Abstract
Both thermally stable states of phytochrome, Pr and Pfr, have been studied by 13C and 15N cross-polarization (CP) magic-angle spinning (MAS) NMR using cyanobacterial (Cph1) and plant (phyA) phytochrome sensory modules containing uniformly 13C- and 15N-labeled bilin chromophores. Two-dimensional homo- and heteronuclear experiments allowed most of the 13C chemical shifts to be assigned in both states. Chemical shift differences reflect changes of the electronic structure of the cofactor at the atomic level as well as its interactions with the chromophore-binding pocket. The chromophore in cyanobacterial and plant phytochromes shows very similar features in the respective Pr and Pfr states. The data are interpreted in terms of a strengthened hydrogen bond at the ring D carbonyl. The red shift in the Pfr state is explained by the increasing length of the conjugation network beyond ring C including the entire ring D. Enhanced conjugation within the π-system stabilizes the more tensed chromophore in the Pfr state. Concomitant changes at the ring C propionate carboxylate and the ring D carbonyl are explained by a loss of hydrogen bonding to Cph1-His-290 and transmittance of conformational changes to the ring C propionate via a water network. These and other conformational changes may lead to modified surface interactions, e.g., along the tongue region contacting the bilin chromophore.
Keywords: photomorphogenesis, photoreceptors, chromophore-protein interaction, phycocyanobilin, solid-state NMR
Phytochrome photoreceptors were first characterized in plants, where they mediate many photomorphogenetic processes (for reviews see refs. 1 and 2). More recently, this family of photoreceptors has been enlarged by the discovery of homologous proteins in cyanobacteria (3, 4) and nonphotosynthetic bacteria (5, 6). A characteristic feature of all phytochromes is the photoreversibility between two states: The absorption of red light initiates photochemical activity of the thermally stable Pr state (λmax ≈ 660 nm), which travels through a series of intermediates (7) and eventually generates the far-red-absorbing Pfr state. The Pfr state, with a moderate thermal stability of several hours to days, is converted back to Pr upon absorption of a far-red photon (λmax ≈ 710 nm). The origin of this red shift is not known. All phytochromes bind an open-chain tetrapyrrole (bilin) as a chromophore, whose photochemistry triggers the conversion between the Pr and Pfr states. The x-ray structures of the chromophore-binding PAS-GAF bidomain of Deinococcus radiodurans (8) and Rhodopseudomonas palustris (9), both assembled with biliverdin, have been reported. The recently solved structure of a complete PAS-GAF-PHY sensory module of Cph1 from Synechocystis sp. PCC 6803 phytochrome in its Pr state demonstrated that the phycocyanobilin (PCB) cofactor is completely sealed from access to bulk solvent (10). Although a comparison between these 3D structures reveals some differences of the chromophore-binding pocket, most of the chromophore–protein interactions are conserved. In all crystal structures, the open-chain tetrapyrrole chromophore has been reported to adopt a ZZZssa geometry [5-Z, 10-Z, 15-Z, 5-syn, 10-syn, 15-anti, see supporting information (SI) Fig. S1] of the three methine bridges. The chromophore in different phytochromes has been intensively studied by various spectroscopic methods. It is commonly accepted that the conversion from Pr to Pfr is initiated by a Z → E photoisomerization of the methine bridge between rings C and D (11–14). However, the exact geometry of the chromophore in the Pfr state and the role of the chromophore-binding pocket in the phototransformation have yet to be established. Moreover, slight rotations around single bonds that are hardly reflected by the crystal structures may cause a more distorted chromophore, a scenario that is supported by a recent investigation using vibrational spectroscopy and density functional theory calculations (15).
Only little is known about the details of the photochemical machinery allowing for intramolecular signal transduction from the chromophore onto the protein surface. Mutational studies on Cph1 (16) have demonstrated the crucial role of Asp-207 (Cph1 numbering of residues is used throughout the article) for intramolecular signal transduction (Fig. S2). It has been proposed that in Cph1 Tyr-176, whose side chain is close to ring D, acts as a molecular gate in the decay of the photochemically excited Pr (17). Moreover, in bacteriophytochrome RpBphP2, two tyrosine residues, Tyr-207 and Tyr-272 (equivalent to Tyr-198 and Tyr-263 in Cph1, respectively), play likewise an important role during the Pr → Pfr photoconversion (9).
Cross-polarization (CP) magic-angle spinning (MAS) NMR has evolved into a uniquely versatile tool for the structure elucidation in systems of high-molecular-mass and solid materials. In conjunction with selective isotope labeling, CP/MAS NMR allows for the study of large protein complexes down to the atomic level (18). In this work, the N-terminal sensory modules of the cyanobacterial phytochrome Cph1 (Cph1Δ2) and the 65-kDa fragment of oat phytochrome A (phyA65) have been studied by 13C and 15N CP/MAS NMR. Holoproteins of these phytochromes were generated by in vitro assembly with uniformly 13C- and 15N-labeled PCB cofactor (u-[13C, 15N]-PCB), thus selective observation of the chromophore in the protein matrix has been achieved. The quality of the 15N data is significantly improved compared with previous work (19), allowing for 2D spectroscopy. Here, we show a full NMR analysis of the cofactor in both states, Pr and Pfr. Dramatic changes around ring D and the propionate side chain of ring C are explained by a model for signal transduction.
Results
Assignments of MAS-NMR Spectra of Pr and Pfr States in Cph1Δ2.
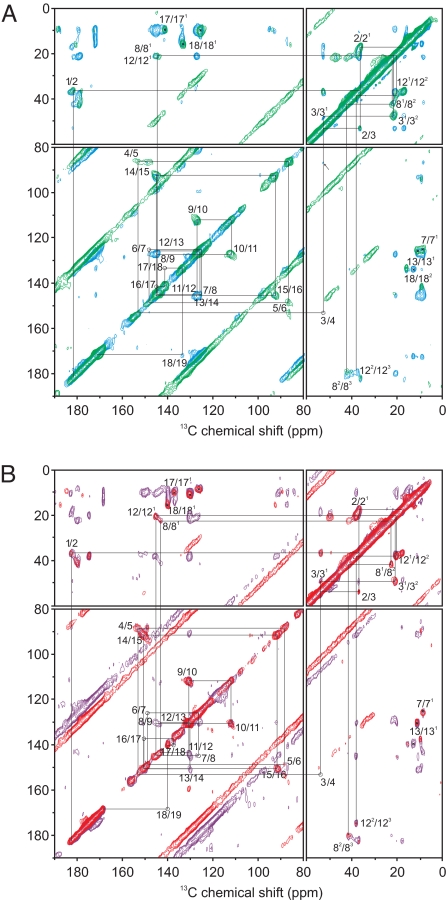
The 2D 13C-13C dipolar-assisted rotational resonance (DARR) (20) spectrum of the u-[13C, 15N]-PCB-Cph1Δ2 in the Pr state was recorded with two 13C homonuclear recoupling mixing times of 5 and 50 ms (Fig. 1A, green and blue, respectively. Enlarged views of the spectra are provided in Fig. S3). Using a 50-ms mixing time, the carbon resonance at 182.7 ppm correlates with two aliphatic carbons, allowing for an unambiguous assignment of carbons 1, 2 and 21 (see Table S1). For carbons 1 and 2, a slight doubling is observed (see below). The well defined correlation peaks in the aliphatic region (60–0 ppm) reveal the correlation network among the 2, 21, 3, 31, and 32 carbon atoms. None of the DARR spectra display the 3/4 cross-peak; however, the 3/5 correlation is visible using a mixing time of 50 ms. The assignment of position 5 has been confirmed by a 1D 13C CP/MAS NMR spectrum of the 13C5-PCB-phyA65 (see Fig. S4). The 4/5 and 5/6 correlations are visible on the spectrum recorded with a short proton mixing time of 5 ms. The two neighbors of carbon 5 are difficult to distinguish, although a weak correlation with 7 suggests that the resonance of 6 is that at 149.5 ppm. The 13C-13C correlation network along the pyrrole rings B and C is shown in Fig. 1A and allows for the assignment of the carbon atoms up to position 16. The 16/17 and 17/18 correlation peaks are weakly visible, and the assignments of 17 and 18 have been obtained from the 18/19 correlation as well as from the methyl and ethyl side chain (171, 181, and 182). The 13C assignment reveals that the central rings B and C of the chromophore are highly symmetrical in the Pr state, which is in line with a recent 15N CP/MAS NMR study (19). The 13C-13C correlations in the aliphatic domain show a single correlation network for one of the propionate side chains (22.9, 41.8, and 180.5 ppm) and a split of the correlation network of the second propionate side chain (22.8, 42.9, and 180.0 ppm and 21.8, 41.4, and 179.3 ppm). Because the 8/81 and 12/121 correlation peaks are overlapping, the two propionate side chains cannot be assigned unambiguously. However, the chemical shift differences occurring in comparison with the Pfr state (see below) make an assignment of the propionate with the single correlation network to carbon 12 more reasonable.
Fig. 1.
Contour plot of the 2D 13C-13C DARR NMR spectra of u-[13C, 15N]-PCB-Cph1Δ2 in the Pr (A) and Pfr (B) states. Proton mixing times of 5 and 50 ms were used for the Pr (green and blue, respectively) and Pfr (red and purple, respectively) states. All DARR spectra were recorded at 233 K and a spinning frequency of 9 or 10 kHz. The lines indicate sequences of nearest-neighbor correlations.
The 2D 13C-13C DARR spectra of Cph1Δ2 in the Pfr state have been recorded with two mixing times (5 and 50 ms) and are depicted in Fig. 1B (red and purple, respectively). The 13C assignments have been obtained in the same manner as for the Pr state. In the Pfr state, the 8/81 and 12/121 correlation peaks do not overlap. Moreover, the 12/13, 13/131, and 131/14 cross-peaks are visible in the 2D spectrum and allow for the unambiguous assignment of the two propionate side chains. Whereas in the side chain of ring B, two sets of chemical shifts appear in the Pr state, only a single set is present in the Pfr state. This is similar to the observation of doubling at the ring A carbons 1 and 2 (see above). Also, because optical spectroscopy, operating on a fast time scale, observes two distinguished forms of the PCB cofactor in the Pr state (21–23), we assume that the two conformers coexist is solution. As the origin of this heterogeneity, mobility of the nearby N-terminal α-helix (Thr-4–Leu-18) that seals parts of the chromophore-binding site along ring A and B can be postulated. The disappearance of the doublings in the Pfr state may be caused by a decreased mobility in this region, thus leading to conformational homogeneity.
A 1H-13C heteronuclear 2D spectrum of the region of the methine carbons of u-[13C, 15N]-PCB in Cph1Δ2 is shown in Fig. S5. The signal labeled with an asterisk originates from the protein and provides an internal reference. The low-field shift of H10 has also been observed previously for the Pr state by solution-state NMR (24) as well as in model compounds in solution (25) and appears to be an intrinsic property of open-chain tetrapyrrole compounds. Upon photoconversion to the Pfr state, the chemical shift of H5 remains unchanged, whereas at H10 and H15 high-field shifts of 0.4 ppm are observed. Such a shift has been expected for H15, where the photoisomerization is initiated. However, its appearance at H10 is surprising and may reflect a change of ring–current interaction with the nearby residue His-260, because the distance between the centroid of its imidazole side chain and the C10 atom accounts for only 3.6 Å in the Pr-state of Cph1.
Pr → Pfr Conversion in Cph1Δ2.
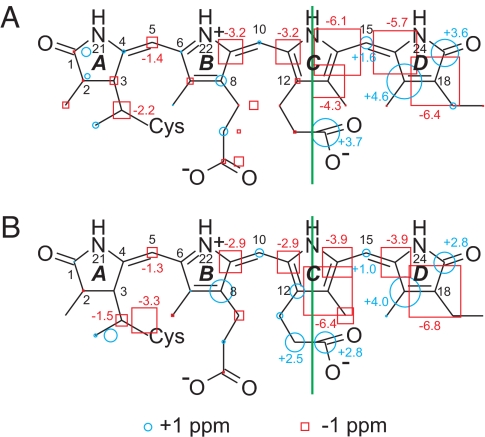
The changes in 13C chemical shifts generated by the Pr → Pfr conversion are illustrated in Fig. 2A. The photoconversion affects mostly the rings C and D, whereas smaller changes are observed along rings A and B. It is generally accepted that the first step of the Pr → Pfr conversion is the photoisomerization of the 15=16 double bond. The striking change in chemical shift at the 14 and 16 positions are in line with this assumption. Furthermore, the entire ring D shows drastic changes in its 13C shifts, denoting a modification of its interaction with the protein surroundings. The symmetry of the two inner rings B and C found in the Pr state is broken in the Pfr state by strong downshifts of the 13 and 14 resonances. Moreover, the Pr → Pfr transformation affects also the two methine bridges between rings A–B and B–C. At the B–C methine bridge, the resonances of 9 and 11 are equally downshifted by 3.2 ppm. The change in chemical shift at position 5 indicates that the A–B methine bridge is affected by the photoconversion. The significant downshift at the 31 position by 2.2 ppm suggests a modification of the chromophore–protein linkage. The 123 carbon atom exhibits a striking 3.7 ppm up-field shift after photoconversion, pointing to a drastic change of the environment of the carboxylate moiety. According to the crystal structures, this carboxyl group of the propionate side chain of ring C interacts via two water molecules with the conserved histidine His-290, which in turn interacts with ring D (9, 10, 26). On the other hand, there are no indications for changes at the propionate side chain of ring B, which forms a salt bridge via two hydrogen bonds to the conserved Arg-254 in the Pr state. Apparently, this interaction remains unaffected in the Pfr state.
Fig. 2.
Schematic representation of the change in 13C chemical shift of the u-[13C, 15N]-PCB chromophore in Cph1Δ2 (A) and phyA65 (B). The Pr state is taken as reference, and the size of the circles and squares refers to the (Pr–Pfr) difference in 13C chemical shift. The few carbons showing two resonances are labeled with two symbols.
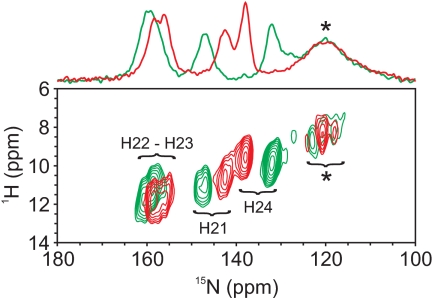
Fig. 3 shows an 1H-15N heteronuclear correlation experiment that indicates good agreement with data obtained in the solution state (27). Based on the 15N assignments of refs. 19 and 24 (Table S2), the strongest changes in 15N chemical shifts are observed for the nitrogens of rings A and D, N21 and N24, respectively. These nitrogen cross-signals show only minor changes in their proton frequency. The hydrogen bonding of the ring D nitrogen is, if at all, weak. The ring B and C nitrogens show clearly hydrogen-bonding interactions. In the Pr state structures of Cph1Δ2 and the bacteriophytochromes, a conserved water molecule is within hydrogen-bonding distance of the ring A, B, and C nitrogens as well as of the side chain of His-260. Accordingly, the 0.8 ppm down-shift of the proton attached to the ring B or C nitrogens may either originate from altered interactions with this water molecule or from a change of ring-current shifts caused by nearby located aromatic residues such as His-260 (N22–imidazole: 4.2 Å, N23–imidazole: 4.6 Å) or Tyr-176 (N22–phenyl: 5.5 Å, N23–phenyl: 6.3 Å).
Fig. 3.
Contour plot of 1H-15N 2D heteronuclear dipolar correlation spectra of u-[13C, 15N]-PCB-Cph1Δ2 in the Pr (green) and Pfr (red) states at a magnetic field of 17.6 T, 8 kHz, and 243 K. Signals marked with an asterisk originate from the protein backbone.
Pr → Pfr Conversion in the Plant Phytochrome phyA.
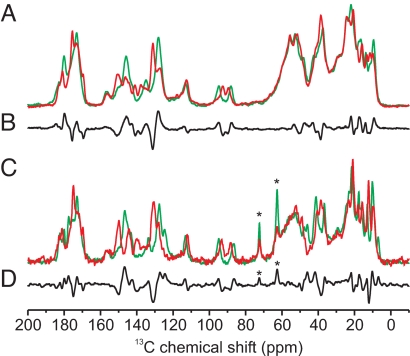
The 1D 13C CP/MAS NMR spectra of u-[13C, 15N]-PCB-Cph1Δ2 in the Pr (green) and Pfr (red) states are shown in Fig. 4A. The Pr-Pfr difference spectrum (Fig. 4B) reveals the changes of chemical shifts generated by the Pr → Pfr photoconversion. Positive signals represent the Pr state, and the negative signals arise from the Pfr state. In comparison, Fig. 4 C and D shows the corresponding spectra for the plant-derived phytochrome u-[13C, 15N]-PCB-phyA65. The Pr (green) and Pfr (red) states are displayed in Fig. 4C, and the Pr-Pfr difference spectrum is presented in Fig. 4D. The two difference spectra (Fig. 4 B and D) show far-reaching similarities. The positions of most of the resonances are almost unchanged, and the pattern of the relative intensities is the same for both species. The overall similarity of the spectra of Cph1Δ2 and phyA65 suggests that the chromophore geometry and the immediate protein environment are similar in both species. A small but clearly detectable difference, however, is noted in the spectral region around 125 ppm (see below).
Fig. 4.
One-dimensional 13C CP/MAS NMR spectra of u-[13C, 15N]-PCB in Cph1Δ2 and phyA65 in the Pr (A and C, green, respectively) and Pfr (A and C, red, respectively) states, and the Pr-Pfr difference spectra in Cph1Δ2 and phyA65 are shown in B and D, respectively. Positive signals represent Pr, and negative signals represent Pfr. The asterisks indicate the position of the glycol signals in natural abundance.
The 2D 13C-13C DARR NMR spectra of the Pr state and of the Pfr/Pr mixture (ratio ≈1:1) of phyA65 (see Fig. S6) lead to the 13C assignment in both Pr and Pfr states (Table S1). The change in chemical shift accompanying the Pr → Pfr conversion is shown in Fig. 2B. Also, the comparison of Fig. 2 A and B demonstrates that the phototransformation affects the chromophore in Cph1Δ2 and phyA65 in a similar manner. In phyA65, however, the positions 13 and 131 show a bigger change in chemical shift than in Cph1Δ2. This explains the difference observed at 125 ppm between the difference spectra (Fig. 4 B and D) and may suggest a different torsion angle around the 14–15 single bond in Cph1Δ2 and phyA65 phytochrome.
As shown by vibrational techniques, the conformations of the bilin chromophore in the Pr and Pfr states in native plant phytochrome, a dimer of two 124 kDa units (phyA124), resemble that in the photosensory modules phyA65 of oat phytochrome (28, 29) and Cph1 of Synechocystis (30). Furthermore, our NMR data add clear evidence that the chromophore and its interactions with the protein are conserved in both states throughout the Cph1/plant phytochrome family. Homology modeling of phyA65 and a comparison of its electrostatic surface properties with Cph1Δ2 (see Fig. S7) indicate that the sensory modules of both phytochromes provide a common electrostatic environment for the PCB chromophore despite significant differences along their solvent-exposed molecular surfaces. These striking similarities justify the following conclusions that may be drawn for the whole Cph1/plant phytochrome family.
Discussion
Chromophore Photoconversion.
As shown in Fig. 2 A and B, the photoconversion of PCB mostly affects rings C and D. The striking change in chemical shifts at the positions 13–19 is in line with photoisomerization occurring along the C15=C16 double bond. During this process, the chromophore is involved throughout its entire structure; hence, the photoisomerization is not a local event but modifies the chromophore–protein interaction dramatically. A significant effect is also seen at the 10-methine carbon that is situated between the almost coplanar rings B and C. In contrast, no major effect occurs around methine carbon 5 that links rings A and B. The observed pattern can be rationalized by the assumption of five effects: (i) The chromophore is tensely fixed in the Pfr state, (ii) the conjugation increases in the Pfr state, (iii) the hydrogen bonding interaction of the ring D carbonyl increases in the Pfr state, (iv) a local change of the electronic structure around ring C is identified, and (v) a significant change of the protein interacting with the ring C carboxylate group takes place.
(i) The loss of conformational heterogeneity at the ring B propionate side chain implies an increase of local tension. Several cross-peaks at rings A and B become sharper in the Pfr spectrum, e.g., carbons 4 and 6. The signal doubling observed for carbons 1 and 2 in the Pr state disappears in the Pfr state. These observations suggest an increase of mechanical tension occurring in the Pfr state, as is also observed by 15N MAS NMR (19) and vibrational spectroscopy (13). Such tension would also explain the change at the 31 carbon atom that links the chromophore via a thioether covalently to Cys-259.
(ii) The entire conjugation pattern undergoes a significant modification. In Fig. 2 on the left side of ring C, even-numbered carbons are marked for a downfield shift (blue), whereas odd-numbered carbons are up-shifted (red). On the right side of ring C, however, the opposite pattern appears. Such pattern change, involving the entire conjugated chain, implies a change in bond order. Assuming enhanced tension in the Pfr state, we would propose an increase of bond order for single bonds, increasing their rotational energy and providing the required stiffness, concomitant with a decrease in bond order for the double bonds.
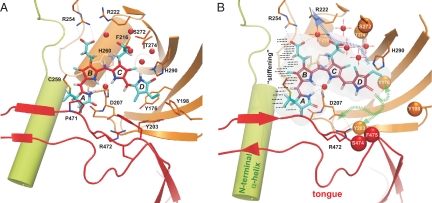
(iii) There is a clear up-shift at the carbonyl of ring D associated with decreased electron density at carbons 17 and 19. This shift has not been observed in previous NMR studies (31); however, a clear change of this group has been shown by FTIR spectroscopy (14, 28). In the Pr state of Cph1 (Fig. 5A), only a weak hydrogen bond is formed between the ring D carbonyl and His-260 due to its energetically unfavorable angle of 102° (O-HE2-NE2) caused by the low tilt of ring D vs. ring C (26.3°). Therefore, an increase of hydrogen-bonding interactions takes place for the ring D carbonyl during Pfr state formation. Such an increased polarization at the terminal group may cause the increased conjugation throughout the entire chain of the chromophore and hence the red shift of Pfr absorption. Because no strong hydrogen bonding is observed for the protonated ring D nitrogen, in neither the Pr nor the Pfr state, it is reasonable to assume that strong hydrogen bonding involving only the carbonyl of ring D stabilizes the chromophore in the Pfr state.
Fig. 5.
The PCB-binding site of Cph1Δ2. (A) Structural view on the protein environment of the PCB-chromophore in the Pr state (10) highlighting the H-bonding network. (B) Overview of putative structural changes caused by Pfr state formation. Blue and green dotted arrows indicate potential paths of signal transmission within the chromophore-binding site. The observed loss of conformational freedom along rings A and B is highlighted by dotted lines. This figure was made by using PyMol (32).
(iv) Whereas chemical shifts of carbons in rings B and C are almost mirror symmetrical in the Pr state, this symmetry is lost in the Pfr state. However, in the other rings, an alternating pattern along the conjugated chain occurs, only in ring C all carbons are labeled in red. This is indicative of an increase of electron density in the Pfr state at this ring. The interruption of the alternating pattern at ring C suggests that the origin of the change of the conjugation pattern is localized here. Conformational rearrangements of the nearby located residues His-260 or Tyr-176 (Fig. 5A) may contribute to a ring-current shift effect in the Pfr state; however, this alone cannot explain the interruption of the alternating pattern. It is possible that, in the Pr state, the conjugation is interrupted at or around ring C because of the interplanar tilt between rings C and D and that the observed alternating pattern is caused by the enlargement of the conjugated system extending beyond ring C. Such an effect could also explain the red-shift of the absorption spectrum upon generating the Pfr state. In any case, ring C appears to be the hotspot for the change of the electronic structure.
(v) At the propionate side chain of ring C, a significant change at the carboxyl group occurs. It can be assumed that this group faces strongly altered interactions with the protein environment. This change may correspond to a modification previously observed by FTIR spectroscopy (14, 28), which has been interpreted as an alteration of either a protein amide or a propionate carboxylate group. In contrast to the ring B propionate, the ring C propionate is well hydrated within a cluster of five water molecules and makes only an indirect interaction with the positive counter charge at Arg-222 via a water molecule (Fig. 5A). Interestingly, a structural comparison between Cph1 and the bacteriophytochromes shows that Arg-222 may either adopt an outward-oriented conformation toward the GAF–PAS interface or point into the core region of the GAF domain itself.
Hence, from our analysis, the following picture of the Pr → Pfr phototransformation emerges: The chromophore forms a strong hydrogen bond via its ring D carbonyl, increasing both the strength and length of the conjugation network and stabilizing the chromophore in a tensed shape (Fig. 5B). Observed changes in 15N chemical shifts in the terminal rings (ref. 19 and Fig. 3) may be explained by such tension linked to traction at the outer rings, leading to a banana-shaped cofactor in the Pfr state. In addition, these changes in 15N chemical shifts may also be linked to the modification of the conjugated system, leading to a flow of electron density from ring A to ring D.
Signal Transduction Pathway.
The question arises at which positions the chromophore dynamics is coupled to the protein environment to allow signal transduction to the protein surface. Important information can be extracted from Fig. 2 A and B. All changes occurring on the left side of the green line can be explained by a change of conjugation only. On the other hand, on the right side of the green line, multiple effects are overlaying, and two of them are clearly due to changed chromophore–protein interaction. There are significant changes localized at the carboxylate group of the propionate side chain of ring C as well as at the carbonyl group of ring D. The x-ray structures of Cph1 and the bacteriophytochromes show that His-290 (His-372 in phyA65 and His-299 in DrBphP) is bridged via two conserved water molecules to the carboxylate group of ring C and forms a hydrogen bond to the carbonyl of ring D (8–10) (Fig. 5). Hence, we assume that this highly conserved His-290 couples the photochemistry of the chromophore to the protein.
Because the 13C signal of the ring A carbonyl and the 1H signal of the ring A nitrogen exhibit only minor changes during the Pr → Pfr photoconversion, only local conformational changes of the protein matrix surroundings are likely to occur. On the other hand, changes of the hydrogen-bonding network by ring D photoisomerization could alter the conserved salt bridge between Asp-207 and Arg-472 and thus transmit the signal to the protein surface, e.g., by rearrangement of the tongue region (Fig. 5B). As possible conserved partners for strong hydrogen bonding to the ring D carbonyl in the Pfr state, nearby H-bonding donors such as Asp-207, Tyr-198, Tyr-203, Tyr-263, and Ser-474 may be considered. Further MAS NMR experiments will be necessary to elucidate such changes of the hydrogen-bonding network within the chromophore-binding site. In particular, MAS NMR distance experiments and selective interface detection spectroscopy (SIDY) (33) will be useful.
Materials and Methods
Sample Preparation for MAS NMR Spectroscopy.
The u-[13C, 15N]-PCB was prepared following published methods (27). Preparation of Cph1Δ2 and phyA65 apo- and holoproteins were performed as described (34, 35). For the measurements of the Pr state, samples were irradiated with light filtered through a far-red cut-off filter (λmax = 730 nm). Pfr/Pr mixture was produced by saturating irradiation at 660 nm by using appropriate LED (Roithner Lasertechnik, Vienna, Austria). Cph1Δ2 phytochrome in its pure Pfr state was obtained by size-exclusion chromatography using Superdex 200 (Amersham Pharmacia/GE) (36).
MAS NMR Spectroscopy.
The 1D 13C CP/MAS spectra were recorded by using a DMX-400 spectrometer, equipped with 4-mm CP/MAS probe. All data were recorded at 243 K with a spinning frequency of 10 kHz. The proton 90° pulse was set to 3.6 μs. The 1H power was ramped 80–100% during CP. During the data acquisition, the protons were decoupled from the carbons by use of the two-pulse phase-modulation (TPPM) decoupling scheme (37). For u-[13C, 15N]-PCB-Cph1Δ2 measurements in the Pr and Pfr states, ≈15 mg of protein were placed in a 4-mm zirconia rotor. Approximately 12 mg of u-[13C, 15N]-PCB-phyA65 was used for the measurements of the Pr state and Pfr/Pr (1:1) mixture.
All 2D 13C-13C DARR experiments were performed at a field of 17.6 T on an Avance-750 WB spectrometer, equipped with a 4-mm triple-resonance CP/MAS probe (Bruker). Typical 1H 90° and 13C 180° pulse lengths were set at 3.1 and 5.0 μs, respectively. Mixing times of 5 and 50 ms were used to maximize homonuclear recoupling between 13C nucleus. The 13C-1H dipolar interaction has been recovered by continuous wave irradiation on 1H radio frequency field intensity to satisfy the n = 1 condition (20). The 1H power was ramped 80–100% during CP. The 1H decoupling was ≈80 kHz TPPM during acquisition. The 2D 13C-13C spectra of Cph1Δ2 were recorded with 1,536 scans and with 8-ms evolution in the indirect dimension, leading to experimental times of 80 h. The spectra of the phyA65 protein were recorded with 2,048 scans in 2.5 days. The data were processed with the Topspin software version 2.0 (Bruker) and subsequently analyzed by using the program Sparky version 3.100 (T. D. Goddard and D. G. Kneller, University of California, San Francisco).
The 1D 15N CP/MAS spectra were recorded by using an AV-750 spectrometer, equipped with 4-mm CP/MAS probe. The proton 90° pulse was set to 3.1 μs, temperature was 243 K, and the spinning frequency was 8 kHz. The 1H-13C and 1H-15N heteronuclear experiments were performed at the DMX-400 and Avance-750 WB spectrometer, respectively, by using a frequency-switched Lee–Goldburg pulse sequence (38).
Supplementary Material
Acknowledgments.
K. Erkelens, F. Lefeber, and J. Hollander are gratefully acknowledged for support during various stages of the experiments. T.R. is very thankful to K. B. Sai Sankar Gupta for practical support and for helpful discussions about the DARR experiment. H. Steffen (MPI for Bioinorganic Chemistry in Mülheim, Germany) is thanked for the purification and preparation of the phyA65 sample. We thank Prof. Huub J. M. de Groot for continuous interest and support. This work was supported by Volkswagen-Stiftung Grant I/79979.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805696105/DCSupplemental.
References
- 1.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schäfer E, Nagy F, editors. Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms. 3rd Ed. Dordrecht, The Netherlands: Springer; 2006. [Google Scholar]
- 3.Hughes J, et al. A prokaryotic phytochrome. Nature. 1997;386:663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- 4.Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 5.Karniol B, Wagner JR, Walker JM, Vierstra RD. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem J. 2005;392:103–116. doi: 10.1042/BJ20050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montgomery BL, Lagarias JC. Phytochrome ancestry: Sensors of bilins and light. Trends Plant Sci. 2002;7:357–366. doi: 10.1016/s1360-1385(02)02304-x. [DOI] [PubMed] [Google Scholar]
- 7.Braslavsky SE, Gärtner W, Schaffner K. Phytochrome photoconversion. Plant Cell Environ. 1997;20:700–706. [Google Scholar]
- 8.Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature. 2005;438:325–331. doi: 10.1038/nature04118. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Stojkovic EA, Kuk J, Moffatt K. Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc Natl Acad Sci USA. 2007;104:12571–12576. doi: 10.1073/pnas.0701737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Essen LO, Maillet J, Hughes J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci USA. 2008;105:14709–14714. doi: 10.1073/pnas.0806477105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rüdiger W, Thümmler F, Cmiel E, Schneider S. Chromophore structure of the physiologically active form (Pfr) of phytochrome. Proc Natl Acad Sci USA. 1983;80:6244–6248. doi: 10.1073/pnas.80.20.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor SPA, Lagarias JC, Mathies RA. Resonance Raman analysis of the Pr and Pfr forms of phytochrome. Biochemistry. 1990;29:11141–11146. doi: 10.1021/bi00502a018. [DOI] [PubMed] [Google Scholar]
- 13.Matysik J, Hildebrandt P, Schlamann W, Braslavsky SE, Schaffner K. Fourier-transform resonance Raman-spectroscopy of intermediates of the phytochrome photocycle. Biochemistry. 1995;34:10497–10507. doi: 10.1021/bi00033a023. [DOI] [PubMed] [Google Scholar]
- 14.Foerstendorf H, Mummert E, Schäfer E, Scheer H, Siebert F. Fourier-transform infrared spectroscopy of phytochrome: Difference spectra of the intermediates of the photoreactions. Biochemistry. 1996;35:10793–10799. doi: 10.1021/bi960960r. [DOI] [PubMed] [Google Scholar]
- 15.Schwinté P, et al. FTIR study of the photoinduced processes of plant phytochrome phyA using isotope-labeled bilins and DFT calculations. Biophys J. 2008;15:1256–1267. doi: 10.1529/biophysj.108.131441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn J, et al. Probing protein-chromophore interactions in Cph1 phytochrome via mutagenesis. FEBS J. 2006;273:1415–1429. doi: 10.1111/j.1742-4658.2006.05164.x. [DOI] [PubMed] [Google Scholar]
- 17.Fischer AJ, Lagarias JC. Harnessing phytochrome's glowing potential. Proc Natl Acad Sci USA. 2004;101:17334–17339. doi: 10.1073/pnas.0407645101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heise H, et al. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohmer T, et al. N-15 MAS NMR studies of Cph1 phytochrome: Chromophore dynamics and intramolecular signal transduction. J Phys Chem B. 2006;110:20580–20585. doi: 10.1021/jp062454+. [DOI] [PubMed] [Google Scholar]
- 20.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 21.Schmidt P, et al. The complexity of the Pr to Pfr phototransformation kinetics is an intrinsic property of native phytochrome. Photochem Photobiol. 1998;68:754–761. [Google Scholar]
- 22.Sineshchekov VA. Extreme dehydration of plant tissues irreversibly converts the major and variable phyA′ into the minor and conserved phyA″. J Photochem Photobiol. 2006;85:85–91. doi: 10.1016/j.jphotobiol.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 23.von Stetten D, et al. Chromophore heterogeneity and photoconversion in phytochrome crystals and solution studied by resonance Raman spectroscopy. Angew Chem Int Ed. 2008;47:4753–4755. doi: 10.1002/anie.200705716. [DOI] [PubMed] [Google Scholar]
- 24.Hahn J, Kühne R, Schmieder P. Solution-state N-15 NMR spectroscopic study of α-C-phycocyanin: Implications for the structure of the chromophore-binding pocket of the cyanobacterial phytochrome Cph1. ChemBioChem. 2007;8:2249–2255. doi: 10.1002/cbic.200700256. [DOI] [PubMed] [Google Scholar]
- 25.Stanek M, Grubmayr K. Protonated 2,3-dihydrobilindiones—Models for the chromophores of phycocyanin and the red-absorbing form of phytochrome. Chem Eur J. 1998;4:1653–1659. [Google Scholar]
- 26.Wagner JR, Zhang JR, Brunzelle JS, Vierstra RD, Forest KT. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J Biol Chem. 2007;282:12298–12309. doi: 10.1074/jbc.M611824200. [DOI] [PubMed] [Google Scholar]
- 27.Strauss HM, Hughes J, Schmieder P. Heteronuclear solution-state NMR studies of the chromophore in cyanobacterial phytochrome Cph1. Biochemistry. 2005;44:8244–8250. doi: 10.1021/bi050457r. [DOI] [PubMed] [Google Scholar]
-
28.Foerstendorf H, et al. FTIR studies of phytochrome photoreactions reveal the C
 O bands of the chromophore: Consequences for its protonation states, conformation, and protein interaction. Biochemistry. 2001;40:14952–14959. doi: 10.1021/bi0156916. [DOI] [PubMed] [Google Scholar]
O bands of the chromophore: Consequences for its protonation states, conformation, and protein interaction. Biochemistry. 2001;40:14952–14959. doi: 10.1021/bi0156916. [DOI] [PubMed] [Google Scholar] - 29.Kneip C, et al. Effect of chromophore exchange on the resonance Raman spectra of recombinant phytochromes. FEBS Lett. 1997;414:23–26. doi: 10.1016/s0014-5793(97)00969-1. [DOI] [PubMed] [Google Scholar]
- 30.Foerstendorf H, Lamparter T, Hughes J, Gartner W, Siebert F. The photoreactions of recombinant phytochrome from the cyanobacterium Synechocystis: A low-temperature UV-Vis and FT-IR spectroscopic study. Photochem Photobiol. 2000;71:655–661. doi: 10.1562/0031-8655(2000)071<0655:tporpf>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 31.van Thor JJ, Mackeen M, Kuprov I, Dwek RA, Wormald MR. Chromophore structure in the photocycle of the cyanobacterial phytochrome Cph1. Biophys J. 2006;91:1811–1822. doi: 10.1529/biophysj.106.084335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. www.pymol.org. [Google Scholar]
- 33.Kiihne SR, et al. Selective interface detection: Mapping binding site contacts in membrane proteins by NMR spectroscopy. J Am Chem Soc. 2005;127:5734–5735. doi: 10.1021/ja045677r. [DOI] [PubMed] [Google Scholar]
- 34.Lamparter T, Esteban B, Hughes J. Phytochrome Cph1 from the cyanobacterium Synechocystis PCC6803—Purification, assembly, and quaternary structure. Eur J Biochem. 2001;268:4720–4730. doi: 10.1046/j.1432-1327.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- 35.Mozley D, Remberg A, Gärtner W. Large-scale generation of affinity-purified recombinant phytochrome chromopeptide. Photochem Photobiol. 1997;66:710–715. doi: 10.1111/j.1751-1097.1997.tb03211.x. [DOI] [PubMed] [Google Scholar]
- 36.Strauss HM, Schmieder P, Hughes J. Light-dependent dimerisation in the N-terminal sensory module of cyanobacterial phytochrome 1. FEBS Lett. 2005;579:3970–3974. doi: 10.1016/j.febslet.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 37.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 38.van Rossum BJ, Förster H, de Groot HJM. High-field and high-speed CP-MAS C-13 NMR heteronuclear dipolar-correlation spectroscopy of solids with frequency-switched Lee–Goldburg homonuclear decoupling. J Magn Res. 1997;124:516–519. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.