Abstract
The atypical cadherin Fat acts as a receptor for a signaling pathway that regulates growth, gene expression, and planar cell polarity (1, 2). Genetic studies in Drosophila identified the four-jointed gene as a regulator of Fat signaling (3–5). We show that four-jointed encodes a protein kinase that phosphorylates Ser or Thr residues within extracellular cadherin domains of Fat and its transmembrane ligand, Dachsous. Four-jointed functions in the Golgi (6), and is the first molecularly-defined kinase that phosphorylates protein domains destined to be extracellular. An acidic sequence motif (DNE) within Four-jointed was essential for its kinase activity in vitro, and for its biological activity in vivo. Our results indicate that Four-jointed regulates Fat signaling by phosphorylating cadherin domains of Fat and Dachsous as they transit through the Golgi.
The Fat and Hippo signaling pathways intersect at multiple points, and influence growth and gene expression through regulation of the transcriptional co-activator Yorkie (7–14). Fat signaling also influences planar cell polarity (PCP) (1). Fat acts as a transmembrane receptor, and is a large (5147 aa) atypical cadherin protein, with 34 extracellular cadherin domains (Fig. 1A) (15). Dachsous (Ds) is also a large (3503 aa) transmembrane protein with multiple cadherin domains (Fig. 1A) (16), and is a candidate Fat ligand because it appears to bind Fat in a cultured cell assay (17), acts non-autonomously to influence Fat pathway gene expression (4, 8), and acts genetically upstream of fat in the regulation of PCP (5). A second protein, Four-jointed (Fj), also acts non-cell autonomously to influence Fat pathway gene expression and acts genetically upstream of fat in the regulation of PCP (4, 5, 8). However, Fj is a type II transmembrane protein that functions in the Golgi (6, 18). Thus, Fj might influence Fat signaling by post-translationally modifying a component of the Fat pathway.
Figure 1. Four-jointed-dependent mobility shift of cadherin domain polypeptides.
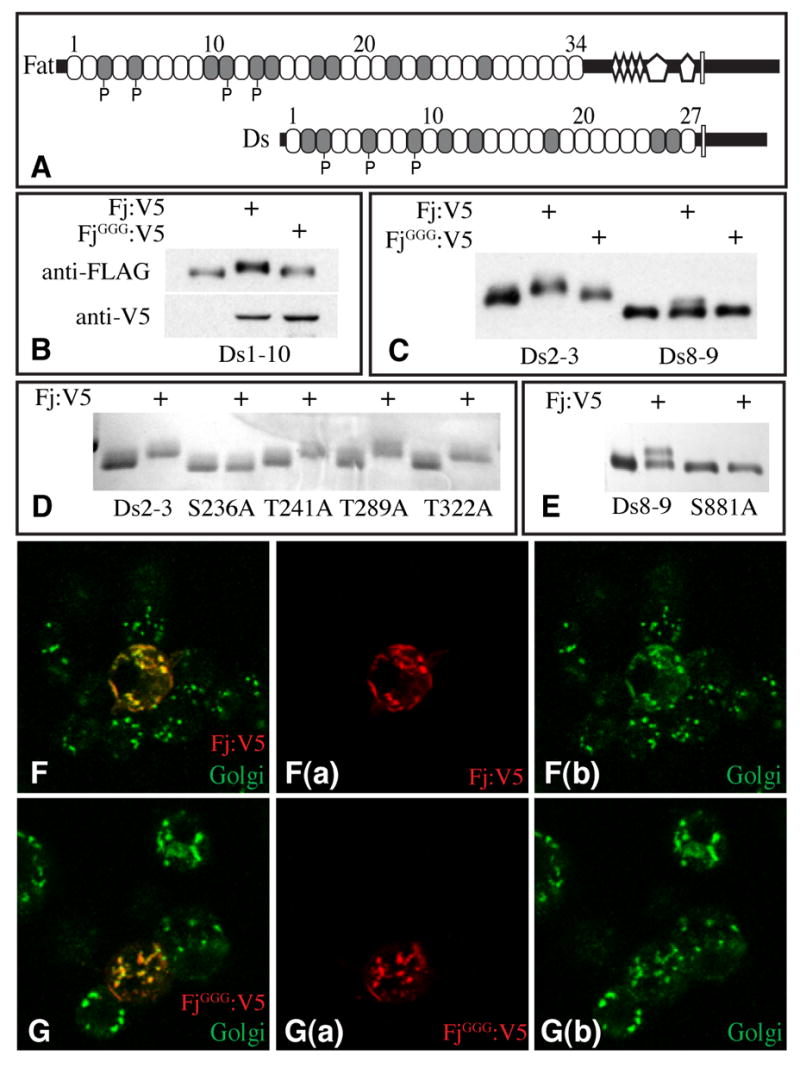
A) Schematics of Fat and Ds. Cadherin domains (ovals), EGF domains (diamonds), LamG domains (pentagons) and transmembrane domains (rectangles) are indicated. Shading identifies cadherin domains with a Ser or Thr as the seventh amino acid, “P” identifies Cadherin domains at which Fj-mediated phosphorylation was detected. B, C, E show Western blots (anti-FLAG) of Ds Cadherin domains isolated from medium of S2 cells; cells were co-transfected with plasmids to express Fj:V5 or FjGGG:V5 as indicated. B) Ds1-10:FLAG. Fj expression is indicated by anti-V5 staining. C) Ds2-3:FLAG and Ds8-9:FLAG. D) Coomasie stain of Ds2-3:FLAG and point mutants immunoprecipitated from the medium of transfected S2 cells, where indicated cells express Fj:V5. E) Ds8-9:FLAG and Ds8-9S881A:FLAG mutant. F-G Show S2 cells transfected to express Fj:V5 (F) or FjGGG:V5 (G) and stained with anti-V5 (red) and anti-Golgi (green); panels to the right show individual stains.
To investigate the possibility of modification of Fat or Ds, we co-expressed FLAG epitope-tagged fragments of their extracellular domains together with Fj in cultured Drosophila S2 cells. When the first 10 cadherin domains of Ds (Ds1-10) were co-expressed with Fj, a shift in mobility was observed (Fig. 1B). A common post-translational modification of secreted and transmembrane proteins as they pass through the Golgi is glycosylation. Most glycosyltransferases contain a conserved sequence motif, DXD (19), which is essential for their activity. As a related sequence motif (DNE at aa 490–492) is present in Fj and its vertebrate homologues (20, 21), we created a mutant form of Fj in which DNE was changed to GGG (FjGGG). The levels and Golgi localization of FjGGG appear normal (Fig. 1B,F,G), but FjGGG expression did not shift Ds1-10 mobility (Fig. 1B).
To identify modified cadherin domains, we expressed smaller fragments of Ds1-10. The smallest fragments whose mobility was shifted in cells expressing Fj were 2-cadherin domain polypeptides: Ds2-3, Ds5-6, and Ds8-9 (Fig. 1C, Fig. S1C in Supplementary material). Ds2-3 and Ds5-6 appeared to be stoichiometrically modified in cells expressing Fj, whereas Ds8-9 was only partially modified. Fat4-5 was also partially shifted by Fj co-expression (Fig S1C). The mobility shifts of these 2-cadherin domain polypeptides were not observed with FjGGG (Fig. 1C, Fig. S1B). To identify potential sites of modification, we aligned their sequences (Fig. S1E). This identified four sites at which a Ser or Thr residue was conserved, whose hydroxyl groups could potentially be sites of post-translational modification. To evaluate their influence, each was mutated in turn to Ala within the Ds2-3 polypeptide. Three of the four mutants had no effect; however, one, Ds2-3S236A, completely eliminated the Fj-dependent mobility shift (Fig. 1D). Introduction of an analogous mutation into Ds8-9 also eliminated its mobility shift (Fig. 1E). Thus, a Ser reside at a specific location within the second of the two cadherin domains was essential for the Fj-dependent mobility shift. This amino acid was a Ser in each of these di-cadherin domains, but Thr was also compatible with the Fj-dependent modification (Fig. S1D). In a structurally solved cadherin domain (22), this Ser is the seventh amino acid, and predicted to be located on the surface near the middle of the cadherin domain (Fig. S2).
To identify post-translational modifications associated with this mobility shift, we purified Ds2-3 from S2 cells expressing or not expressing Fj, digested the proteins with trypsin, and analyzed the resulting peptides by mass spectrometry. One peptide from Fj-expressing cells was stoichiometrically shifted by 80 daltons relative to the same peptide from cells not expressing Fj, and also eluted earlier on HPLC (Fig. 2A, B and S3A). Mass and tandem mass spectrometry (MS/MS) fragmentation patterns identified this peptide as amino acids 215 to 237 of Ds, and refined the site of modification to within amino acids 232 to 237 (Fig. 2A, B). The mass of the equivalent peptide from Ds2-3S236A was not altered by Fj expression (Fig. S3D, E). We identified the majority of peptides corresponding to Ds2-3 cadherin domains, and none of the others were detectably modified in cells expressing Fj (Fig. S3A–C). Thus, the Fj-dependent modification of Ds2-3 comprises an addition of 80 daltons, which is attached to Ser236. An 80 dalton mass does not correspond to any known glycans, but does correspond to the mass associated with addition of a phosphate group. Incubation of Fj-modified Ds fragments with either calf intenstinal alkaline phosphatase (CIP) or Antarctic phosphatase (AnP) reversed the Fj-dependent mobility shifts of Ds2-3, Ds8-9, and Fat 4–5 (Fig. 2C, D). Thus, Ds and Fat cadherin domains are subject to Fj-dependent phosphorylation at a specific Ser residue.
Figure 2. Four-jointed dependent phosphorylation of Ds and Fat cadherin domains.
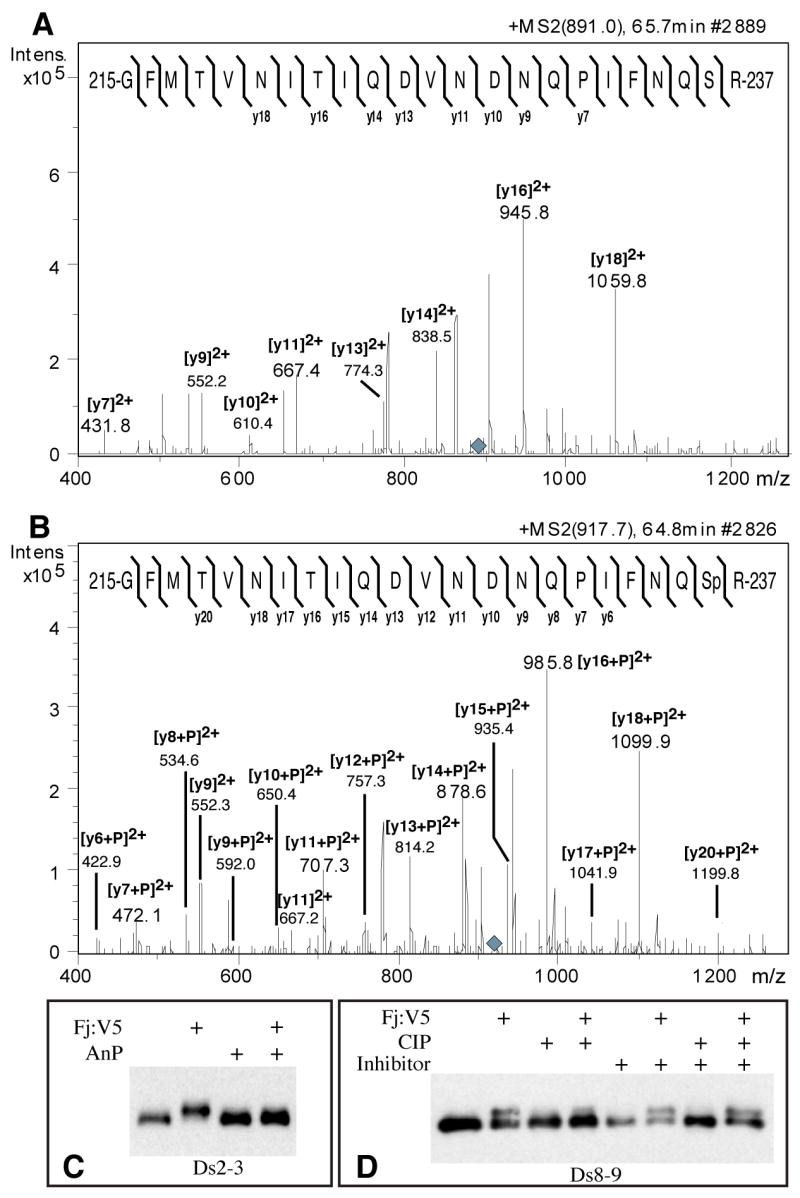
A) MS/MS fragmentation of a tryptic peptide from Ds2-3:FLAG isolated from S2 cells, eluting on HPLC at 65.7 minutes (Supplementary Fig. S3A). The parental ion is triply charged (m/z 891.0, grey diamond). The mass and fragmentation pattern identify this as aa 215-237; the positions of identified y ions from peptide fragmentation are indicated. B) MS/MS fragmentation of a tryptic peptide from Ds2-3:FLAG isolated from Fj:V5-expressing S2 cells, eluting on HPLC at 64.8 minutes (parent ion m/z 917.7, grey diamond). The positions of identified y ions from peptide fragmentation are indicated. C) Western blot on Ds2-3:FLAG isolated from S2 cell media, indicated cells expressed Fj:V5, and/or were treated with antarctic phosphatase (AnP). D) Western blot on Ds8-9:FLAG isolated from S2 cell media, indicated cells expressed Fj:V5, and/or treated with calf intestinal alkaline phosphatase (CIP), and/or treated with phosphatase inhibitor cocktail.
To investigate whether Fj itself has kinase activity, a secreted, epitope-tagged Fj (sFj:V5) was purified from the medium of cultured S2 cells (Fig. S4). Purified sFj:V5 was then incubated with affinity purified Ds2-3 and [γ-32P]ATP in buffer. Transfer of 32P onto Ds2-3 was observed in the presence of sFj, but not in its absence, and not when sFjGGG was used as the enzyme (Fig. 3A). Moreover, Ds2-3S236A was not detectably phosphorylated by sFj. We also characterized the activity of Fj expressed in a heterologous system by expressing a GST:Fj fusion protein in E. coli and partially purifying it on glutathione beads. GST:Fj, but not GST:FjGGG, catalyzed the transfer of 32P onto Ds2-3 (Fig 3B). Thus, Fj is a protein kinase.
Figure 3. Kinase activity of Four-jointed.
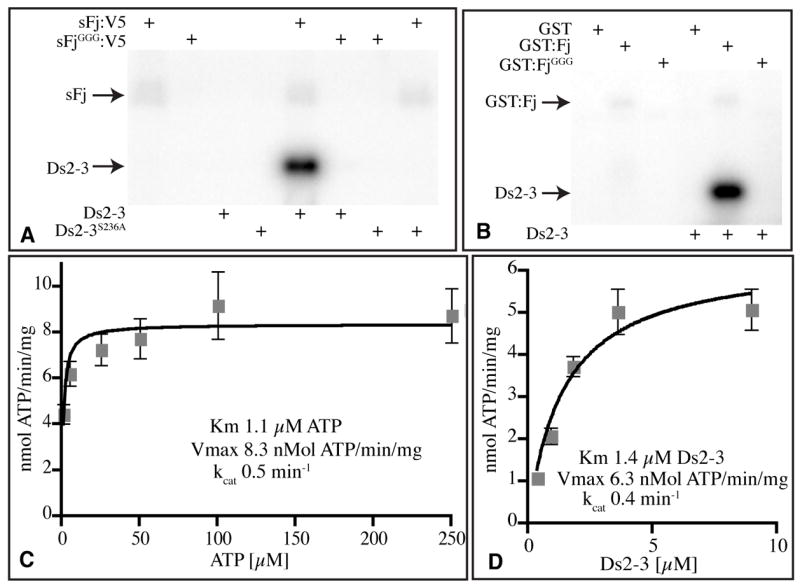
A, B) Autoradiograms of protein gels dispalying the products of in vitro kinase reactions with the indicated enzymes (top) and substrates (bottom). The positions of Ds2-3:FLAG and sFj:V5 or GST:Fj are indicated. A) Phosphorylation of Ds2-3, but not Ds2-3S236A, by Fj in vitro. B) Kinase activity of GST:Fj isolated from E. coli. C) Reaction velocity versus ATP concentration, using 2.5 μg Ds2-3:FLAG, 10 ng sFj:V5, and 1, 5, 25, 50, 100, or 250 μM ATP for 15 min. G) Reaction velocity versus Ds2-3 concentration, using 500 μM ATP, 10 ng sFj:V5, and 0.36, 0.90, 1.8, 3.6, or 9.0 μM Ds2-3:FLAG for 15 min. Plots in C and D show average values from four experiments, error bars indicate s.e.m.
The generic kinase substrates, Myelin basic protein and Casein were not detectably phosphorylated by sFj (Fig. S4C). Thus, Fj appears to have a limited substrate specificity. Only a few proteins have been identified as being phosphorylated in the secretory pathway, and none of the responsible kinase(s) have been molecularly identified. A Golgi kinase activity, referred to as Golgi Casein kinase, preferentially phosphorylates Ser or Thr residues within a S/T-X-E/D/S(Phos) consensus sequence (23, 24). Because Fj does not phosphorylate Casein (Fig. S4C), and the Ser residues within Cadherin domains targeted by Fj do not conform to Golgi Casein kinase sites (Fig. S6B), Fj is not Golgi Casein kinase. Fj autophosphorylation was detected, but this reaction was weak compared to phosphorylation of Ds2-3 (Fig. 3A, B). The autophosphorylation reaction is apparently unimolecular, because GST:Fj and sFj:V5 did not phosphorylate each other, and the fraction of Fj phosphorylated was independent of concentration (Fig. S5).
Some cadherin domain polypeptides that include a Ser as the seventh amino acid were not detectably shifted, but the mobility shift on Ds2-3 might reflect a conformational effect. To examine the ability of Fj to phosphorylate other cadherin domains, we performed in vitro kinase reactions with [γ-32P]ATP. This identified phosphorylation sites on polypeptides that were not gel shifted, including Fat2-3, Fat10-11, and Fat12-13 (Fig S6). The in vitro kinase reactions also identified differences in the efficiency with which different Cadherin domains were phosphorylated by Fj, with Ft3, Ds3, and Ds6 the best substrates (Fig. S6).
If we take the presence of a Ser or Thr at the seventh amino acid of a cadherin domain as the minimal requirement for Fj-mediated phosphorylation, there are 9 potential sites in Ds and 11 in Fat (Fig. 1A). However, Fat10, Ds2, Ds11, Ds13 and Ds18 were not detectably phosphorylated, despite the presence of Ser or Thr at this position (Fig. S6). Presumably, there are other structural features important for recognition by Fj. This was also emphasized by the detection of phosphorylation of our Ds2-3 polypeptide, but not our Ds3-4 polypeptide, even though both contain Ser236. Our di-cadherin constructs were based on published annotations (16), but in comparing Ds cadherin domains to structurally solved cadherin domains (22), we realized that these mis-position the inter-cadherin domain boundary, and consequently these constructs lacked three amino acids of the first cadherin domain. Addition of these amino acids, together with the inter-cadherin domain linker sequence, enabled phosphorylation of a Ds3 single cadherin domain construct (Fig. S6D).
Parameters of Fj kinase activity were examined using purified sFj:V5 and Ds2-3:FLAG. The reaction rate was linear over short times, and dependent on enzyme and substrate concentrations (Figs 3, S7). The turnover number (kcat) was 0.5 min−1, confirming that Fj acts catalytically. Fj exhibited a strong preference for Mn++ as a co-factor, and was active over a wide pH range (Fig. S7). The estimated Km for Ds2-3 was 1.4 μM (Fig. 3D), and the Km for ATP was approximately 1μM (Fig. 3C). [γ-32P]guanosine triphosphate (GTP) could also be used as a phosphate donor, but was several fold less effective than ATP (Fig S7E). In a competition assay, unlabelled ATP efficiently competed with [γ-P32]ATP, but GTP, cytidine triphosphate, thymidine triphosphate, and uridine triphosphate did not (Fig S7F). Thus, ATP is the preferred phosphate donor.
A weak similarity between Fj and the bacterial kinase HipA, and between Fj and the mammalian lipid kinase PI 4-kinase II, have been suggested previously based on bioinformatic analyses in which HipA or PI4KII were used as the starting point for PSI-BLAST searches (25, 26). Asp residues play critical roles in catalysis and in the coordination of Mg++ in these and other kinases (26, 27), and the loss of Fj kinase activity associated with mutation of the conserved DNE motif is thus consistent with the inference that Fj is related to other kinases. A single Fj orthologue, Fjx1, is present in a range of vertebrate species, including humans (20).
To investigate the biological requirement for Fj kinase activity, we assayed the catalytically inactive fjGGG mutant in vivo. A V5 epitope-tagged form of this gene was expressed in transgenic Drosophila. At the same time, we constructed V5-tagged wild-type fj. To ensure that both forms were expressed in similar amounts, we used a site-specific integration method to insert transgenes at the same chromosomal location (28). Immunostaining confirmed that FjGGG:V5 and Fj:V5 both exhibited normal Golgi localization and were expressed similarly (Fig. S8). Uniform over-expression of fj reduces the growth of legs and wings and interferes with normal PCP (4, 5, 29). Fj:V5 exhibited phenotypes consistent with prior studies, but FjGGG:V5 was completely inactive (Fig. 4). Thus, mutation of the DNE motif in Fj abolishes its biological activity.
Figure 4. Requirement for Four-jointed kinase activity in vivo.
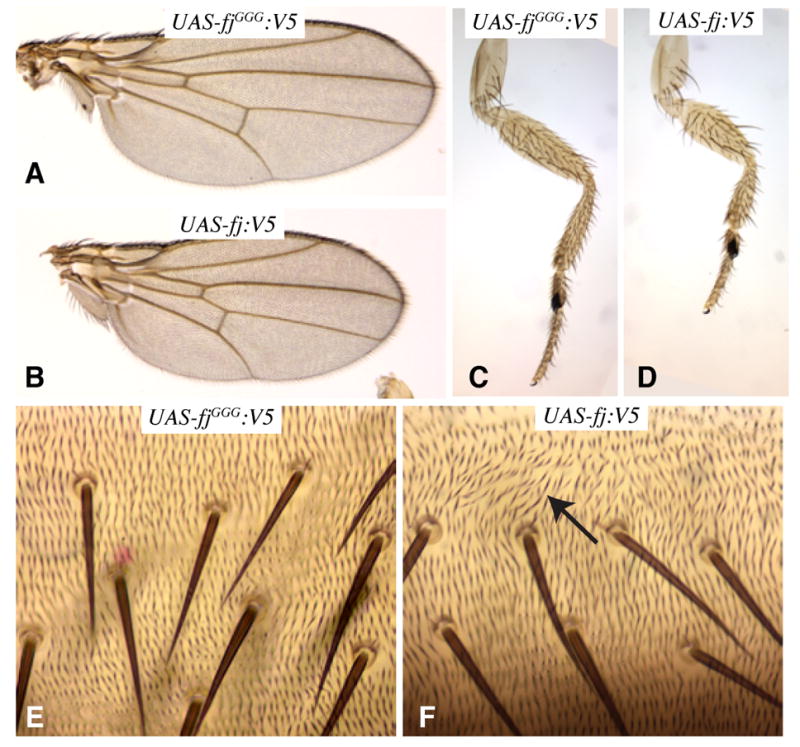
A–F Show portions of adult flies, expressing either wild-type fj:V5 or the fjGGG:V5 mutant. A) Wing from tub-Gal4 UAS-fjGGG:V5. B) Wing from tub-Gal4 UAS-fj:V5. C) Leg from tub-Gal4 UAS-fjGGG:V5. D) Leg from tub-Gal4 UAS-fj:V5. E) Abdomen from tub-Gal4 UAS-fjGGG:V5 animal. F) Abdomen from tub-Gal4 UAS-fj:V5 animal with disturbed hair polarity (arrow).
The identification of Fj’s cadherin domain kinase activity provides a biochemical explanation for the influence of Fj on Fat signaling, and supports a model in which Fj directly phosphorylates Fat and Ds as they transit through the Golgi to influence their activity, presumably by modulating interactions between their cadherin domains. As there was a substantial difference in the efficiency with which individual cadherin domains could be modified by Fj, both in cell-based and in vitro assays, it is also possible that differences in the extent of Fat and Ds phosphorylation normally occur in vivo, and might differentially modify their binding or activity. (30)
Supplementary Material
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. Their manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Strutt H, Strutt D. Bioessays. 2005;27:1218. doi: 10.1002/bies.20318. [DOI] [PubMed] [Google Scholar]
- 2.Pan D. Genes Dev. 2007;21:886. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 3.Casal J, Struhl G, Lawrence P. Curr Biol. 2002;12:1189. doi: 10.1016/s0960-9822(02)00974-0. [DOI] [PubMed] [Google Scholar]
- 4.Cho E, Irvine KD. Development. 2004;131:4489. doi: 10.1242/dev.01315. [DOI] [PubMed] [Google Scholar]
- 5.Yang C, Axelrod JD, Simon MA. Cell. 2002;108:675. doi: 10.1016/s0092-8674(02)00658-x. [DOI] [PubMed] [Google Scholar]
- 6.Strutt H, Mundy J, Hofstra K, Strutt D. Development. 2004;131:881. doi: 10.1242/dev.00996. [DOI] [PubMed] [Google Scholar]
- 7.Bennett FC, Harvey KF. Curr Biol. 2006;16:2101. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 8.Cho E, et al. Nat Genet. 2006;38:1142. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 9.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. Curr Biol. 2006;16:2081. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Willecke M, et al. Curr Biol. 2006;16:2090. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Irvine KD. Proc Natl Acad Sci U S A. 2007;104:20362. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong J, et al. Cell. 2007;130:1120. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Wu S, Barrera J, Matthews K, Pan D. Cell. 2005;122:421. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Oh H, Irvine KD. Development. 2008;135:1081. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahoney PA, et al. Cell. 1991;67:853. doi: 10.1016/0092-8674(91)90359-7. [DOI] [PubMed] [Google Scholar]
- 16.Clark HF, et al. Genes Dev. 1995;9:1530. doi: 10.1101/gad.9.12.1530. [DOI] [PubMed] [Google Scholar]
- 17.Matakatsu H, Blair SS. Development. 2006;133:2315. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- 18.Villano JL, Katz FN. Development. 1995;121:2767. doi: 10.1242/dev.121.9.2767. [DOI] [PubMed] [Google Scholar]
- 19.Wiggins CA, Munro S. Proc Natl Acad Sci U S A. 1998;95:7945. doi: 10.1073/pnas.95.14.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rock R, Heinrich AC, Schumacher N, Gessler M. Dev Dyn. 2005;234:602. doi: 10.1002/dvdy.20553. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi K, Parish J, Akita K, Francis-West P. Dev Dyn. 2006;235:3085. doi: 10.1002/dvdy.20946. [DOI] [PubMed] [Google Scholar]
- 22.Patel SD, et al. Cell. 2006;124:1255. doi: 10.1016/j.cell.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 23.Christensen B, Nielsen MS, Haselmann KF, Petersen TE, Sorensen ES. Biochem J. 2005;390:285. doi: 10.1042/BJ20050341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasa-Benito M, Marin O, Meggio F, Pinna LA. FEBS Lett. 1996;382:149. doi: 10.1016/0014-5793(96)00136-6. [DOI] [PubMed] [Google Scholar]
- 25.Barylko B, et al. J Biol Chem. 2001;276:7705. doi: 10.1074/jbc.C000861200. [DOI] [PubMed] [Google Scholar]
- 26.Correia FF, et al. J Bacteriol. 2006;188:8360. doi: 10.1128/JB.01237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groth AC, Fish M, Nusse R, Calos MP. Genetics. 2004;166:1775. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeidler MP, Perrimon N, Strutt DI. Dev Biol. 2000;228:181. doi: 10.1006/dbio.2000.9940. [DOI] [PubMed] [Google Scholar]
- 30.We thank M. Simon for communicating unpublished observations, the Developmental Studies Hybridoma Bank and the Bloomington stock center for antibodies and Drosophila stocks, C. Rauskolb and T. Miller for comments on the manuscript, E. Cho and A. Xu for initial Fj, Fat, and Ds expression plasmids, and C. Rauskolb for assistance in generating images for figure panels. Supported by the Howard Hughes Medical Institute (K.D.I.), NIH grants CA123071, GM061126 (R.S.H) and GM078620 (K.D.I.), and a fellowship from the Yamada Science Foundation (H.O.I.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


