/C18:2-PC | 758.6 | 758.7 |
| Positive | 766.7, [M+ - H + Li+]+ | C16:0/C18:1-PC | 760.6 | 760.7 |
| Positive | 780.7, [M+ - H + Na+]+ | C16:0/C18:2-PC | 758.6 | 758.7 |
| Positive | 782.7, [M+ - H + Na+]+ | C16:0/C18:1-PC | 760.6 | 760.7 |
| Positive | 786.7, [M + H]+ | C18:1/C18:2-PS | 785.5 | 785.7 |
| Positive | 790.7, [M+ - H + Li+]+ | C18:1/C18:2-PC | 784.6 | 784.7 |
| Positive | 804.7, [M+ - H + Na+]+ | C16:0/C20:4-PC | 782.6 | 782.7 |
| Positive | 806.7, [M+ - H + Na+]+ | C18:1/C18:2-PC | 784.6 | 784.7 |
| Positive | 808.7, [M+ - H + Na+]+ | C18:1/C18:1-PC | 786.6 | 786.7 |
Deduced from the MS/MS spectrum (see Supplemental Figure 1).
Proteomic analysis of the H. capsulatum extracellular vesicles
After vesicle purification, proteins were enzymatically digested and resulting peptides were fractionated by cation exchange chromatography and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). All generated MS/MS spectra were searched against a database assembled with H. capsulatum predicted sequences and randomly generated sequences. After estimating the false-positive rate (FPR), 283 proteins were validated and 206 identified by sequence analysis. Table 2 summarizes the identified proteins with associated biological function(s). A comprehensive list of all identified proteins and detailed parameters of the LC-MS/MS analysis are provided in Supplemental Table 1. Some of these proteins, such as chaperones (Hsp70, Hsp30, and Hsp60 precursors), superoxide dismutase, and catalase B, are involved in H. capsulatum pathogenesis and host immune responses. Others (e.g., Rab GDP-dissociation inhibitor, Rab1a, GTP-binding nuclear protein GSP1/Ran) are involved in signal transduction pathways and vesicle formation. We also identified several proteins implicated in cell wall architecture, cell growth, sugar, lipid, and amino acid metabolism, as well as cytoskeleton-related proteins. Several peroxisomal, nuclear, proteasomal, and ribosomal proteins and proteins with additional localization/function were also identified. Many of these proteins were recently described in the proteome of vesicles from C. neoformans (Rodrigues et al., 2008) as well as in mammalian vesicles (Aoki et al., 2007; Potolicchio et al., 2005). Table 3 shows the distribution of the identified H. capsulatum vesicle proteins according to their functions.
Table 2.
Protein components of H. capsulatum vesicles.
| Protein hit number | H. capsulatum genome database accession number | Protein identification | Function* |
|---|---|---|---|
| Chaperone-like proteins | |||
| 1 | HCAG_06961.1 | Heat shock protein 60, mitochondrial precursor | Chaperone |
| 2 | HCAG_01398.1 | Heat shock 70 kDa protein 7 | Chaperone |
| 3 | HCAG_00783.1 | Heat shock protein Hsp88 | Chaperone |
| 4 | HCAG_08176.1 | Heat shock protein SSC1, mitochondrial precursor | Chaperone |
| 5 | HCAG_05805.1 | Heat shock 70 kDa protein C precursor | Chaperone |
| 6 | HCAG_04111.1 | Heat shock protein 30 kDa | Chaperone |
| 7 | HCAG_04686.1 | ATP-dependent molecular chaperone HSC82 | Chaperone |
| 8 | HCAG_05524.1 | Hypothetical protein similar to chaperonin subunit 7 | Chaperone |
| Cell wall architecture | |||
| 9 | HCAG_06565.1 | Endochitinase 1 precursor | Cell wall assembly |
| 10 | HCAG_02853.1 | Hypothetical protein similar to beta-glucosidase 5 | Cell wall assembly |
| 11 | HCAG_01828.1 | Beta-glucosidase 4 | Cell wall assembly |
| 12 | HCAG_05285.1 | Beta-1,3-glucanosyltransferase 3 | Cell wall assembly |
| 13 | HCAG_02309.1 | Hypothetical protein similar to exo-1,3-beta-D-glucanase | Cell wall assembly |
| 14 | HCAG_08883.1 | Chitin synthase B | Cell wall assembly |
| 15 | HCAG_01250.1 | Endochitinase 1 precursor | Cell wall assembly |
| 16 | HCAG_04277.1 | Hypothetical protein similar to mannosidase II | Cell wall assembly |
| 17 | HCAG_05925.1 | Hypothetical protein similar to N-acetyl-beta-glucosaminidase | Hydrolysis of chitin oligomers |
| 18 | HCAG_00683.1 | Woronin body major protein | Septal pore sealing in response to cellular damage |
| 19 | HCAG_07031.1 | CPC2 protein | Putative receptor for protein kinase C in the regulation of actin cytoskeleton organization during cell wall synthesis and morphogenesis |
| Cell signaling | |||
| 20 | HCAG_07830.1 | Rab GDP-dissociation inhibitor | Regulation of the secretory pathway |
| 21 | HCAG_06999.1 | Rab1a | GTP binding; small GTPase mediated signal transduction |
| 22 | HCAG_05187.1 | GTP-binding nuclear protein GSP1/Ran | GTP-binding protein involved in nucleocytoplasmic transport. |
| 23 | HCAG_05560.1 | Rho GTPase | GTP binding; regulation of multiple signaling pathways |
| 24 | HCAG_01447.1 | Hypothetical protein similar to GTP binding protein | GTP binding |
| 25 | HCAG_04173.1 | 14-3-3 protein epsilon (308 aa) | Adapter protein implicated in the regulation of a large spectrum of both general and specialized signaling pathways |
| 26 | HCAG_04527.1 | 14-3-3 family protein ArtA | Cell signaling |
| 27 | HCAG_04840.1 | GTP-binding protein sarA | Regulation of transport from the endoplasmic reticulum to the Golgi apparatus |
| Sugar metabolism | |||
| 28 | HCAG_08808.1 | Phosphoglucomutase | Breakdown and synthesis of glucose |
| 29 | HCAG_03803.1 | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor | Ketogenesis, production of acetyl-CoA and from (S)-3-hydroxy-3-methylglutaryl-CoA |
| 30 | HCAG_06184.1 | Isocitrate lyase | Glyoxylate cycle; production of L-malate from isocitrate |
| 31 | HCAG_02260.1 | 3-isopropylmalate dehydrogenase A | Oxidation of 3-carboxy-2-hydroxy-4-methylpentanoate (3-isopropylmalate) to 3-carboxy-4-methyl-2-oxopentanoate |
| 32 | HCAG_07619.1 | Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor | Conversion of pyruvate to acetyl-CoA and CO |
| 33 | HCAG_04910.1 | Glyceraldehyde-3-phosphate dehydrogenase | Glycolysis; pyruvate formation from D-glyceraldehyde 3-phosphate |
| 34 | HCAG_06981.1 | Citrate synthase, mitochondrial precursor | Tricarboxylic acid cycle |
| 35 | HCAG_00638.1 | Transaldolase | Balance of metabolites in the pentose-phosphate pathway |
| 36 | HCAG_04227.1 | Hypothetical protein similar to pyruvate carboxylase | Tricarboxylic acid cycle |
| 37 | HCAG_03969.1 | Malate dehydrogenase, mitochondrial precursor | Conversion of malate into oxaloacetate |
| 38 | HCAG_08720.1 | Mannitol-1-phosphate dehydrogenase | Mannitol synthesis |
| 39 | HCAG_05084.1 | Hypothetical protein similar to malate synthase 2 | 2-Isopropylmalate formation from the condensation of the acetyl group of acetyl-CoA with 3-methyl-2-oxobutanoate |
| 40 | HCAG_00626.1 | Sorbitol utilization protein SOU1 | NADP dependent reduction of L-sorbose to D-glucitol |
| 41 | HCAG_00064.1 | Hypothetical protein similar to phosphoacetylglucosamine mutase | Interconverts N-acetylglucosamine-6-P and N-acetylglucosamine -1-P |
| 42 | HCAG_03323.1 | Hypothetical protein similar to fumarate reductase | Unidirectional fumarate reduction |
| 43 | HCAG_08493.1 | Fumarate hydratase class II | Conversion of malate into fumarate, tricarboxylic acid cycle |
| 44 | HCAG_03385.1 | Phosphoglycerate kinase | Conversion of 3-phospho-D-glycerate into 3-phospho-D-glyceroyl phosphate; glycolysis. |
| 45 | HCAG_07781.1 | Hypothetical protein similar to CDC19 Also known as: Pyruvate kinase 1 | Glycolysis, conversion of pyruvate to phosphoenolpyruvate |
| 46 | HCAG_05884.1 | Hypothetical protein similar to 6-Phosphogluconate dehydrogenase | Formation of D-ribulose 5-phosphate; pentose phosphate pathway |
| 47 | HCAG_08202.1 | Glucose-6-phosphate isomerase | Conversion of D-glucose 6-phosphate into D-fructose 6-phosphate; glycolysis |
| 48 | HCAG_01552.1 | Mannose-1-phosphate guanyltransferase | Synthesis of GDP-mannose; protein glycosylation |
| 49 | HCAG_04139.1 | Phosphomannomutase | Synthesis of the GDP-mannose |
| 50 | HCAG_06317.1 | Succinate dehydrogenase flavoprotein subunit, mitochondrial precursor | Tricarboxylic acid cycle |
| 51 | HCAG_00010.1 | Fructose-bisphosphate aldolase | Carbohydrate degradation; glycolysis |
| 52 | HCAG_07697.1 | Succinyl-CoA ligase beta-chain, mitochondrial precursor | Tricarboxylic acid cycle |
| 53 | HCAG_03263.1 | Succinate dehydrogenase iron-sulfur protein, mitochondrial precursor | Tricarboxylic acid cycle |
| 54 | HCAG_05090.1 | 2-methylcitrate synthase, mitochondrial precursor | Propionate catabolism; 2-methylcitric acid cycle. |
| 55 | HCAG_01535.1 | 2-oxoglutarate dehydrogenase E1 component, mitochondrial precursor | Conversion of 2-oxoglutarate to succinyl-CoA and CO2; tricarboxylic acid cycle. |
| 56 | HCAG_04416.1 | UDP-N-acetylglucosamine pyrophosphorylase | De novo biosynthetic pathway for UDP-GlcNAc |
| 57 | HCAG_03522.1 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial precursor | Conversion of 2-oxoglutarate to succinyl-CoA and CO2; tricarboxylic acid cycle. |
| 58 | HCAG_05266.1 | Aconitate hydratase, mitochondrial precursor | Conversion of citrate to isocitrate; tricarboxylic acid cycle. |
| 59 | HCAG_06641.1 | Hypothetical protein similar to UDP-galactopyranose mutase | Conversion of UDP-galactopyranose into UDP-galactofuranose |
| 60 | HCAG_02511.1 | Triose phosphate isomerase | Carbohydrate degradation; glycolysis |
| 61 | HCAG_03191.1 | Glucokinase | Phosphorylation of aldohexoses and glucose uptake |
| 62 | HCAG_03322.1 | Fructose-1,6-bisphosphatase | Gluconeogenesis; D-fructose 6-phosphate formation |
| 63 | HCAG_05681.1 | Phosphoenolpyruvate carboxykinase | Formation of phosphoenolpyruvate, gluconeogenesis |
| Lipid metabolism | |||
| 64 | HCAG_00413.1 | Farnesyl pyrophosphate synthetase | Isoprene biosynthesis, sterol biosynthesis. |
| 65 | HCAG_01596.1 | Hypothetical protein similar to 3-ketoacyl-CoA thiolase B | Formation of 3-oxoacyl-CoA, fatty acid beta-oxidation |
| 66 | HCAG_08039.1 | Hypothetical protein similar to peroxisomal hydratase-dehydrogenase-epimerase | Beta-oxidation pathway for fatty acids; converts trans-2-enoyl-CoA via D-3-hydroxyacyl-CoA to 3-ketoacyl-CoA. |
| 67 | HCAG_04933.1 | Hypothetical protein similar to ATP-citrate-lyase | Cleavage of citrate to yield acetyl CoA, oxaloacetate, ADP, and orthophosphate; fatty acid biosynthesis |
| 68 | HCAG_01606.1 | Acetyl-coenzyme A synthetase 2 | Acetyl-CoA synthesis |
| 69 | HCAG_08621.1 | Acetyl-CoA acetyltransferase | Short-chain fatty acid metabolism |
| 70 | HCAG_04934.1 | ATP-citrate synthase subunit 1 | Formation of cytosolic acetyl-CoA, fatty acid synthesis |
| 71 | HCAG_07725.1 | 3-hydroxybutyryl-CoA dehydrogenase | Formation of an hydroxyacyl-CoA, fatty acid oxidation |
| 72 | HCAG_07637.1 | Fatty acid synthase beta subunit dehydratase | Fatty acid biosynthesis |
| Amino acid and protein metabolism | |||
| 73 | HCAG_06019.1 | Ubiquitin | Pos translational protein modification |
| 74 | HCAG_03166.1 | Ubiquitin-conjugating enzyme E2-17 kDa | Catalyzes the attachment of ubiquitin to other proteins |
| 75 | HCAG_05565.1 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase (764 aa) | Methionine formation |
| 76 | HCAG_05651.1 | Glutamate dehydrogenase | N-acetyl-L-glutamate 5-phosphate formation; L-arginine biosynthesis |
| 77 | HCAG_07571.1 | Glycine dehydrogenase, mitochondrial precursor | Degradation of glycine |
| 78 | HCAG_06102.1 | Aspartate aminotransferase, mitochondrial precursor | Conversion of L-aspartate + 2-oxoglutarate into oxaloacetate + L-glutamate. |
| 79 | |||
| 80 | HCAG_09000.1 | Delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial precursor | L-glutamate biosynthesis |
| 81 | HCAG_00677.1 | Isovaleryl-CoA dehydrogenase, mitochondrial precursor | Leucine catabolism |
| 82 | HCAG_08890.1 | Ketol-acid reductoisomerase, mitochondrial precursor | L-isoleucine biosynthesis |
| 83 | HCAG_00676.1 | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial precursor | Leucine catabolism |
| 84 | HCAG_00029.1 | Mitochondrial methylglutaconyl-CoA hydratase | Leucine catabolism |
| 85 | HCAG_04215.1 | Peptidyl-prolyl cis-trans isomerase A | Protein folding |
| 86 | HCAG_04485.1 | Peptidyl-prolyl cis-trans isomerase | Protein folding |
| 87 | HCAG_08833.1 | Peptidyl-prolyl cis-trans isomerase | Protein folding |
| 88 | HCAG_07972.1 | FK506-binding protein | Protein folding |
| 89 | HCAG_00035.1 | Arginase | Formation of urea from arginine |
| 90 | HCAG_02357.1 | Histidinol dehydrogenase | L-histidine biosynthesis |
| 91 | HCAG_06901.1 | Malate dehydrogenase | Oxidation of 3-carboxy-2-hydroxy-4-methylpentanoate (3-isopropylmalate) to 3-carboxy-4-methyl-2-oxopentanoate |
| 92 | HCAG_08058.1 | Enoyl-CoA hydratase, mitochondrial precursor | conversion of 3-methylglutaconyl-CoA to 3-hydroxy-3-methylglutaryl-CoA, leucine metabolism |
| 93 | HCAG_03650.1 | NAD-specific glutamate dehydrogenase | Amino acid catabolism; oxoglutarate production |
| 94 | HCAG_07418.1 | Serine hydroxymethyltransferase | Interconversion of serine and glycine |
| 95 | HCAG_05988.1 | Hypothetical protein similar to elongation factor 2 | Protein biosynthesis; polypeptide chain elongation |
| 96 | HCAG_08798.1 | Translation elongation factor 1-alpha | Protein biosynthesis; polypeptide chain elongation |
| 97 | HCAG_04297.1 | Hypothetical protein similar to aspartyl aminopeptidase | Release of an N-terminal aspartate or glutamate from a peptide, with a preference for aspartate |
| 98 | HCAG_03371.1 | Adenosylhomocysteinase | Homocysteine biosynthesis |
| 99 | HCAG_01212.1 | Histoplasma capsulatum sulfate adenylyltransferase | Biosynthesis of sulfur-containing amino acids |
| 100 | HCAG_04206.1 | Eukaryotic translation initiation factor 2 gamma subunit | Protein synthesis |
| 101 | HCAG_04356.1 | eukaryotic translation initiation factor 3 39 kDa subunit | Protein synthesis |
| 102 | HCAG_00267.1 | Hypothetical protein similar to eukaryotic translation initiation factor 3 subunit 7 | Protein synthesis |
| 103 | HCAG_00678.1 | Hypothetical protein similar to 3-methylcrotonyl-CoA carboxylase biotin-containing subunit | Catabolism of leucine and isovalerate |
| 104 | HCAG_08678.1 | Aspartate aminotransferase | Oxaloacetate production, amino acid metabolism |
| 105 | HCAG_03630.1 | Disulfide-isomerase precursor | Rearrangement of -S-S- bonds in proteins |
| 106 | HCAG_06021.1 | Translation initiation factor 5A-2 | Protein synthesis |
| 107 | HCAG_00131.1 | Nuclear transport factor 2 | Protein synthesis |
| 108 | HCAG_00635.1 | Subtilase-type proteinase psp3 precursor | Peptidase |
| 109 | HCAG_07805.1 | Argininosuccinate lyase | Fumarate and L-arginine synthesis |
| Ribosomal proteins | |||
| 110 | HCAG_01850.1 | 60S ribosomal protein L10a | Ribosomal protein |
| 111 | HCAG_00055.1 | 60S ribosomal protein L27-A | Ribosomal protein |
| 112 | HCAG_04185.1 | 60S ribosomal protein L36 | Ribosomal protein |
| 113 | HCAG_02703.1 | 60S acidic ribosomal protein P2 | Ribosomal protein |
| 114 | HCAG_04231.1 | 60S ribosomal protein L11-1 | Ribosomal protein |
| 115 | HCAG_03695.1 | 60S ribosomal protein L10-B | Ribosomal protein |
| 116 | HCAG_07463.1 | 60S ribosomal protein L18-B | Ribosomal protein |
| 117 | HCAG_08515.1 | 60S ribosomal protein L2 | Ribosomal protein |
| 118 | HCAG_03923.1 | 60S ribosomal protein L3 | Ribosomal protein |
| 119 | HCAG_08444.1 | 60S ribosomal protein L5 | Ribosomal protein |
| 120 | HCAG_00468.1 | 60S ribosomal protein L4-A | Ribosomal protein |
| 121 | HCAG_02430.1 | 40S ribosomal protein S5 | Ribosomal protein |
| 122 | HCAG_06613.1 | 40S ribosomal protein S7 | Ribosomal protein |
| 123 | HCAG_06914.1 | 40S ribosomal protein S3A | Ribosomal protein |
| 124 | HCAG_07773.1 | 40S ribosomal protein S11 | Ribosomal protein |
| 125 | HCAG_06308.1 | 40S ribosomal protein S12 | Ribosomal protein |
| 126 | HCAG_01666.1 | ribosomal protein S6 | Ribosomal protein |
| 127 | HCAG_08667.1 | 40S ribosomal protein S18 | Ribosomal protein |
| 128 | HCAG_02272.1 | 40S ribosomal protein S22 | Ribosomal protein |
| 129 | HCAG_07237.1 | ribosomal protein S4 | Ribosomal protein |
| 130 | HCAG_07961.1 | ribosomal protein S5 | Ribosomal protein |
| 131 | HCAG_03504.1 | ribosomal protein L22 | Ribosomal protein |
| 132 | HCAG_08821.1 | ribosomal protein S20 | Ribosomal protein |
| 133 | HCAG_04498.1 | ribosomal protein S21e | Ribosomal protein |
| 134 | HCAG_06425.1 | ribosomal protein L32 | Ribosomal protein |
| 135 | HCAG_00214.1 | Hypothetical protein similar to ribosomal protein S3 | Ribosomal protein |
| 136 | HCAG_03415.1 | Hypothetical protein similar to ribosomal protein L35 | Ribosomal protein |
| 137 | HCAG_04856.1 | ribosomal protein P0 | Ribosomal protein |
| 138 | HCAG_07708.1 | Hypothetical protein similar to ribosomal protein L13 | Ribosomal protein |
| 139 | HCAG_06198.1 | Hypothetical protein similar to QDE2 protein | Structural constituent of ribosome |
| 140 | HCAG_03694.1 | Large ribosomal subunit protein L30 | Ribosomal protein |
| Proteasome components | |||
| 141 | HCAG_04090.1 | Proteasome component C1 | Proteasome Component |
| 142 | HCAG_05739.1 | Proteasome component Y13 | Proteasome Component |
| 143 | HCAG_04101.1 | Proteasome component C5 | Proteasome Component |
| 144 | HCAG_00053.1 | Proteasome component C7-alpha | Proteasome Component |
| 145 | HCAG_07121.1 | Proteasome component PUP2 | Proteasome Component |
| 146 | HCAG_03737.1 | Proteasome component Pup1 | Proteasome Component |
| 147 | HCAG_04198.1 | Proteasome component PRE1 | Proteasome Component |
| 148 | HCAG_05910.1 | Proteasome component PRE2 precursor | Proteasome Component |
| 149 | HCAG_06342.1 | Proteasome component PRE3 precursor | Proteasome Component |
| 150 | HCAG_04107.1 | Proteasome component PRE4 | Proteasome Component |
| 151 | HCAG_08215.1 | Proteasome component PRE5 | Proteasome Component |
| 152 | HCAG_00347.1 | Proteasome component PRE6 | Proteasome Component |
| 153 | HCAG_03939.1 | Proteasome component Pre8 | Proteasome Component |
| Nuclear proteins | |||
| 154 | HCAG_03885.1 | Histone H4.2 | DNA assembly |
| 155 | HCAG_03524.1 | Histone H2A | DNA assembly |
| 156 | HCAG_03525.1 | Histone H2B | DNA assembly, cell surface component in H. capsulatum |
| 157 | HCAG_02608.1 | Guanine nucleotide-binding protein beta subunit | Nucleotide binding |
| 158 | HCAG_01433.1 | Adenylosuccinate lyase | Purine metabolism; AMP biosynthesis via de novo pathway |
| 159 | HCAG_04273.1 | eukaryotic initiation factor 4A | ATP-dependent RNA helicase |
| 160 | HCAG_06523.1 | Curved DNA-binding protein 42 kDa protein | DNA-binding protein. |
| 161 | HCAG_00544.1 | Nucleoside-diphosphate kinase | Phosphate exchange between different nucleoside diphosphates |
| 162 | HCAG_02553.1 | Small nuclear ribonucleoprotein Sm D2 | Pre-mRNA splicing |
| 163 | HCAG_01986.1 | Poly(A)+ RNA export protein | Nuclear mRNA export |
| 164 | HCAG_00018.1 | Nucleosome binding protein | Nucleosome binding |
| Cell growth/division | |||
| 165 | HCAG_00717.1 | Hypothetical protein similar to septin-1 | Cytokinesis |
| 166 | HCAG_02452.1 | Cell division cycle protein 48 | Regulation of cell growth |
| 167 | HCAG_06283.1 | AAC1 protein | Spore germination |
| 168 | HCAG_08831.1 | Formamidase | Formamide hydrolysis with the production of ammonia, to be used as nitrogen source |
| Plasma membrane proteins | |||
| 169 | HCAG_06977.1 | Plasma membrane ATPase | Hydrogen-exporting ATPase activity, phosphorylative mechanism |
| 170 | HCAG_06935.1 | Hypothetical protein similar to aminopeptidase B | Single-pass type II membrane protein |
| 171 | HCAG_07210.1 | Glycolipid-anchored surface protein 5 precursor | Cell membrane lipid anchor |
| Cytoskeleton proteins | |||
| 172 | HCAG_04706.1 | ARP2/3 complex 34 kDa subunit | Regulation of actin polymerization |
| 173 | HCAG_04115.1 | Arp2/3 complex chain sop2 | Regulation of actin polymerization |
| 174 | HCAG_00848.1 | Arp2/3 complex subunit | Regulation of actin polymerization |
| 175 | HCAG_08210.1 | Actin | Cytoskeleton assembly |
| Anti-oxidant proteins | |||
| 176 | HCAG_06210.1 | Thiol-specific antioxidant protein | Antioxidant defense |
| 177 | HCAG_08064.1 | Catalase B | Antioxidant defense |
| 178 | HCAG_03448.1 | Superoxide dismutase, mitochondrial precursor | Antioxidant defense |
| 179 | HCAG_01543.1 | Superoxide dismutase, mitochondrial precursor | Antioxidant defense |
| 180 | HCAG_07098.1 | Ascorbate peroxidase | Antioxidant defense |
| 181 | HCAG_08190.1 | Oxidoreductase 2-nitropropane dioxygenase | Antioxidant defense |
| Miscellaneous | |||
| 182 | HCAG_03815.1 | ATP synthase subunit 5, mitochondrial precursor | Purine metabolism; AMP biosynthesis via de novo pathway |
| 183 | HCAG_06944.1 | ATP synthase beta chain, mitochondrial precursor | Proton-dependent ATP production |
| 184 | HCAG_00404.1 | Vacuolar ATP synthase catalytic subunit A | ATP synthesis |
| 185 | HCAG_06929.1 | NADH-ubiquinone oxidoreductase 78 kDa subunit, mitochondrial precursor | Core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase |
| 186 | HCAG_04307.1 | Inorganic pyrophosphatase | Phosphate supply |
| 187 | HCAG_00437.1 | Hypothetical protein similar to cytochrome-c oxidase chain VI precursor | Mitochondrial electron transport |
| 188 | HCAG_05938.1 | Cytochrome c | Mitochondrial electron transport |
| 189 | HCAG_02342.1 | Mitochondrial processing peptidase beta subunit | Mitochondrial protein processing |
| 190 | HCAG_02570.1 | Cytochrome P450 55A3 | Electron transfer component. |
| 191 | HCAG_04096.1 | Allergen Asp f 4 | Induction of allergic reactions |
| 192 | HCAG_02813.1 | ATP synthase alpha chain isoform, mitochondrial precursor | Hydrogen ion transporting ATP synthase activity |
| 193 | HCAG_06539.1 | Hypothetical protein similar to succinate:fumarate antiporter | Transmembrane transport |
| 194 | HCAG_03543.1 | Glutamate carboxypeptidase | Hydrolysis of reduced and non-reduced folates to pteroates and L-glutamate |
| 195 | HCAG_05000.1 | Transketolase 1 | Assimilation of formaldehyde |
| 196 | HCAG_06026.1 | Polyadenylate-binding protein | Binding of the poly(A) tail of mRNA |
| 197 | HCAG_04999.1 | Spermidine synthase | Amine and polyamine biosynthesis; spermidine biosynthesis |
| 198 | HCAG_02994.1 | Pyridoxal biosynthesis lyase pdxS | Pyridoxal phosphate production |
| 199 | HCAG_02035.1 | Imidazole glycerol phosphate synthase hisHF | Synthesis of Imidazole glycerol phosphate |
| 200 | HCAG_07004.1 | O-acetylhomoserine | Catalysis of trans-sulfuration. |
| 201 | HCAG_08367.1 | Aldehyde dehydrogenase | Aldehyde oxidation |
| 202 | HCAG_03605.1 | Zinc-binding dehydrogenase | Ethanol metabolism |
| 203 | HCAG_08561.1 | Alcohol dehydrogenase I | Interconversion between alcohols and aldehydes or ketones |
| 204 | HCAG_07206.1 | S-(hydroxymethyl)glutathione dehydrogenase | Interconversion between alcohols and aldehydes or ketones |
| 205 | HCAG_05094.1 | 2-Methylcitrate dehydratase | dehydration of 2-methylcitrate to 2-methyl-cis-aconitate |
| 206 | HCAG_03008.1 | Sulfur metabolite repression control protein C | Regulation of sulfur metabolism |
Biological functions of the proteins detected by proteomics were obtained from the ExPASy Proteomics Server (http://ca.expasy.org/). Proteins with unknown functions were also identified, but are not present in this table. For detailed information, see Supplemental material.
Table 3.
Distribution of the identified H. capsulatum vesicle proteins according to their functions.
| Protein association | Percent total (%) |
|---|---|
| Amino acid/protein metabolism | 37 |
| Sugar metabolism | 36 |
| Ribosomal | 31 |
| Proteasome component | 13 |
| Nuclear | 11 |
| Cell wall architecture | 11 |
| Lipid metabolism | 9 |
| Cell signaling | 8 |
| Chaperone-like | 8 |
| Anti-oxidant | 6 |
| Cytoskeletal | 4 |
| Cell growth/division | 4 |
| Plasma membrane | 3 |
| Miscellaneous | 25 |
TEM of C. albicans, C. parapsilosis, S. schenckii, and S. cerevisiae vesicles
TEM of the material recovered by ultracentrifugation from culture supernatants of C. albicans, C. parapsilosis, S. schenckii and S. cerevisiae revealed that other ascomycetes similarly produce extracellular vesicles (Fig. 5). The structures identified were similar to vesicles produced by C. neoformans (Rodrigues et al., 2008; Rodrigues et al., 2007) and H. capsulatum, consisting of bilayered membranes and largely spherical morphologies. Although significant differences in size were found for the ascomycetes studied, they all predominantly produced vesicles ≤ 50 nm in diameter. Only 4% of S. cerevisiae vesicles were larger than 50 nm though none were more than 100 nm in diameter. For S. schenckii, 11% were between 51–100 nm, but none were larger. For C. albicans and C. parapsilosis, 13% and 36% of vesicles were 50–100 nm, respectively. Vesicles larger than 100 nm comprised 1% and 18% of total vesicles for C. albicans and C. parapsilosis, respectively.
Fig. 5.
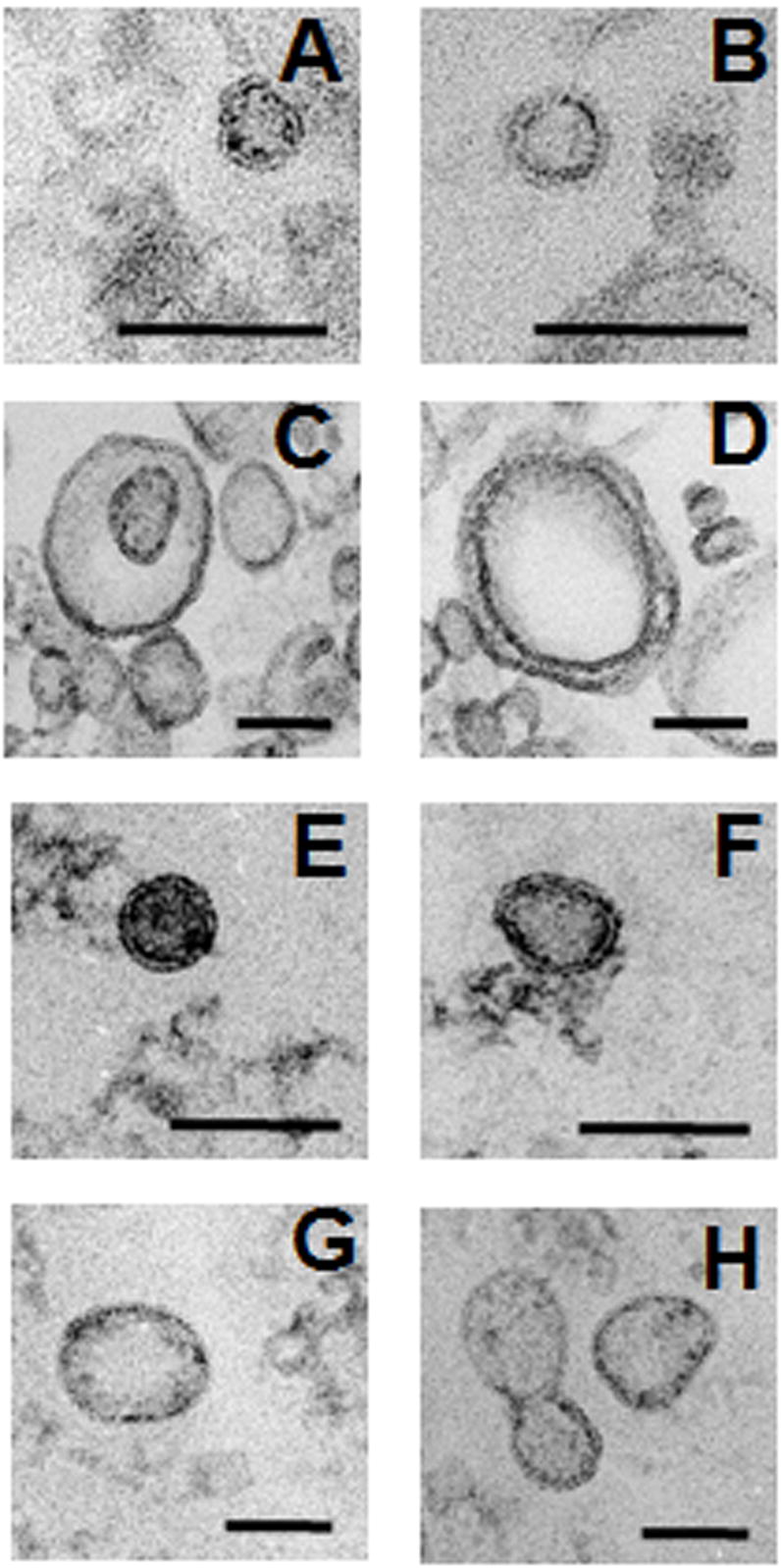
TEM of extracellular vesicles from S. cerevisiae (A, B), C. parapsilosis (C, D), S. schenckii (E, F) and C. albicans (G, H). The structures identified were similar to vesicles produced by C. neoformans and H. capsulatum. Bars 100 nm.
Sera of patients recognized proteins from the vesicles
Pooled sera from patients with histoplasmosis were used in immunoblots against protein extracts of H. capsulatum (Fig. 6). Extracts of H. capsulatum vesicles reacted with serum from patients with histoplasmosis. Immunogenic proteins with diverse molecular masses were observed (Figure 6 B). To confirm the identification of certain proteins identified in the proteomic analysis for which reagents for H. capsulatum are available, immunoblots were performed with mAbs to histone 2B and heat shock protein 60 (Figure 6 D and E, respectively). The identified bands corresponded to bands recognized by the pooled histoplasmosis sera. These proteins were identified in the proteomic analysis described above. Non-immune sera did not react with proteins from the vesicles (Figure 6 C).
Fig. 6.
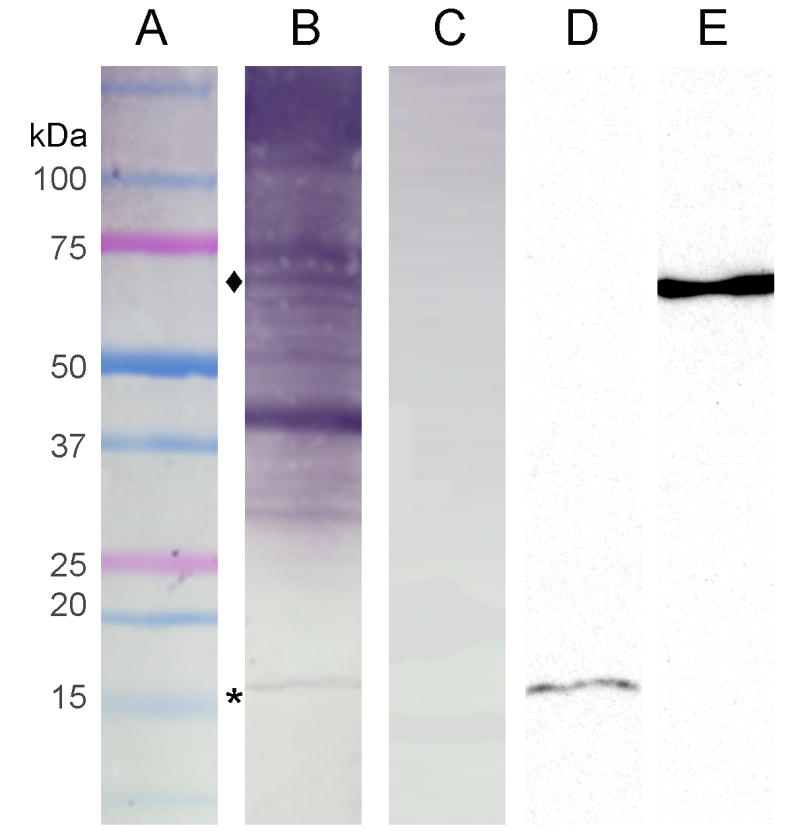
H. capsulatum vesicles contain immunoreactive proteins. Pooled serum from patients with histoplasmosis reacts with proteins from extracellular H. capsulatum vesicles. Lane A shows the molecular weight marker. Lane B shows H. capsulatum pooled hyperimmune sera reacting with extracts from H. capsulatum vesicles, whereas the non-immune serum in lane C does not interact with the extracted proteins. Lanes D and E demonstrate the binding of monoclonal antibodies to histone 2B (17 kDa; corresponding to *, lane B) and heat shock protein 60 (62 kDa; corresponding to ♦, lane B), respectively, in the vesicular protein preparations.
Discussion
We recently showed that secretory vesicles are involved in the extracellular release of virulence determinants in the fungal pathogen C. neoformans (Rodrigues et al., 2008; Rodrigues et al., 2007). We now describe that H. capsulatum, C. albicans, C. parapsilosis, S. schenckii and S. cerevisiae also produce extracellular vesicles. Furthermore, we show that H. capsulatum produces vesicles containing key molecules related to virulence, stress response and fungal physiology. By microscopic and mass spectrometric approaches, H. capsulatum vesicles were identified as lipid bodies with bilayered membrane containing proteins of diverse functions and a number of phospholipids. The findings of this study, combined with the recent reports on C. neoformans (Rodrigues et al., 2008; Rodrigues et al., 2007; Yoneda and Doering, 2006; Garcia-Rivera et al., 2004; Feldmesser et al., 2001), indicate that vesicular secretion is an important mechanism for fungi to deliver intracellular molecules to the extracellular space.
For H. capsulatum, vesicular bodies were observed in association with the cell wall and in the extracellular environment, suggesting the active use of vesicular transport for secretory processes. Microscopic analysis of the samples obtained after differential centrifugation of culture supernatants revealed intact vesicles ranging in size from approximately 10 to 350 nm (Fig 2). Despite this heterogeneity, the vesicles all had an ovoid appearance and displayed lipid bilayered membranes. Differences in electron density were observed, suggesting heterogeneity in vesicular contents (Fig. 1). Vesicles were not released from dead cells and the yeast were studied in log phase growth during which there is negligible cell death.
C. albicans continues to be the leading opportunistic pathogen involved in oral, vaginal, and systemic infections resulting in high mortality, and Candida spp. are the fourth most common cause of bloodstream infection in the United States (Wisplinghoff et al., 2004). C. parapsilosis is currently the second most common cause of invasive candidiasis worldwide (Fridkin et al., 2006) and is particularly associated with disease in premature infants, immunocompromised adults, and patients in intensive care units (Clerihew et al., 2007). The dimorphic fungus S. schenckii has a worldwide distribution and causes disease primarily after traumatic inoculation of colonized materials and less commonly by inhalation of spores through the respiratory tract (Almeida-Paes et al., 2007a). Rarely pathogenic, S. cerevisiae is a well-established model organism for understanding fundamental cellular processes relevant to higher eukaryotic organisms (Botstein and Fink, 1988). Microscopic analysis of the additional fungal species studied revealed intact vesicles of varied morphology, yet the vesicles shared a common ovoid appearance and all displayed lipid bilayered membranes. Interestingly, vesicular transport has been proposed previously in C. albicans in an opaque variant of strain WO-1 in which electron microscopy analysis of intact cells revealed “pimples” in the cell wall with vesicles within channels or emerging from the “pimples” (Anderson et al., 1990). Future studies are required to assess the contents of the vesicles produced by these ascomycetes, and it will be imperative to assess whether vesicles of different sizes transport specific compounds. For instance, it will be important to determine whether virulence associated products (ie. heat shock proteins, catalases, superoxide dismutases, etc) are transported in the larger vesicles previously described in C. neoformans (Rodrigues et al., 2008) and herein identified for Candida spp. and H. capsulatum but are lacking in the smaller vesicles of less pathogenic fungi such as S. cerevisiae.
In our analyses of H. capsulatum vesicles, phospholipids were characterized as lipid components of vesicle membranes. The major phospholipids found were PC, PE and PS, which are building blocks for cellular membranes (lipid bilayers). These lipids also perform a diverse number of other functions, from compartmentalization of cytoplasm to the housing of proteins involved in cell signaling, intercellular adhesion, and cytoskeletal support (16). Previous studies have shown that cell communication might not be limited to soluble agonists, but that various types of vesicles also participate in the process (Denzer et al., 2000). It is notable that mammalian exosome membranes display a similar content of phospholipids and are also formed as lipid bilayers with a random distribution analogous to H. capsulatum vesicle phospholipids (Laulagnier et al., 2004). Hence, this similarity to mammalian exosomes supports the supposition that the vesicles from H. capsulatum are exosome-like structures. Exosomes are part of the family of bioactive vesicles and appear to be involved in distal communications between cells. They transport bioactive lipids and lipolytic enzymes and their biogenesis requires specific lipids and membrane reorganization (Subra et al., 2007). Bioactive vesicles are receiving increasing interest since they are important in enhancing biodiversity and are the only type of bioactive vesicles originating from intracellular compartments, namely multi-vesicular bodies (MVBs, or late endosomes) (Fevrier and Raposo, 2004). MVBs participate in the eradication of obsolete proteins, but they can also be released into extracellular space where they can potentially affect intercellular communication (van Niel et al., 2006).
We used a proteomics approach to analyze the protein contents of vesicles. H. capsulatum survives and replicates within host macrophages (Allendoerfer and Deepe, 1997), denoting the necessity of fungal mechanisms to escape the antimicrobial armory of phagocytes. The secretion of virulence factors is a mechanism used by different pathogens to cause damage to host cells. In this context, the presence of anti-oxidant proteins in secreted vesicles, such as catalase B (M antigen) (Zancope-Oliveira et al., 1999), superoxide dismutase precursors (Brummer and Stevens, 1995), and a thiol-specific antioxidant protein (Demasi et al., 2006), could represent an effective mechanism of fungal defense. The proteomic analysis of the H. capsulatum vesicles identified proteins involved in vesicular transport and fusion, especially proteins from the Rab family. In mammals, Rabs define a family of almost 70 proteins that play critical roles in the trafficking of vesicles that mediate transport between compartments of the exocytic and endocytic pathways (Pfeffer, 2005; Pfeffer, 2001). Several of the identified H. capsulatum Rab proteins have been characterized to have similar functions, such as H. capsulatum Rab GDP-dissociation inhibitor that plays a key role in the recycling of Rabs (Ma et al., 2006)and H. capsulatum Rab1a that regulates antegrade transport between the ER and the Golgi apparatus (Sannerud et al., 2006). The presence of H. capsulatum endochitinase and glucanases in the vesicles is also consistent, since these molecules are membrane proteins and the vesicles may originate from membrane invagination, similar to exosome formation (Sannerud et al., 2006). The mechanisms by which fungal vesicles traverse the cell wall are still unknown. In this context, the existence of vesicular enzymes with the ability to hydrolyze cell wall components is particularly interesting, since these molecules have the potential to promote cell wall reassembly for vesicle passage.
The extracellular H. capsulatum vesicles also contained chaperone and nucleus-associated proteins. Several heat-shock proteins were present in the vesicles. H. capsulatum Hsp60 is particularly noteworthy since this immunodominant molecule is key to the engagement of the yeast with CD18 receptors on host macrophage (Long et al., 2003) and vaccination with this protein is protective (Gomez et al., 1995). H. capsulatum Hsp70 is also immunogenic, though it induces non-protective cellular responses (Allendoerfer et al., 1996). H. capsulatum nuclear proteins, such as H2B, can be displayed on the fungal cell surface where they can be targeted by antifungal antibodies (Nosanchuk et al., 2003). An H. capsulatum glyceraldehyde-3-phosphate dehydrogenase was also identified and a homologous protein is present in the cell wall of the fungal pathogen Paracoccidioides brasiliensis, where it mediates the adhesion of yeast cells to host cells and extracellular matrixes (Barbosa et al., 2006). These examples of single proteins with multiple activities are consistent with the emerging understanding that many proteins have ‘moonlighting’ functions enabling cells to efficiently perform diverse tasks despite limited genomes (Jeffery, 2003b; Jeffery, 2003a; Jeffery, 1999). Moonlighting proteins described from S. cerevisiae to humans have included enzymes, chaperones, ribosomal protein, receptors, and transmembrane channels.
In order to assess whether vesicles have a biological effect on the host, we tested the immunoreactivity of extracted vesicular proteins with patients’ sera. The recognition of diverse proteins by pooled hyperimmune patient sera indicates that these vesicularly transported proteins could be important in the pathogenesis of these mycoses. For example heat shock protein 60 from H. capsulatum has been associated with virulence (Deepe and Gibbons, 2002; Scheckelhoff and Deepe, 2002; Deepe and Gibbons, 2001a; Allendoerfer et al., 1996; Gomez et al., 1995). The findings are consistent with vesicular transport playing a role in host-pathogen interactions.
In summary, we report the trans-cell wall vesicular transport of several important components of virulence, signaling and recycling in H. capsulatum. The vesicles appear to be similar to mammalian exosomes. We also show that other ascomycetes produce vesicles that can function in the transport of macromolecules. The products of the vesicles are immunoreactive with serum from patients, which supports our hypothesis that the vesicles are involved in fungal pathogenesis. Hence, we propose that fungal extracellular vesicle secretion is an important mechanism in fungal biology.
Materials and Methods
Fungal strains and growth conditions
H. capsulatum strain G217B (ATCC 26032) was obtained from the American Type Culture Collection (ATCC, Rockville, Maryland, USA). G217B yeast cells were grown in 500 mL Ham’s F-12 medium with rotary shaking (150 rpm) at 37 °C for 48 h in Erlenmeyer flasks as described previously (Nosanchuk et al., 2003). Thimerosal- and heat-killed H. capsulatum yeast cells were used as a negative control. Candida albicans SC5314 (ATCC MYA-2876 (Gillum et al., 1984)), Candida parapsilosis strain GA1 (a clinical isolate (Gacser et al., 2005)) and S. schenckii strain 23508 (a clinical isolate (Almeida-Paes et al., 2007b)) were grown in Sabouraud dextrose broth (Difco Laboratories, Detroit, MI) with rotary shaking (150 rpm) at 30°C for 48 hours for Candida spp. or at 37°C for 3 days in the case of S. schenckii. S. cerevisiae strain KFY 471 (BY4741; ATTC 201388 (Winzeler et al., 1999)) was provided by Dr. Michael Keogh (Albert Einstein, New York), and was grown in YPD broth (Difco Laboratories, Detroit, MI) in the same conditions used for Candida strains.
Isolation of vesicles
Vesicle isolation was performed according to our previously described protocol (Rodrigues et al., 2007). Briefly, the fungal cells were separated from culture supernatants by centrifugation at 4,000 g for 15 min at 4°C. Supernatants were collected and again centrifuged at 15,000 g (4°C) for 30 min to remove smaller debris. The pellets were discarded, and the resulting supernatant was concentrated approximately 20-fold using an Amicon ultrafiltration system (cutoff, 100 kDa). To ensure the removal of cells and cellular debris, the concentrated culture fluid was again centrifuged as described above and the resulting supernatant was then centrifuged at 100,000 g for 1 h at 4°C. The supernatants were discarded and the pellets suspended in 3 mL of 0.1 M Phosphate-buffered saline (PBS) and centrifuged at 100,000 g for 1 h at 4°C. The supernatant was again removed from the pellets and a fixative solution (as described below), was added for electron microscopy analysis. Additionally, pellets from H. capsulatum were used for proteomic analysis or extracted with a chloroform-methanol mixture for lipidomic analysis as described below.
Transmission electron microscopy (TEM)
TEM was used to visualize vesicles in intact H. capsulatum yeast cells and vesicles isolated from culture supernatants of H. capsulatum and the other fungi by ultracentrifugation. Samples were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer at room temperature for 2 h and then incubated overnight in 4% paraformaldehyde, 1% glutaraldehyde, and 0.1% phosphate-buffered saline (PBS). The samples were incubated for 90 min in 2% osmium tetroxide, serially dehydrated in ethanol, and embedded in Spurr’s epoxy resin. Thin sections were obtained on a Reichert Ultracut and stained with 0.5% uranyl acetate and 0.5% lead citrate. Samples were observed in a JEOL 1200EX transmission electron microscope operating at 80 kV.
Mass spectrometry analysis of phospholipids
The H. capsulatum vesicle fraction was suspended in 100 μL of ultrapure water and extracted 3x by addition of 1.5 ml chloroform:methanol (2:1, v/v) solution followed by vigorous vortexing and then centrifugation for 10 min at 1000 g. After drying under nitrogen stream, the sample was dissolved in 500 μL chloroform and loaded onto a silica gel 60 column, equilibrated with pure chloroform. The silica column was manufactured in a Pasteur pipette, using a very fine glass wool and about 500 mg of silica gel 60 resin (pore size 60 Å, 200–400 μm mesh) (Sigma-Aldrich, St. Louis. MO). After washing the column with chloroform, followed by acetone, the phospholipids were eluted with methanol and dried under nitrogen stream. The phospholipid fraction was dissolved in chloroform:methanol (1:1, v/v) and diluted either in chloroform:methanol (1:1, v/v) containing 10 mM LiOH (for positive-ion mode analysis) or chloroform:methanol (1:1, v/v) containing 0.1% formic acid (FA) and 0.1% NH4OH (for negative-ion mode analysis), and analyzed in an electrospray ionization time-of-flight mass spectrometer (ESI-Q-TOF-MS) (Qtof-1, Waters). The spectra were collected in a range from 400 to 1500 m/z and each ion with intensity higher than 10 counts was automatically submitted to collision-induced dissociation (CID) (22–60 eV, 50–1500 m/z range). MS/MS spectra were analyzed manually for the identification of phospholipid species.
Protein identification by liquid chromatography tandem mass spectrometry
Protein digestion was performed as described previously (Stone and Williams, 1996). Purified vesicles were suspended in 40 μl 400 mM NH4HCO3 containing 8 M urea and the protein disulfide bounds were reduced by the addition of 10 μl 50 mM dithiotreitol for 15 min at 50°C. Cysteine residues were alkylated by the addition of 10 μl 100 mM iodoacetamide and incubation for an additional 15 min at room temperature protected from light. The reaction was diluted with HPLC-grade water (Sigma-Aldrich) to obtain a final concentration of 1 M urea, and the digestion was performed overnight at 37°C with 4 μg sequencing-grade trypsin (Promega). Each sample was desalted in a reverse phase ziptip (POROS R2 50, Applied Biosystems) as described by Jurado et al. (Jurado et al., 2007), and peptides were fractionated in a strong cation-exchange (SCX) ziptip, manufactured in a 200-μl micropipette tip with glass fiber filter and POROS HS 50 resin (Applied Biosystems). After equilibrating the SCX ziptip with 25% acetonitrile (ACN)/0.5% FA, peptides were loaded and eluted with increasing NaCl concentration (0, 10, 20, 40, 60, 80, 100, 150, 200, and 500 mM NaCl in 25% ACN/0.5% FA). Each SCX fraction was dried in a vacuum centrifuge (Eppendorf), purified in POROS R2 50 ziptip and redissolved in 30 μl 0.05% trifluoroacetic acid (TFA). Eight μl of fractionated peptides were loaded onto a C18 trap column (1 yL C18, OPTI-PAK) and washed for 10 min with 2% ACN/0.1% FA. The separation was performed in a capillary reverse-phase column (Acclaim, 3 μm C18, 75 μm x 25 cm, LC Packings) connected to a nanoHPLC system (nanoLC 1D plus, Eksigent). Peptides were eluted in a 0–40% gradient of solvent B (solvent A: 2%ACN/0.1%FA, solvent B: 80%ACN/0.1% FA) during 100 min and directly analyzed in an electrospray ionization-linear ion trap-mass spectrometer (ESI-LIT-MS) equipped with a nanospray source (LTQ XL, Thermo Fisher Scientific, San Jose CA). MS spectra were collected in centroid mode in a range from 400 to 1700 m/z and the five most abundant ions were submitted twice to CID (35% normalized collision energy), before they were dynamically excluded for 120 sec.
All MS/MS spectra were obtained from peptides with 600–3500 Da and at least 15 fragments were converted into DTA files using Bioworks v.3.3.1 (Thermo Fisher Scientific). The DTA files were subjected to a database search using TurboSequest (Eng et al., 1994) (available in Bioworks software) against a database assembled with H. capsulatum (protein database, version of 05/11/2006, available at http://www.broad.mit.edu/annotation/genome/histoplasma_capsulatum/Downloads.html;jsessionid=A347F284A23BE3CC423191220E09A48D), common contaminant sequences (retrieved from GenBank -http://www.ncbi.nlm.nih.gov/ and International Protein Index -http://www.ebi.ac.uk/IPI) and 100,000 randomly generated sequences. The database search parameters were: trypsin cleavage in both peptide termini with allowance for one missed cleavage site, carbamidomethylation of cysteine residues as a fixed modification, oxidation of methionine residues as a variable modification, and 2.0 Da and 1.0 Da for peptide and fragment mass tolerance, respectively. To ensure the quality of our identifications, we estimated the false-positive rate (FPR) from the TurboSequest output. This estimation was done using the following formula:
A FPR was obtained after applying the following filters in Bioworks: distinct peptides, consensus score ≥ 10.1, DCn ≥ 0.1, protein probability ≤ 1×10−3, and Xcorr ≥ 1.5, 2.2, and 2.7 for singly-, doubly-, and triply-charged peptides, respectively. Using these parameters, the FPR was estimated as 3.7%.
Western Blot Analysis
H. capsulatum vesicles pellets were subjected to acetone precipitation. The precipitate was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 10% gels. Separated proteins were transferred to nitrocellulose membranes and blocked (1% BSA in 0.1M PBS) for 1 h at 37°C. The membranes were then incubated in the presence of pooled sera from patients with culture proven histoplasmosis (Fiocruz- IPEC). Positive reactions were observed after incubation of blotted proteins with alkaline phosphatase-conjugated goat anti-human antibodies in blocking buffer for 1 h at 37°C followed by development with NBT-BCIP. Alternatively, the membranes were blocked and then incubated with monoclonal antibody to H2B (Nosanchuk et al., 2003) or heat shock protein 60 (Guimarães et al., 2006), washed in TBST, and incubated with goat anti-mouse Ig conjugated to horse raddish peroxidase. The samples were developed with ECL substrate (SuperSignal; Pierce Chemical Co.) and exposed on X-Omat AR film (Eastman Kodak Co., Rochester, New York, USA).
Supplementary Material
Acknowledgments
PCA was supported in part by an Interhemispheric Research Training Grant in Infectious Diseases, Fogarty International Center (NIH D43-TW007129). JDN is supported by NIH AI52733 and AI056070-01A2, and the Einstein MMC Center for AIDS Research [NIH NIAID AI51519]. MLR is supported by grants from the Brazilian agencies FAPERJ, CNPq and CAPES. R.M.Z.-O. is in part supported by CNPq 306288/2006-0 and by a grant from FAPERJ 306288/2006-0. AC is supported by NIH grants AI033142, AI033774, AI052733, and HL059842. ICA is supported by NIH grant 5G12RR008124 to the Border Biomedical Research Center (BBRC)/University of Texas at El Paso (UTEP). We are thankful to the Biomolecule Analysis Core Facility/BBRC/UTEP, supported by NIH/NCRR grant 5G12RR008124, for the use of the LC-MS instruments. The authors thank Dr. Fabio Gozzo (Laboratório Nacional de Luz Sincrontron, Campinas, Brazil) for kindly providing the 100,000 randomly generated sequences and Dr. Michael Keogh for providing the S. cerevisiae strain.
We also acknowledge that JE McEwen and CH Johnson postulated during the ASM Conference on Dimorphic Fungal Pathogens (Denver, Colorado, March 2006) that vesicles may be produced by H. capsulatum.
Footnotes
Conflict of interest: The authors do not have any conflicts of interest.
References
- Allendoerfer R, Deepe GS., Jr Intrapulmonary response to Histoplasma capsulatum in gamma interferon knockout mice. Infect Immun. 1997;65:2564–2569. doi: 10.1128/iai.65.7.2564-2569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendoerfer R, Maresca B, Deepe GS., Jr Cellular immune responses to recombinant heat shock protein 70 from Histoplasma capsulatum. Infect Immun. 1996;64:4123–4128. doi: 10.1128/iai.64.10.4123-4128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida-Paes R, Pimenta MA, Monteiro PC, Nosanchuk JD, Zancope-Oliveira RM. Immunoglobulins G, M and A against Sporothrix schenckii exoantigens in patients with sporotrichosis before and during treatment with itraconazole. Clin Vaccine Immunol. 2007a;14:1149–1157. doi: 10.1128/CVI.00149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida-Paes R, Pimenta MA, Pizzini CV, Monteiro PC, Peralta JM, Nosanchuk JD, Zancope-Oliveira RM. Use of mycelial-phase Sporothrix schenckii exoantigens in an enzyme-linked immunosorbent assay for diagnosis of sporotrichosis by antibody detection. Clin Vaccine Immunol. 2007b;14:244–249. doi: 10.1128/CVI.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Mihalik R, Soll DR. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J Bacteriol. 1990;172:224–235. doi: 10.1128/jb.172.1.224-235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N, Jin-no S, Nakagawa Y, Asai N, Arakawa E, Tamura N, et al. Identification and characterization of microvesicles secreted by 3T3-L1 adipocytes: redox- and hormone-dependent induction of milk fat globule-epidermal growth factor 8-associated microvesicles. Endocrinology. 2007;148:3850–3862. doi: 10.1210/en.2006-1479. [DOI] [PubMed] [Google Scholar]
- Barbosa MS, Bao SN, Andreotti PF, de Faria FP, Felipe MS, dos Santos Feitosa L, et al. Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect Immun. 2006;74:382–389. doi: 10.1128/IAI.74.1.382-389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohse ML, Woods JP. Surface localization of the Yps3p protein of Histoplasma capsulatum. Eukaryot Cell. 2005;4:685–693. doi: 10.1128/EC.4.4.685-693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohse ML, Woods JP. RNA interference-mediated silencing of the YPS3 gene of Histoplasma capsulatum reveals virulence defects. Infect Immun. 2007;75:2811–2817. doi: 10.1128/IAI.00304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Fink GR. Yeast: an experimental organism for modern biology. Science. 1988;240:1439–1443. doi: 10.1126/science.3287619. [DOI] [PubMed] [Google Scholar]
- Brummer E, Stevens DA. Antifungal mechanisms of activated murine bronchoalveolar or peritoneal macrophages for Histoplasma capsulatum. Clin Exp Immunol. 1995;102:65–70. doi: 10.1111/j.1365-2249.1995.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnie JP, Carter TL, Hodgetts SJ, Matthews RC. Fungal heat-shock proteins in human disease. FEMS Microbiol Rev. 2006;30:53–88. doi: 10.1111/j.1574-6976.2005.00001.x. [DOI] [PubMed] [Google Scholar]
- Clerihew L, Lamagni TL, Brocklehurst P, McGuire W. Candida parapsilosis infection in very low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 2007;92:F127–129. doi: 10.1136/fnn.2006.097758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepe GS, Jr, Gibbons R. V beta 6+ T cells are obligatory for vaccine-induced immunity to Histoplasma capsulatum. J Immunol. 2001a;167:2219–2226. doi: 10.4049/jimmunol.167.4.2219. [DOI] [PubMed] [Google Scholar]
- Deepe GS, Jr, Gibbons R. Protective efficacy of H antigen from Histoplasma capsulatum in a murine model of pulmonary histoplasmosis. Infect Immun. 2001b;69:3128–3134. doi: 10.1128/IAI.69.5.3128-3134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepe GS, Jr, Gibbons RS. Cellular and molecular regulation of vaccination with heat shock protein 60 from Histoplasma capsulatum. Infect Immun. 2002;70:3759–3767. doi: 10.1128/IAI.70.7.3759-3767.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demasi AP, Pereira GA, Netto LE. Yeast oxidative stress response. Influences of cytosolic thioredoxin peroxidase I and of the mitochondrial functional state. Febs J. 2006;273:805–816. doi: 10.1111/j.1742-4658.2006.05116.x. [DOI] [PubMed] [Google Scholar]
- Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113 Pt 19:3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- Feldmesser M, Kress Y, Casadevall A. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology. 2001;147:2355–2365. doi: 10.1099/00221287-147-8-2355. [DOI] [PubMed] [Google Scholar]
- Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Fisher KL, Woods JP. Determination of beta-glucosidase enzymatic function of the Histoplasma capsulatum H antigen using a native expression system. Gene. 2000;247:191–197. doi: 10.1016/s0378-1119(00)00099-8. [DOI] [PubMed] [Google Scholar]
- Fridkin SK, Kaufman D, Edwards JR, Shetty S, Horan T. Changing incidence of Candida bloodstream infections among NICU patients in the United States: 1995–2004. Pediatrics. 2006;117:1680–1687. doi: 10.1542/peds.2005-1996. [DOI] [PubMed] [Google Scholar]
- Gacser A, Salomon S, Schafer W. Direct transformation of a clinical isolate of Candida parapsilosis using a dominant selection marker. FEMS Microbiol Lett. 2005;245:117–121. doi: 10.1016/j.femsle.2005.02.035. [DOI] [PubMed] [Google Scholar]
- Garcia-Rivera J, Chang YC, Kwon-Chung KJ, Casadevall A. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot Cell. 2004;3:385–392. doi: 10.1128/EC.3.2.385-392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Gomez FJ, Allendoerfer R, Deepe GS., Jr Vaccination with recombinant heat shock protein 60 from Histoplasma capsulatum protects mice against pulmonary histoplasmosis. Infect Immun. 1995;63:2587–2595. doi: 10.1128/iai.63.7.2587-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães AJ, Williams DA, Zancope-Oliveira RM, Nosanchuk JD. Protective antibodies to Histoplasma capsulatum cell surface antigens: heat shock protein 60 and M antigen (Abstract B35). ASM Conference on Dimorphic Fungal Pathogens; Denver, CO, ASM. 2006. p. 42. [Google Scholar]
- Jeffery CJ. Moonlighting proteins. Trends Biochem Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- Jeffery CJ. Multifunctional proteins: examples of gene sharing. Ann Med. 2003a;35:28–35. doi: 10.1080/07853890310004101. [DOI] [PubMed] [Google Scholar]
- Jeffery CJ. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 2003b;19:415–417. doi: 10.1016/S0168-9525(03)00167-7. [DOI] [PubMed] [Google Scholar]
- Jurado JD, Rael ED, Lieb CS, Nakayasu E, Hayes WK, Bush SP, Ross JA. Complement inactivating proteins and intraspecies venom variation in Crotalus oreganus helleri. Toxicon. 2007;49:339–350. doi: 10.1016/j.toxicon.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kauffman CA. Histoplasmosis: a clinical and laboratory update. Clin Microbiol Rev. 2007;20:115–132. doi: 10.1128/CMR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux JF, et al. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem J. 2004;380:161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KH, Gomez FJ, Morris RE, Newman SL. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J Immunol. 2003;170:487–494. doi: 10.4049/jimmunol.170.1.487. [DOI] [PubMed] [Google Scholar]
- Ma Y, Kuno T, Kita A, Nabata T, Uno S, Sugiura R. Genetic evidence for phospholipid-mediated regulation of the Rab GDP-dissociation inhibitor in fission yeast. Genetics. 2006;174:1259–1271. doi: 10.1534/genetics.106.064709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosanchuk JD, Steenbergen JN, Shi L, Deepe GS, Jr, Casadevall A. Antibodies to a cell surface histone-like protein protect against Histoplasma capsulatum. J Clin Invest. 2003;112:1164–1175. doi: 10.1172/JCI19361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S. A model for Rab GTPase localization. Biochem Soc Trans. 2005;33:627–630. doi: 10.1042/BST0330627. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- Ponnambalam S, Baldwin SA. Constitutive protein secretion from the trans-Golgi network to the plasma membrane. Mol Membr Biol. 2003;20:129–139. doi: 10.1080/0968768031000084172. [DOI] [PubMed] [Google Scholar]
- Potolicchio I, Carven GJ, Xu X, Stipp C, Riese RJ, Stern LJ, Santambrogio L. Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J Immunol. 2005;175:2237–2243. doi: 10.4049/jimmunol.175.4.2237. [DOI] [PubMed] [Google Scholar]
- Rattray JB, Schibeci A, Kidby DK. Lipids of yeasts. Bacteriol Rev. 1975;39:197–231. doi: 10.1128/br.39.3.197-231.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell. 2008;7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, et al. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 2007;6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R, Marie M, Nizak C, Dale HA, Pernet-Gallay K, Perez F, et al. Rab1 defines a novel pathway connecting the pre-Golgi intermediate compartment with the cell periphery. Mol Biol Cell. 2006;17:1514–1526. doi: 10.1091/mbc.E05-08-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheckelhoff M, Deepe GS., Jr The protective immune response to heat shock protein 60 of Histoplasma capsulatum is mediated by a subset of V beta 8.1/8.2+ T cells. J Immunol. 2002;169:5818–5826. doi: 10.4049/jimmunol.169.10.5818. [DOI] [PubMed] [Google Scholar]
- Sebghati TS, Engle JT, Goldman WE. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science. 2000;290:1368–1372. doi: 10.1126/science.290.5495.1368. [DOI] [PubMed] [Google Scholar]
- Stone KL, Williams KR. The Protein Protocols Handbook. Totowa, NJ: Humana Press; 1996. Enzymatic digestion of proteins in solution and in SDS polyacrylamide gels. [Google Scholar]
- Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- van Meer G, Sprong H. Membrane lipids and vesicular traffic. Curr Opin Cell Biol. 2004;16:373–378. doi: 10.1016/j.ceb.2004.06.004. [DOI] [PubMed] [Google Scholar]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- Woods JP. Histoplasma capsulatum molecular genetics, pathogenesis, and responsiveness to its environment. Fungal Genet Biol. 2002;35:81–97. doi: 10.1006/fgbi.2001.1311. [DOI] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell. 2006;17:5131–5140. doi: 10.1091/mbc.E06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zancope-Oliveira RM, Reiss E, Lott TJ, Mayer LW, Deepe GS., Jr Molecular cloning, characterization, and expression of the M antigen of Histoplasma capsulatum. Infect Immun. 1999;67:1947–1953. doi: 10.1128/iai.67.4.1947-1953.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


