Abstract
The myotubularin (MTM) enzymes are phosphotidylinositol-3-phosphate and phosphatidylinositol 3,5-bisphosphate phosphatases. Mutation of MTM1, the founder member of this family, is responsible for X-linked myotubular myopathy in humans. Here, we have isolated and characterized a Caenorhabditis elegans homology of the enzymes designated ceMTM3. ceMTM3 preferably dephosphorylates phosphotidylinositol-3-phosphate and contains a FYVE lipid-binding domain at its C-terminus which binds phosphotidylinositol-3-phosphate. Immunoblotting analyses revealed that the enzyme is expressed during the early development and adulthood of the animal. Immunofluorescent staining revealed predominant expression of the enzyme in eggs and muscles. Knockdown of the enzyme by using feeding-based RNA interference resulted in an increased level of phosphotidylinositol-3-phosphate and caused severe impairment of body movement of the worms at their post-reproductive ages and significantly shortened their lifespan. This study thus reveals an important role of the MTM phosphatases in maintaining muscle function, which may have clinical implications in prevention and treatment of sarcopenia.
Keywords: Tyrosine phosphatase, myotubularin, phosphoinositide, aging, locomotion
Introduction
The tyrosine phosphatase super-family consists of highly diverse members of enzymes characterized by the presence of a conserved (H/V)C(X)5R(S/T) signature motif [Alonso et al, 2004; Tonks, 2006]. The classic tyrosine phosphatases, also known as PTP1B-like enzymes, are tyrosine-specific, while others are dual specificity phosphatases (DSP) that can act on both serine/threonine and tyrosine. Interestingly, some of the dual specificity enzymes preferably dephosphorylate phosphoinositides. Tumor suppressor PTEN is a well-known example which dephosphorylates phosphatidylinositol 3, 4-bisphosphate (PI3,4P2) and phosphatidylinositol 3,4,5-trisphosphate (PI3,4,5P3) [Maehama and Dixon, 1998; Myers et al, 1998]. MTMs stand for another subfamily of the dual specificity phosphatases that act on phosphoinositide substrates [Clague and Lorenzo, 2005; Robinson and Dixon, 2006]. Unlike PTEN, MTMs specifically dephosphorylate phosphatidylinositol 3-phosphate (PI3P) and phosphatidylinositol 3,5-bisphosphate (PI3,5P2). Myotubularin (hMTM1) is the founding member of the MTM subfamily enzymes. It was isolated by positional cloning from the Xq28 region of human chromosomes [Laporte et al, 1996]. Over 81 mutations have been found in unrelated patients with X-linked recessive myotubular myopathy, which is characterized by severe hypotonia and generalized muscle weakness with impaired maturation of muscle fibers. Loss of MTM1 function has a profound impact on the organization of muscle cells during myogenesis. Two other members of the MTM subfamily, namely, hMTMR2 and hMTMR13, were found mutated in recessive forms of Charcot-Marie-Tooth neuropathy type 4B [Bolino et al, 2000; Senderek et al, 2003]. How the absence of these MTM enzymes causes diseases is not well understood. The MTM enzymes have diverse structural features, and 14 family members have been identified in the human genome [Robinson and Dixon, 2006]. Some of them contain FYVE domains [Zhao et al, 2000; Zhao et al, 2001; Walker et al, 2001]. The FYVE domain (also known as the FYVE finger) is named after the first four proteins (Fab1, YOTB, Vac1p, and EEA1) that were shown to contain this domain [Stenmark et al, 1996]. These domains specifically bind PI3P thereby regulating membrane trafficking and signal transduction [Stenmark and Aasland, 1999]. Binding of FYVE domains with PI3P presumably targets FYVE domain-containing MTMs to PI3P-rich cell compartments, thereby dephosphorylating PI3P and PI3,5P2 more efficiently. PI3P is known to play an important role in membrane trafficking [Vanhaesebroeck, 2001].
Materials and Methods
Molecular cloning of ceMTM3
By searching the WormBase database with the cDNA sequences of human FYVE-DSPs [Zhao et al, 2000; Zhao et al, 2001], we found a homologous gene in the C. elegans genome with designation of T24A11.1 and T24A11.1b. To verify the cDNA sequence of the C. elegans enzyme, we employed the rapid amplification of cDNA ends (RACE) strategy by using the Smart-RACE kit from Clontech following the manufacture’s directions. First, a RACE cDNA library was synthesized from the total RNA of N2 C. elegans. The cDNA library was then amplified by PCR with adaptor primers provided in the kit and two gene specific primers with the sequences of 5′ CCCATCCATCAGAACAATGAACCA and 5′ CAACTGGATGGCTTCTCAATCTGAG 3′. The PCR products were cloned into the pCRII vector, and multiple clones selected for sequencing analysis. Based on the sequence of the RACE PCR products, primers corresponding to the 5′ and 3′ ends of the open reading frame were synthesized and used to amplify the complete coding regions of the cDNA which were cloned into the pBluescript KS vector. Sequencing analyses revealed two complete cDNAs which we designated ceMTM3a and ceMTM3b,
Generation of anti-ceMTM3 antibody
A GST fusion protein (GST-ceMTM3CT) containing the C-terminal segment (aa 701 to 961, FYVE domain-included) of ceMTM3b was purified from an E. coli expression system with the pGex-2T vector. The purified protein was used to immunize a rabbit to generate anti-serum. The antibody was purified through negative selection on GST-agarose and then positive selection on a GST-ceMTM3CT-agarose.
Knockdown of ceMTM3 expression by feeding-based RNAi
The full-length coding sequence of ceMTM3b (−6 to 2910 with translation starting codon ATG starting from 1) and two fragments F1 (27– 876) and F2 (877–2910) were cloned into the pPD129.36 vector (provided by Dr. Andrew Fire, Stanford University). The vector has a bi-directional promoter configuration consisting of two T7 promoters flanking a single copy of a cDNA insert to facilitate synthesis of both sense and anti-sense RNAs [Timmons et al, 2001]. Plain p129.36 vector was used as control throughout the study. The HT115(DE3) E. coli cells were employed as a host for expression of double strand RNAs, and induction and feeding were as performed following the standard procedures [Timmons et al, 2001]. The Bristol N2 strain of C. elegans hermaphrodite was used in the study, and the worms were cultured on NGM plates at 20 °C according to standard protocols.
Western blotting analyses and phosphatase activity assays
C. elegans worms in mixed population or at different stages were collected and extracted by sonication in a whole cell extraction buffer containing 25 mM 2-glycerol phosphatase (pH 7.3), 5 mM EDTA, 5 mM EGTA, 0.1 M NaCl, 1% Triton X-100, 10mM β –mercaptolethanol, and a cocktail of protease inhibitors (Roche Applied Science). Samples containing 15 – 20 μg of total proteins were separated on SDS gel and transferred to polyvinylidene difluoride (PVDF) membrane for Western blotting analysis using the enhanced chemiluminescence method. For phosphatase activity assays, extracts containing 200–300 μg of total proteins were subjected to immunoprecipitation with anti-ceMTM3. Activity assays were performed with the immunoprecipitates in the presence of 50 μM PI3P lipid vesicles at pH 6.0 according to procedure described by Taylor et al. [2000].
Lipid-membrane overlay assays
Nitrocellulose membranes spotted with various lipid compounds were incubated with 0.5 μg/ml GST or GST-ceMTM3CT in a buffer containing 10 mM Tris-HCl (pH 8.0), 0.15 M NaCl, 0.1% Tween-20, and 3% fatty acid-free bovine serum albumin [Zhao et al, 2003]. After extensive washing with the buffer, proteins bound to the membranes were probed with anti-GST antibodies and then with horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibodies. Detection was made by enhanced chemoluminescence reactions. To access the contents of PI3P in control and RNAi-treated worms, total phospholipids were extracted from the worms with methanol/chloroform (2:1, v/v) containing 0.25% concentrated HCl and quantified by using the Malachite green phosphate method following ashing of the lipids with sulfuric acid and perchloric acid. Equivalent amounts of crude phospholipids were then resolved by thin layer chromatography on silica plates [Sui et al, 2000], and the sections corresponding to standard PI3P (1,2-Dioleoyl-sn-Glycero-3-Phosphoinositol from Avanti Polar Lipids, Alabaster, AL) was scraped off and extracted in the aforementioned chloroform/methanol/HCl solution. The partially purified lipids were spotted on nitrocellulose membranes together with standard PI3P and then incubated with GST-ceMTM3CT as described above.
Immunofluorescent staining
Whole mount worms were fixed onto glass slides by cracking freezing following standard procedures, and the animals were treated in methanol for 5 min and then in acetone for 5 min at −20°C. For paraffin embedding, worms were fixed with 3.7% formaldehyde for 4 hr at 4°C and then put into melted soft agar. The solidified agar pieces were then processed in a paraffin embedding machine. Three-micron slices were cut and fixed onto glass slides. Immunofluorsecent staining was performed with primary antibodies and Cy3-conjugated donkey anti-rabbit and FITC-conjugated goat anti-mouse secondary antibodies. Immunofluorescent micrographs were obtained by using an Olympus BX51 microscope equipped with a DP71 digital camera.
Results
Molecular cloning of ceMTM3
We have previously isolated two FYVE domain-containing members of the MTM family enzymes designated FYVE-DSP1 and FYVE-DSP2 [Zhao et al, 2000; Zhao et al, 2001], also known as MTMR3 and MTMR4 [Walker et al, 2001; Robinson and Dixon, 2006], respectively. Searching of the WormBase database by using the BLAST program with the cDNA sequences of the human enzymes revealed a homologous gene in the C. elegans genome with designation of T24A11.1 and T24A11.1b. To verify the cDNA sequence of the C. elegans enzyme, we employed the rapid amplification of cDNA ends (RACE) strategy by using the Smart-RACE kit from Clontech. This resulted in the isolation of two cDNA sequences. One has an open reading frame encoding a protein of 1006 amino acid residues, while the other gives rise to a 961 amino acid protein. The two cDNAs resulted from alternative splicing; the former matches C. elegans MTM3 previously reported (Xue et al, 2003; Genbank accession NM_065365) which we re-designated ceMTM3a here, while the latter represents a new isoform and was named ceMTM3b. The cDNA sequence of ceMTM3b has been deposited into the GenBank database (accession number DQ988041). Both ceMTM3 and ceMTM3b have differences from the predicted coding sequences of T24A11.1 and T24A11.1b defined in WormBase. The ceMTM3 gene is located on chromosome III (−5.21) of the C. elegans genome and spans over 10 kb. ceMTM3a has 10 exons while ceMTM3b contains a 12-bp-shorter exon 3 and skips the entire 335 bp exon 9. A schematic diagram of the ceMTM3 protein structure is shown in Fig. S1 of Online Supplementary Materials. ceMTM3 has conserved catalytic domain and a C-terminal FYVE domain. The FYVE domain is preceded by a coiled coil structure of 30–40 aa. ceMTM3b differs from ceMTM3a by missing 4 amino acid residues at the N-terminus and having a different, shorter C-terminal segment after the FYVE domain. The human and C. elegans enzymes share ~30 % overall sequence identity in their catalytic domains and the FYVE domain regions. Similar structures suggest similar functions and regulatory mechanisms. Therefore, finding ceMTM3 in C. elegans provides an excellent model system to study the function and regulation of FYVE domain-containing MTM enzymes.
Expression and distribution of ceMTM3 in C. elegans
To analyze the expression of ceMTM3 in C. elegans, we first generated a rabbit polyclonal antibody against GST-ceMTM3CT, a GST fusion molecule containing the C-terminal region (aa 701 to 961) of ceMTM3b expressed in E. coli cells. This antibody was purified through negative selection on GST-agarose and positive selection on an antigen affinity column. On Western blotting analysis, ceMTM3 appears as a relatively broad band of ~105 kDa (Fig. 1, upper panel). It is expressed at the egg and young larva (L1 and L2) stages. It decreases to near absence in the L3 to young adult stages, and then regains high expression in the egg-laying period and post-reproductive age. We further employed immunofluorescent cell staining to detect the tissue expression of the enzyme in C. elegans. Staining of whole-mount gravid worms showed expression of the enzyme in body wall muscle and in eggs (Fig. 1, middle panel). This was further verified by staining paraffin-embedded cross-sections of adult worms (Fig. 1, lower panel). Note that the staining of paraffin-embedded cross-sections also revealed partial expression of the enzyme in the intestine. The muscle expression was further verified by co-staining the paraffin-embedded sections with an antibody against myosin, a muscle marker (Fig. S2). Interestingly, in an earlier study, by using a green fluorescent protein (GPF) reporter construct, Xue et al [2003] demonstrated a predominant expression of ceMTM3 in head neurons and a low level of expression in posterior intestinal cells. However, the authors cautioned that the reporter constructs may not show all the tissues in which the genes are expressed and that expression in the posterior intestinal cells is a common artifact for GFP expression vectors [Xue et al, 2003]. In this study, we employed a specific antibody to detect the expressed protein directly, and specificity of the method was further verified by knockdown studies (see latter in Fig. 3C).
Fig. 1. Expression and distribution of ceMTM3 in C. elegans.
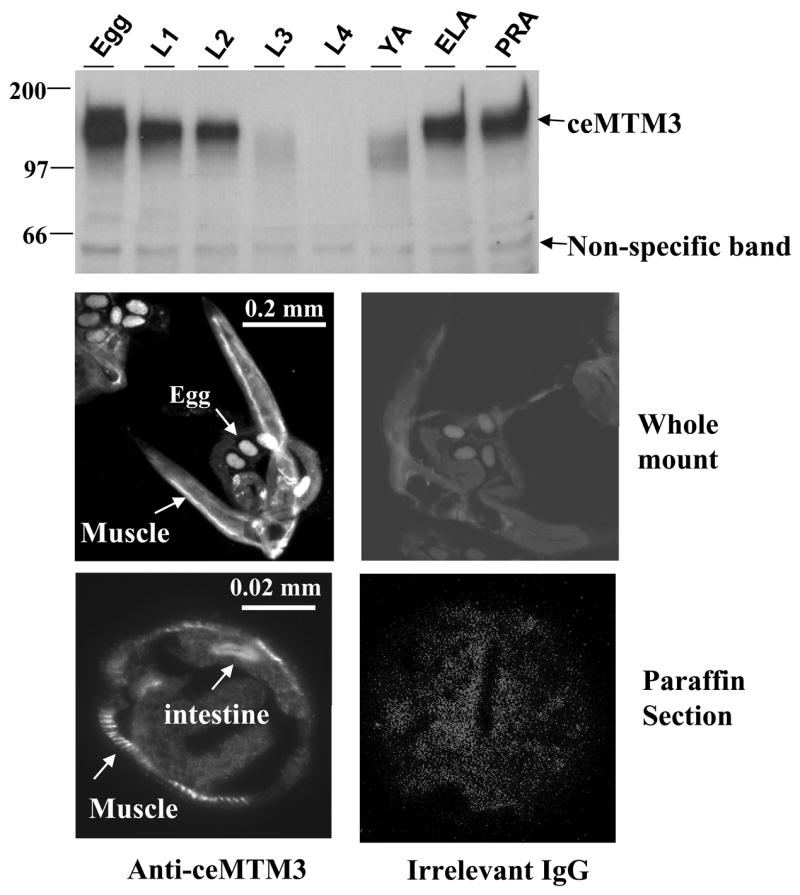
Top panel: C. elegans at different stages including egg, L1, L2, L3, L4, YA (young adult), egg-laying adult (ELA), and post-reproductive adult (PRA) were collected according to synchronized culture procedures. Cell extracts containing equal amounts of total proteins were subjected to Western blotting analysis with anti-ceMTM3 antibody. The position of ceMTM3 is indicated. A weak, non-specific band of ~60 kDa essentially serves as an internal loading control. Middle and bottom panels: whole mount (middle panel) and paraffin-embedded cross sections (bottom panel) of adult C. elegans were subjected to indirect immunofluorescent staining with anti-ceMTM3 antibody or irrelevant rabbit IgG. Arrows highlight positive staining at body wall muscle, eggs, and intestine.
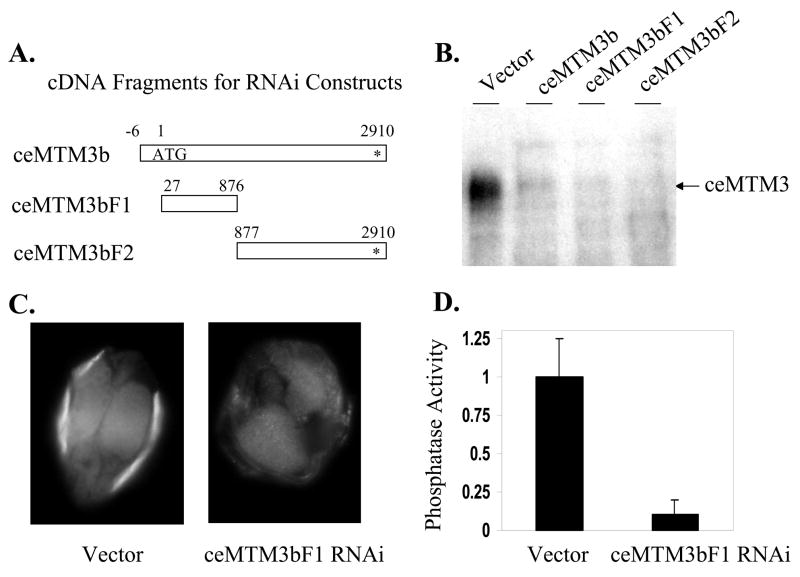
Fig. 3. Knockdown of ceMTM3 expression by RNAi.
Normal N2 worms were fed with E. coli cells carrying vector control, full-length ceMTM3b, and two ceMTM3b cDNA fragments (ceMTM3bF1 and ceMTM3bF2) as indicated in A. B. Crude cell extracts containing equal amounts of total proteins were subjected to Western blotting analyses. C. Paraffin-embedded worm sections were stained with anti-ceMTM3 antibody. D. The phosphatase activity of immunoprecipitated ceMTM3 was determined with 5 μM PI3P substrate at pH 6.0 as described in Fig. 2A. Error bars denote standard deviation (n = 4).
Biochemical characterization of ceMTM3
By using immuno-purified ceMTM3, we analyzed the substrate specificity of ceMTM3 (Fig. 2A). As expected, ceMTM3 preferably dephosphorylated PI3P. It also displayed a significant activity toward PI3,5P2 but essentially no activity to PI4P and PI5P. Like other members of the tyrosine phosphatase superfamily, ceMTM3 was effectively inhibited by sodium vanadate and peroxide. It is generally believed that FYVE domain specifically binds PI3P [Stenmark and Aasland, 1999]. To determine the binding specificity of the FYVE domains to phosphoinositides, we employed the commonly used lipid membrane overlay assays. The PIP array membranes that were spotted with different doses of various phosphoinositides (Echelon Research Lab, Salt Lake City, UT) were incubated with GST or GST-ceMTM3CT, a GST fusion protein containing the FYVE domain of ceMTM3. As shown in Fig. 2B, GST-ceMTM3CT binds specifically to PI3P, whereas no binding was found with the GST control. The data indicate that ceMTM contains a typical FYVE domain.
Fig. 2. Biochemical characterization of ceMTM3.
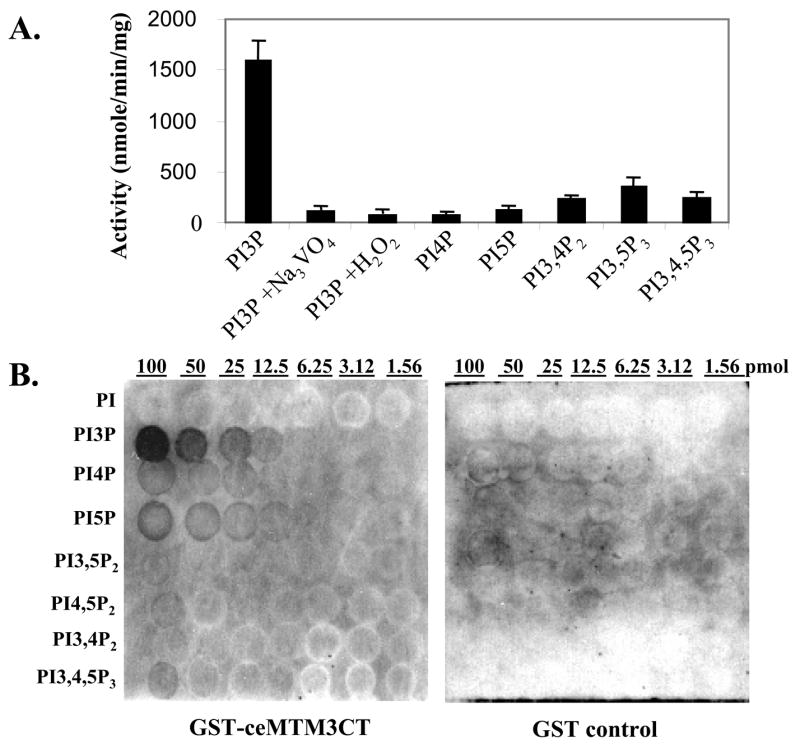
A. Substrate specificity of ceMTM3. ceMTM3 was immuno-purified from worm extracts. Phosphatase activity assays with various phosphoinositides were performed at pH 6.0 with 5 μM each of lipids (Echelon Research Lab, Salt Lake City, UT). For PI3P, assays were also performed in the presence of 1 mM sodium vanadate or 0.1 mM hydrogen peroxide. Error bars denote standard deviation (n = 3). B. Binding specificity of the FYVE domain of ceMTM3. PIP arrays (Echelon Research Lab, Salt Lake City, UT) with indicated amounts (1.56 – 100 pmol) of various phopshoinositides were probed with 0.5 μg/ml GST-ceMTM3CT or GST in a washing buffer containing 10 mM Tris-HCl (pH 8.0), 0.15 M NaCl, 0.1% Tween-20, and 3% fatty acid-free BSA. Bound proteins were detected by immunoblotting with anti-GST antibody and HRP-conjugated secondary antibody.
Knockdown of ceMTM by feeding-based RNAi
To study the biological function of ceMTM3, feeding-based RNA interference (RNAi) was employed to knockdown the expression of enzyme in C. elegans. For this purpose, the full-length coding sequence of ceMTM3b and two of its fragments were cloned into the pPD129.36 vector that contains the T7 promoter at both 5′ and 3′ ends to drive transcription (Fig. 3A). As shown in Fig. 3B, and 3C, treatment of wild type N2 worms with E. coli cells expressing double-strand RNA derived from the full-length or either of the two truncated fragments of ceMTM3b cDNA resulted in nearly total deletion of ceMTM3 as revealed by Western blotting analysis and immunofluoresecent staining of paraffin-embedded sections. These data not only indicate the efficiency of the feeding-based RNAi in knocking down the gene expression of ceMTM3 but also verified the specificity of our anti-ceMTM3 antibody. As expected, knockdown of ceMTM3 by RNAi also greatly diminished its phosphatase activity measured with PI3P as a substrate (Fig. 3D). We obtained similar results with all three cDNA fragments shown in Fig. 3A. This essentially rules out the possibility that phenotype is due to the knockdown of non-specific genes. For simplicity, results observed with the ceMTM3bF1 fragment are described below.
As a PI3P phosphatase, knockdown is expected to increase the level of intracellular PI3P. To access the level of PI3P, we took advantage of GST-ceMTM3CT which binds specifically to PI3P. First, phospholipids were extracted by acidified chloroform/methanol from worms untreated or treated with ceMTM3bf1 RNAi. The crude lipid extracts were quantified based on phosphate contents and then resolved by thin layer chromatography on silica plates. Sections corresponding to standard PI3P were scraped off and extracted in acidified chloroform/methanol. Equivalent amounts of partially purified lipids were spotted on nitrocellulose membranes together with standard PI3P and then incubated with GST-ceMTM3CT as described above. Fig. 4 demonstrates that knockdown of ceMTM3 caused an approximately 2-fold increase in the binding of GST-ceMTMCT, representing a 2-fold increase in the PI3P contents in the RNAi-treated worms. It should be noted that the standard curve was non-linear and that the separation of crude lipids by thin-layer chromatography on silica plates is necessary to reduce background binding.
Fig. 4. Increased levels of PI3P in C. elegans caused by knockdown of ceMTM3 expression by RNAi.
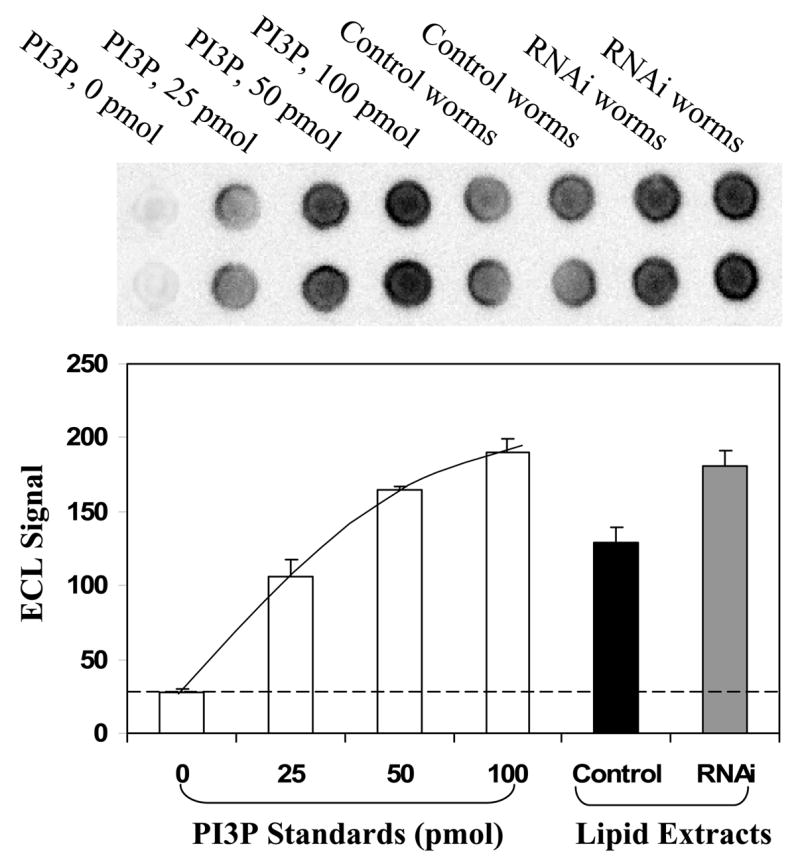
PI3P was extracted from N2 worms cultured on NGM plates containing E. coli cells carrying vector control or ceMTM3bF1 and then fractionated on thin layer chromatography plates as described in Methods and Materials. Equivalent amounts of samples together with standard PI3P were spotted on a nitrocellulose membrane. The membrane was incubated with a GST-ceMTM3CT followed by detection with anti-GST antibody and HRP-conjugated secondary antibody. Capture of immunoblot images and quantification of band signals were carried out by using FluorChem SP imaging system from Alpha Innotech. Error bars denote standard deviation (n ≥ 4).
Morbidity and mortality of C. elegans worms caused by knockdown of ceMTM3
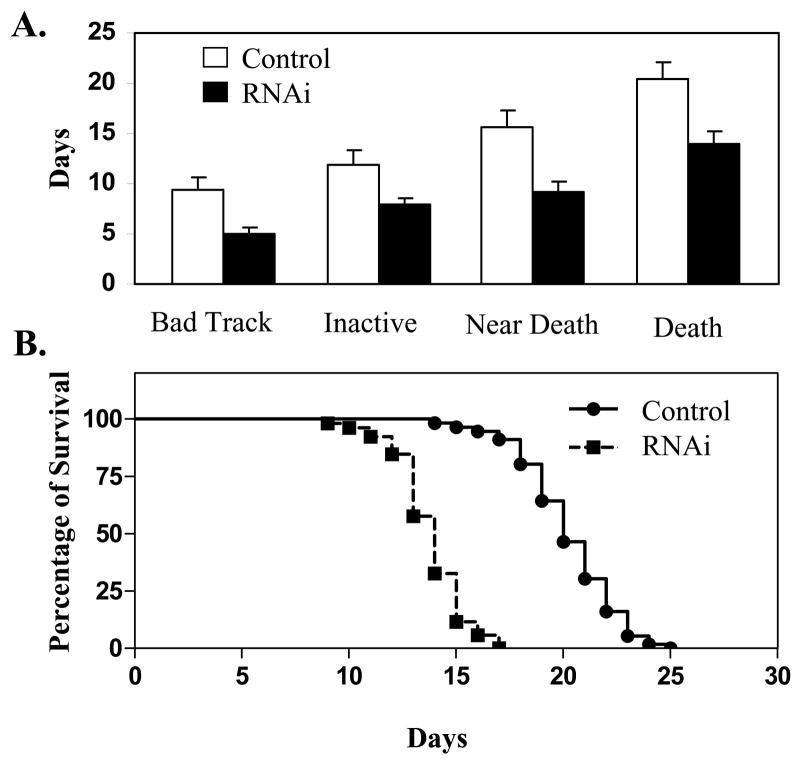
A clear phenotype associated with knocking-down of ceMTM3 expression was the impaired locomotion of the worms when they late and post reproductive stages (day 5 and thereafter). This is clearly reflected in the moving tracks of the animal as shown in Fig. 5. C. elegans worms move by propagating waves of alternating dorsal and ventral flexions along its body length, producing regular sinusoidal tracks on a bacterial lawn. Normal worms fed on vector control E. coli cells kept such a moving track until an average age of 9 days. However, RNAi-treated worms started to show very sluggish body movement by day 5. Their tracks had shorter wavelengths, and their traveling distance at fixed period of time was much shorter than that exhibited by the control worms. The body movement gradually worsened as the animal aged. By day 8, they became very inactive and failed to move out of a 1 cm diameter cycle within 2 hr, and by day 9, their body movement was essentially impaired. Consequently, they died, on average, on day 14. In contrast, normal worms have an average of lifespan of around 20 days. Fig. 6 compares the morbidity and mortality of control and ceMTM3F1 RNAi-treated C. elegans worms. It should be noted that the poor body movement with RNAi-treated worms occurs after day 5 in the late reproductive period. At the larva and young adult stages, their locomotion appeared normal. These RNAi-treated worms developed normally and grew to normal body size although there was a 5–7% reduction in brood size. In addition, ~4% of the RNAi-treated worms formed “worm bag” caused by hatching of young larva in the womb. These worms were not included in our morbidity and mortality analyses (Fig. 6). It should be pointed out that a mild egg laying defect caused RNAi-mediated knockdown of ceMTM3 has also been noted in an earlier study by Xue et al [2003]. However, in that study, the effects on locomotion and lifespan were either not detected or not investigated.
Fig. 5. Impaired locomotion of C. elegans worms caused by knockdown of ceMTM3.
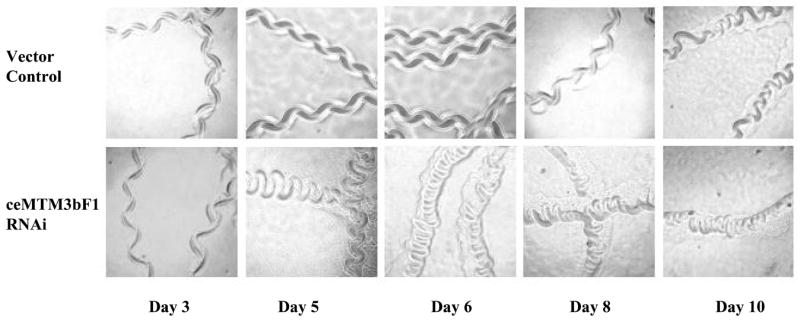
Normal N2 C. elegans worms were cultured on NGM plates containing E. coli cells carrying vector control or ceMTM3bF1. Data show moving tracks of the worms at different ages. Note that sluggish moving tracks were seen with RMAi-treated worms from day 5.
Fig. 6. Morbidity and mortality of C. elegans worms caused by knockdown of ceMTM3.
Normal N2 C. elegans worms were cultured on NGM plates containing E. coli cells carrying vector control or ceMTM3bF1. A. Statistical analysis of morbidity and mortality. B. Survival curves. Data (Mean ± SD, n ≥50) represent the ages at which the indicated symptoms occurred. Back Track, loss of sinusoidal moving tracks; Inactive, failure to move 0.5 cm in 2 hr; Near Death, failure to move 0.5 cm in 24 hr; Death, no pharyngeal pumping and no response to prodding. Borderline cases were usually resolved clearly by the next day of scoring. About 4% of ceMTM1F1-treated worms died of formation of larvae in the womb (worm bagging), and were excluded from this analysis.
Discussion
In the present study, we have cloned and characterized a FYVE domain-containing member of the MTM tyrosine phosphatase subfamily. More importantly, by using the RNAi technique, we found that loss of ceMTM3 expression impaired locomotion of the C. elegans worm as the animal aged. Considering the predominant expression of the enzyme in the muscle of the animal, we believe that ceMTM3 may be required for maintenance of muscle function in the C. elegans worm. The function of ceMTM3 in muscle is not unexpected since mutation of hMTM1, the founding member of the MTM subfamily enzymes, is responsible for X-linked recessive myotubular myopathy [Laporte et al, 1996], which is characterized by severe hypotonia and generalized muscle weakness with impaired maturation of muscle fibers in affected newborn males. However, mutation of hMTM1 appears to affect organization of muscle cells during myogenesis since the disease was believed to result from an arrest in the normal development of muscle fibers at the myotubular stage [Spiro et al, 1966; van Wijngaarden et al, 1969]. Interestingly, MTM1-deficient mice develop a progressive centronuclear myopathy during postnatal life that severely reduces their life expectancy [Buj-Bello et al, 2002]. The data indicate that MTM1 plays a role in muscle maintenance in mice rather than in myogenesis seen in human X-linked recessive myotubular myopathy. Apparently, ceMTM3 may be similar to mouse MTM1 in muscle maintenance. The loss of muscle mass, referred to as sarcopenia, is a normal phenomenon in animals as a consequence of aging [Deschenes, 2004]. This is apparent for normal worms after day 15. Decrease in muscle tissue begins around the age of 50 years for human, but becomes more dramatic beyond the 60th year of life. Loss of muscle mass among the aged directly results in diminished muscle function. Loss of muscle fiber number is the principal cause of sarcopenia. The mechanism for this loss is unknown. Our data suggest that reduced function of the MTM family enzymes may play a role.
Our study suggests that the function of ceMTM3 is aging-related. The MTM family enzymes were initially identified as dual specificity phosphatases, but subsequent studies demonstrated that these enzymes more preferentially dephosphorylate PI3P and PI3,5P2 [Clague and Lorenzo, 2005; Robinson and Dixon, 2006]. By specifically dephosphorylating PI3P and PI3,5P2, the MTM family enzymes must be crucial in regulating the level of phosphoinositides, which are important signal transducers, and loss of their expression naturally causes accumulation of PI3P and PI3,5P2, which may indirectly affect the level of other phosphoinositides. The role of phosphoinositides has been well documented. In fact, disruption of the age-1/PI(3) kinase, a downstream signaling component in the DAF-2 insulin-like signaling pathway is known to extend the lifespan of C. elegans and to cause the constitutive dauer formation (Daf-c) phenotype, which is considered a nonaging stage [Friedman and Johnson, 1988]. Conversely, loss-of-function of daf-18/PTEN, the counterpart of age-1/PI3 kinase, suppresses the life extension and constitutive dauer formation associated with daf-2 or age-1 mutants [Rouault et al, 1999; Mihaylova et al, 1999]. Of course, the products of age-1/PI(3) kinase is PI3,4,5P and PI3,4P, which are primary substrate of daf-18/PTEN [Vanhaesebroeck et al, 2001]. As PI3P phosphatase, ceMTM3 may not directly dephosphorylate PI3,4,5P and PI3,4P but may affect their levels indirectly through altering the pool of phosphoinositides. The MTM enzymes mainly dephosphorylate the D-3 position of PI3P and PI3,5P2 [Clague and Lorenzo, 2005; Robinson and Dixon, 2006; see also Fig. 2], and they may not directly antagonize age-1/PI(3) kinase. Instead, the counterpart of ceMTM3 may be the class III PI3 kinase LET-512/Vps34 which is a PI3 kinase responsible for production of PI3P in C. elegans. LET-512/Vps34 is an essential protein required for membrane trafficking and endocytosis [Takacs-Vellai et al, 2005]. The let-512/Vps34 mutant worms display lethality and molting defects, as well as alterations in the outer nuclear membrane and in the endoplasmic reticulum [Roggo et al, 2002]. However, earlier studies indicate that knockdown of two other C. elegans MTM enzymes, namely, MTM-6 and MTM-9, but not that of ceMTM3, was able to rescue larval lethality of let-512/Vps34 mutants [Xue et al, 2003; Dang et al, 2004]. LET-512/Vps34 is also required for autophagy [Takacs-Vellai et al, 2005]. Autophagy is a form of programmed cell death. Like apoptosis, it has major implications in various diseases, including cancer and degenerative disorders [Kroemer and Jaattela, 2005; Maiuri et al, 2007]. It is to be determined if loss-of-function of ceMTM3 is equivalent to gain-of-function of let-512/VPS34 and thereby leads to autophagy of worm cells.
Like all the other members of the tyrosine phosphatase superfamily, ceMTM3 is inactivated by oxidation because the key cysteinyl residues at the catalytic center are highly sensitive to oxidation (see Fig. 2A). Reactive oxygen species (ROS), such as the superoxide radical (O2−•), hydrogen peroxide (H2O2), and the hydroxyl radical (OH•), are generated during cellular metabolism, especially during mitochondrial energy production [Beckman and Ames, 1998]. Oxidative damage is considered to be the main cause of aging. It was postulated that a slowing in the rate of accumulation of oxidative damage is at least partly responsible for the life extension phenotype seen in C. elegans with daf-2 and age-1 mutations [Honda and Honda, 2002]. Our data showed that ceMTM3 is sensitive to peroxide and loss of ceMTM3 function causes muscle deterioration in C. elegans. Therefore, inactivation of ceMTM3 by oxidation may be attributable to loss of muscle fibers in normal worms at their later ages. We believe that studying the MTM enzymes may have clinical implications in the prevention and treatment of human sarcopenia.
Supplementary Material
Acknowledgments
Grant sponsors: National Natural Science Foundation of China No. 30470391 (to X Fu) and NIH HL076309 and HL079441 (to ZJ Zhao).
Abbreviations used
- PI3P
phosphotidylinositol 3-phosphate
- PI4P
phosphotidylinositol 4-phosphate
- PI5P
phosphotidylinositol 5-phosphate
- PI3
4P2, phosphotidylinositol 3,4-bisphosphate
- PI3
5P2, phosphotidylinositol 3,5-bisphosphate
- PI3
4,5P3, phosphotidylinositol 3,4,5-trisphosphate
References
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- Bolino A, Muglia M, Conforti FL, LeGuern E, Salih MA, Georgiou DM, Christodoulou K, Hausmanowa-Petrusewicz I, Mandich P, Schenone A, Gambardella A, Bono F, Quattrone A, Devoto M, Monaco AP. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nat Genet. 2000;25:17–19. doi: 10.1038/75542. [DOI] [PubMed] [Google Scholar]
- Buj-Bello A, Laugel V, Messaddeq N, Zahreddine H, Laporte J, Pellissier JF, Mandel JL. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc Natl Acad Sci U S A. 2002;99:15060–15065. doi: 10.1073/pnas.212498399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Lorenzo O. The myotubularin family of lipid phosphatases. Traffic. 2005;6:1063–1069. doi: 10.1111/j.1600-0854.2005.00338.x. [DOI] [PubMed] [Google Scholar]
- Dang H, Li Z, Skolnik EY, Fares H. Disease-related myotubularins function in endocytic traffic in Caenorhabditis elegans. Mol Biol Cell. 2004;15:189–196. doi: 10.1091/mbc.E03-08-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschenes MR. Effects of aging on muscle fibre type and size. Sports Med. 2004;34:809–824. doi: 10.2165/00007256-200434120-00002. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda Y, Honda S. Oxidative stress and life span determination in the nematode Caenorhabditis elegans. Ann N Y Acad Sci. 2002;959:466–474. doi: 10.1111/j.1749-6632.2002.tb02117.x. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- Laporte J, Hu LJ, Kretz C, Mandel JL, Kioschis P, Coy J, Klauck SM, Poustka A, Dahl N. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Mihaylova VT, Borland CZ, Manjarrez L, Stern MJ, Sun H. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc Natl Acad Sci U S A. 1999;96:7427–7432. doi: 10.1073/pnas.96.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson FL, Dixon JE. Myotubularin phosphatases, policing 3-phosphoinositides. Trends Cell Biol. 2006;16:403–412. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Roggo L, Bernard V, Kovacs AL, Rose AM, Savoy F, Zetka M, Wymann MP, Muller F. Membrane transport in Caenorhabditis elegans: an essential role for VPS34 at the nuclear membrane. EMBO J. 2002;21:1673–1683. doi: 10.1093/emboj/21.7.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault JP, Kuwabara PE, Sinilnikova OM, Duret L, Thierry-Mieg D, Billaud M. Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr Biol. 1999;9:329–332. doi: 10.1016/s0960-9822(99)80143-2. [DOI] [PubMed] [Google Scholar]
- Senderek J, Bergmann C, Weber S, Ketelsen UP, Schorle H, Rudnik-Schoneborn S, Buttner R, Buchheim E, Zerres K. Mutation of the SBF2 gene, encoding a novel member of the myotubularin family, in Charcot-Marie-Tooth neuropathy type 4B2/11p15. Hum Mol Genet. 2003;12:349–356. doi: 10.1093/hmg/ddg030. [DOI] [PubMed] [Google Scholar]
- Spiro AJ, Shy GM, Gonatas NK. Myotubular myopathy. Persistence of fetal muscle in an adolescent boy. Arch Neurol. 1966;14:1–14. doi: 10.1001/archneur.1966.00470070005001. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Aasland R. FYVE-finger proteins--effectors of an inositol lipid. J Cell Sci. 1999;112:4175–4183. doi: 10.1242/jcs.112.23.4175. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Toh BH, D'Arrigo A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J Biol Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- Sui X, Krantz SB, Zhao ZJ. Stem cell factor and erythropoietin inhibit apoptosis of human erythroid progenitor cells through different signalling pathways. Br J Haematol. 2000;110:63–70. doi: 10.1046/j.1365-2141.2000.02145.x. [DOI] [PubMed] [Google Scholar]
- Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, Kovacs AL, Muller F. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- Taylor GS, Maehama T, Dixon JE. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci U S A. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- van Wijngaarden GK, Fleury P, Bethlem J, Meijer AE. Familial "myotubular" myopathy. Neurology. 1969;19:901–908. doi: 10.1212/wnl.19.9.901. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- Walker DM, Urbe S, Dove SK, Tenza D, Raposo G, Clague MJ Characterization of MTMR3. An inositol lipid 3-phosphatase with novel substrate specificity. Curr Biol. 2001;11:1600–1605. doi: 10.1016/s0960-9822(01)00501-2. [DOI] [PubMed] [Google Scholar]
- Xue Y, Fares H, Grant B, Li Z, Rose AM, Clark SG, Skolnik EY. Genetic analysis of the myotubularin family of phosphatases in Caenorhabditis elegans. J Biol Chem. 2003;278:34380–6. doi: 10.1074/jbc.M303259200. [DOI] [PubMed] [Google Scholar]
- Zhao R, Fu X, Li Q, Krantz SB, Zhao ZJ. Specific interaction of protein tyrosine phosphatase-MEG2 with phosphatidylserine. J Biol Chem. 2003;278:22609–22614. doi: 10.1074/jbc.M301560200. [DOI] [PubMed] [Google Scholar]
- Zhao R, Qi Y, Zhao ZJ. FYVE-DSP1, a dual-specificity protein phosphatase containing an FYVE domain. Biochem Biophys Res Commun. 2000;270:222–229. doi: 10.1006/bbrc.2000.2417. [DOI] [PubMed] [Google Scholar]
- Zhao R, Qi Y, Zhao ZJ. FYVE-DSP2, a FYVE Domain-Containing Dual Specificity Protein Phosphatase That Dephosphorylates Phosphotidylinositol 3-Phosphate. Exp Cell Res. 2001;265:329–338. doi: 10.1006/excr.2001.5185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.