Abstract
Species-area relationships (SARs) are fundamental to the study of key and high-profile issues in conservation biology and are particularly widely used in establishing the broad patterns of biodiversity that underpin approaches to determining priority areas for biological conservation. Classically, the SAR has been argued in general to conform to a power-law relationship, and this form has been widely assumed in most applications in the field of conservation biology. Here, using nonlinear regressions within an information theoretical model selection framework, we included uncertainty regarding both model selection and parameter estimation in SAR modeling and conducted a global-scale analysis of the form of SARs for vascular plants and major vertebrate groups across 792 terrestrial ecoregions representing almost 97% of Earth's inhabited land. The results revealed a high level of uncertainty in model selection across biomes and taxa, and that the power-law model is clearly the most appropriate in only a minority of cases. Incorporating this uncertainty into a hotspots analysis using multimodel SARs led to the identification of a dramatically different set of global richness hotspots than when the power-law SAR was assumed. Our findings suggest that the results of analyses that assume a power-law model may be at severe odds with real ecological patterns, raising significant concerns for conservation priority-setting schemes and biogeographical studies.
Keywords: conservation biology, ecoregions, model selection, vascular plants, vertebrates
Species-area relationships (SARs), the change in species numbers with increasing area, are fundamental to the present understanding of many key and high-profile issues in conservation biology. They have, for example, variously been used to predict regional species extinction rates after habitat loss, as a consequence of such pressures as deforestation and climate change (1–4) and to predict species extinction rates in blocks of remnant habitat, including protected areas, as a consequence of their isolation (5). More fundamentally, the SAR is an essential tool used to estimate broad patterns and to identify hotspots of species richness when regions differ in area (6–13).
In the main, applications of SARs have assumed that these relationships take the classical form of a log-linearizable power function, S = cAz, where S is species richness, A is area, and c and z are constants (14). Depending on the objectives and opportunities, the parameters of this function (notably the exponent, or rate, z) are derived from theory (15–18), from particular datasets or from broad collations of datasets (19–22). However, although the power function has been applied extremely widely, in practice there is much variation in the basic form of SARs (23, 24). Attention has focused foremost on how this form changes with spatial scale (25–27) or assemblage properties (28). Other kinds of systematic variation may also exist, but analyses have principally only rather narrowly addressed these by comparing the parameter values estimated from fitting a power function relationship (e.g., space, refs. 21, 29, 30; environment, ref. 31; and anthropogenic threats, ref. 22).
Given that a single generic form for SARs is widely assumed to pertain, of particular concern for conservation biology would be if the underlying form actually differed markedly between major taxonomic groups and/or biomes (global-scale biogeographic regions distinguished by unique collections of ecosystems and species assemblages; ref. 32). Whether such variation was systematic, it could have significant implications particularly for the fundamental understanding of the distribution of biodiversity that underlies much of the prioritization of lands for conservation investment and action (33). For example, studies have variously sought to incorporate the effects of variation in area on species richness at large spatial scales (often ecoregions) when considering the concordance of spatial variation in richness of different higher taxa (13, 34), patterns of protected area coverage (35), the impacts of urbanization on biodiversity (36), and the allocation of conservation resources (37, 38).
In this article, we conduct an analysis of global-scale SARs with two aims. First, we investigate the uncertainty about the best-fitting SAR model by quantifying the relative probabilities that different models best describe SARs and determine whether those probabilities vary systematically for the same higher taxon in different biomes and for different higher taxa in the same biome. Second, we conduct a global identification of hotspots of richness, incorporating the uncertainty about the best-fit SAR model, and compare these results with those obtained when it is assumed that the power model is the best-fitting SAR model. We use data on the species richness of vascular plants and vertebrates across the world's terrestrial ecoregions (13, 39) [supporting information (SI) Text and Table S1]. Ecoregions are large units of land containing geographically distinct species assemblages and experiencing geographically distinct environmental conditions and have proven valuable for addressing a range of issues in conservation prioritization (13, 40, 41).
Results
Taxonomic and Regional Uncertainty in Species-Area Relationships.
The relative fit of eight different potential forms for SARs (Table S2) was evaluated for each combination of higher taxon and biome. These forms encompassed convex, sigmoid, asymptotic, and nonasymptotic models, with the fit being evaluated using nonlinear regressions in the so-called model selection framework (42). This emerging approach in the context of SARs (43) aims to evaluate, for a given dataset, the strength of evidence for alternative explanatory models (44). Furthermore, by averaging across statistically valid models, this framework allows the construction of robust inferences incorporating uncertainty regarding both model selection and parameter estimation (multimodel SARs; see Materials and Methods for details).
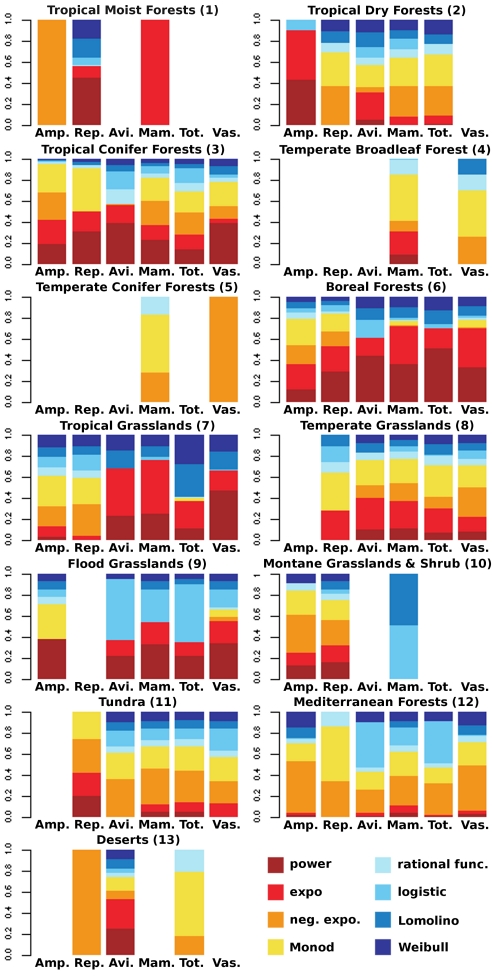
Surprisingly, given the apparent generality of the SAR, the analysis revealed substantial variation in the strength of the effect of area on species richness. Although the R2 for multimodel SARs had an overall mean of 0.30, values for different combinations of higher taxa and biomes ranged from 0.02 for amphibians in Tropical Dry Forests to 0.69 for total vertebrates in Tropical Grasslands (Table S3). Furthermore, for several datasets (21 of 78), the SAR cannot be adequately described by any of the candidate models (Fig. 1, Table S4). This latter tendency was not limited to those datasets with narrower ranges of variation in species richness or area but is more obvious for biomes than for higher taxa. For example, SARs were statistically validated across temperate forest ecoregions only for mammals and vascular plants.
Fig. 1.
SAR model selection patterns. Patterns of model selection are presented for each biome for amphibians (Amp.), reptiles (Rep.), birds (Avi.), mammals (Mam.), total vertebrates (Tot.), and vascular plants (Vas.). The height of each fraction of the colored band is proportional to the probability (Akaike weight) that each model [see color legend, exponential (expo.), negative exponential (neg. expo.), rational function (rational func.)] is the best in explaining the dataset. A lack of colored band means that none of the eight SAR models was statistically valid for the corresponding dataset.
The best-fitting model varied markedly across biomes for all higher taxa and across higher taxa for each biome (Fig. 1, Table S4). It was the asymptotic negative exponential (convex) and the Monod (convex) models in 18 and 13 cases, respectively, the nonasymptotic power and exponential models in 10 cases each, and the logistic and Lomolino models in five and one case, respectively. The rational function and the cumulative Weibull models never provided the best fit. However, with the exception of four datasets (amphibians and mammals in Tropical and Subtropical Moist Broadleaf Forests, vascular plants in Temperate Conifer Forests, and reptiles in Deserts), there was a substantial degree of uncertainty about the best-fitting SAR model (Fig. 1, Table S4). For most of the datasets, no single model was clearly superior.
Furthermore, for almost all higher taxa, model probabilities differed markedly across biomes (Fig. 1, Table S4). Although for almost all biomes, model probabilities also differed markedly across higher taxa (Fig. 1, Table S4), summing these probabilities across the different models revealed some coarse tendencies. Indeed, for Boreal Forests, except for amphibians, the sum of the probabilities of nonasymptotic models that best describe the SAR was always >0.5. In contrast, for the Tundra and Mediterranean Forests, the SAR was likely to be asymptotic for most higher taxa (Fig. 1, Table S4).
Hotspot Detection.
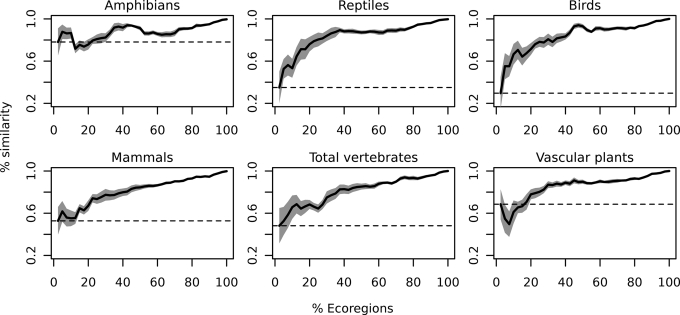
Using multimodel SARs could result in a rather different set of species richness hotspots being recognized than was the case when it was assumed that the power model was the best-fitting SAR model (Fig. 2). For example, using the frequent (but arbitrary) cut-off of distinguishing the richest 2.5% of ecoregions as hotspots, between 30% (birds) and 78% (amphibians) of the hotspots identified by the two approaches were the same. Inevitably, the similarity in the composition of the hotspots increased as the cut-off was increased, but it remained quite variable even when this cut-off was rather high (Fig. 2).
Fig. 2.
Relationship between the criterion used to define hotspots (% of ecoregions) and the similarity between hotspots identified assuming a power SAR and those identified when using multimodel SARs. The percentage similarity among the two methods was determined as the number of ecoregions identified as hotspots by both, divided by the total number of ecoregions in a group. For example, the highest 2.5% of ranks in a dataset consisting of 200 ecoregions comprises five ecoregions. If three ecoregions occur in the two groups, then the percentage similarity among the two methods is 60%. Dark plain line represents mean percentage similarity averaged across biomes for each higher taxon, gray polygon is the associated standard error of the mean, and dashed horizontal line indicates the percentage similarity at the 2.5% cut-off.
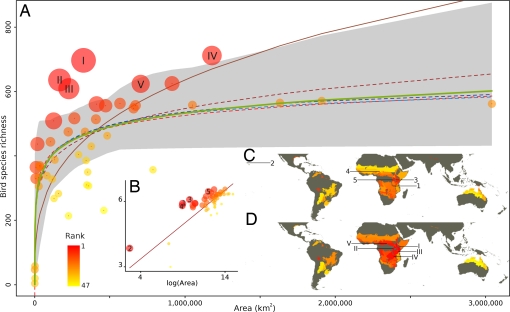
The differences in the hotspots recognized were especially marked when focusing on particular combinations of higher taxa and biomes. In one of many possible examples, for birds in Tropical Grasslands, the five richest ecoregions (approximately the richest 10%) were, with one exception, entirely different when determined using multimodel SARs and when using a power model (Fig. 3, Table 1).
Fig. 3.
Ecoregions of Tropical grasslands, birds SAR, and richness hotspots maps. SAR for the birds of Tropical grasslands (A and B) and maps of ecoregion ranks according to bird species richness in Tropical grasslands (C and D). (A and D) Nonlinear multimodel analysis; dashed lines are the fitted model predictions (brown: power, red: exponential, light-blue: Lomolino, dark-blue: Weibull), and the green solid curve is the result of model averaging, gray shading is the nonparametric bootstrap confidence interval used to rank ecoregions (see Materials and Methods) and the brown solid curve is a log linear fit on the arithmetic scale (B). (B and C) Log-linear power analysis. On all subplots the color of an ecoregion (A, B: points; C, D: regions) represents a rank (see color chart) according to the corresponding analysis. On subplots A and B, the size of a point is inversely proportional to its rank according to the corresponding analysis. On all subplots, the five richest ecoregions (corresponding to an ≈10% cutoff of higher rank hotspot criterion) are presented (A and D: Roman numerals, B and C: Arabic numerals). Ecoregions are Itigi–Sumbu thicket (1), Northwestern Hawaii scrub (2), Serengeti volcanic grasslands (3), Mandara Plateau mosaic (4), Victoria Basin forest-savanna mosaic (5, II), Northern Acacia-Commiphora bushlands and thickets (I), Southern Acacia-Commiphora bushlands and thickets (III), Central Zambezian Miombo woodlands (IV), and Northern Congolian forest-savanna mosaic (V).
Table 1.
Five leading bird richness hotspot ecoregions of Tropical grasslands
| Rank | Ecoregion names | Area, km2 | Bird species richness | Rank | Ecoregion names | Area, km2 | Bird species richness |
|---|---|---|---|---|---|---|---|
| 1 | Itigi-Sumbu thicket | 7,809.2 | 365 | I | Northern Acacia-Commiphora bushlands and thickets | 324,481.6 | 697 |
| 2 | Northwestern Hawaii scrub | 14.7 | 47 | II | Victoria Basin forest-savanna mosaic | 165,041.8 | 636 |
| 3 | Serengeti volcanic grasslands | 17,947.6 | 437 | III | Southern Acacia-Commiphora bushlands and thickets | 226,769.7 | 609 |
| 4 | Mandara Plateau mosaic | 7,478.5 | 330 | IV | Central Zambezian Miombo woodlands | 1,179,319.1 | 712 |
| 5 | Victoria Basin forest-savanna mosaic | 165,041.8 | 636 | V | Northern Congolian forest-savanna mosaic | 705,005.8 | 624 |
| Totals | 198,291.8 | 1,815 | Totals | 2,600,618 | 3,278 |
Discussion
Although it has long been apparent that the assumption of a single generic form for SARs was potentially problematic (19, 23, 43), the practice has remained widespread. In part, this has been because of the understandable demand to address important, and often urgent, conservation issues in circumstances for which information on the actual form of SARs is wanting and difficult to obtain. The results of the analyses reported here highlight several key issues that result from such an approach.
First, the assumption that SARs follow a single generic form overlooks the fact that the effect of area on species richness can differ dramatically among datasets. At one extreme, the majority of variation in richness can be explained by area, and individual models can provide excellent fits; and, at the other extreme, no single model may adequately describe the relationship between species richness and area (19, 43). Although such lacks of fit have been reported (43), our study highlights that this circumstance may not always be a rare one, pertaining in 27% of the cases (Fig. 1, Table S4, combinations of higher taxon and biome) that we studied, despite our using a particularly wide range of possible models (embracing most forms that have been discussed in the literature).
Second, where one or more of the models tested did fit datasets, that which fit best was extremely variable (Fig. 1, Table S4); a power model was the best fit in only 10 of 57 cases. Although it has been suggested that the most appropriate model may depend on scale and the nature of the organisms or of the environment (19, 22, 23, 43, 45), no simple tendencies in these regards seem to emerge from our analyses. Indeed, all of the different shapes of SARs represented by the set of models used (convex, sigmoid, asymptotic, and nonasymptotic) were selected at least once for the different datasets. This suggests that none of a wide range of potential SAR models can a priori be ignored, and that a universal model does not emerge. The applied implications of this observation could be further complicated if, as some have suggested, the form of SARs can be influenced by human activities, although thus far this influence has principally been explored in terms of power models (22, 46).
Third, where more than one of the models tested fitted a dataset, there was often substantial uncertainty as to which of these provided the best fit (Fig. 1, Table S4). Again this highlights the importance of considering multiple models when making inferences about SARs. It also draws attention to the need to remember there is commonly substantial spatial variation in species richness not attributable to variation in area, even when, as here, comparisons are constrained to the same biome. This said, summing the probabilities across the different types of models allowed us to infer coarse tendencies about the shape of SARs. Globally, intrabiome SARs are more likely to be convex than sigmoid (mean summed probabilities: 0.7 vs. 0.3 ± 0.18) and more likely to be asymptotic than nonasymptotic (mean summed probabilities: 0.68 vs. 0.32 ± 0.26); this does not imply that species richness generally tends actually to saturate when areas are large (Fig. 1, ref. 47, and Table S4). This suggests the possibility that there may be some general patterns in the circumstances under which different kinds of models tend to prevail. Extensive metaanalyses of large numbers of datasets (with a wide range of average area sizes), and building on the approaches developed here, could be used to explore this issue to obtain more definitive conclusions.
Finally, given the above, assuming that a power model is the most appropriate description of SARs can make a substantial difference to the outcome of analyses and the conservation recommendations that may follow (6, 12). Certainly, a rather different set of hotspots would be identified than is the case when alternative models are considered (Figs. 2 and 3). Moreover, there will tend to be systematic biases in these hotspots. For example, in the case of birds in Tropical grasslands (Fig. 3, Table 1), those hotspots recognized using a linearized power model tend to be smaller than when using multimodel SARs (summed areas of hotspots using power model is 198,292 km2 and incorporating uncertainty is 2,600,618 km2, Table 1). We anticipate that such variation in outcomes will be very common, and that the conclusions of a number of studies of the distribution of species richness and its consequences for conservation prioritization will need to be revisited to ascertain their sensitivity to assumptions about the underlying form of SARs.
In conclusion, we recommend that, particularly in the context of studies whose outcomes may be of significance for conservation decision making, (i) in empirical analyses involving SARs, the relative fit of different models is examined, and uncertainty in this fit is accounted for; and (ii) in more theoretical studies involving SARs, the consequences of assuming different underlying forms of these relationships are examined. Failing to do so may well lead to conclusions at odds with real patterns of spatial variation in species richness, as exemplified in the identification of hotspots of richness among areas of differing size.
Materials and Methods
Data.
Analyses were based on the numbers of species of vascular plants, amphibians, reptiles, birds, and mammals in each terrestrial ecoregion of the world as delimited by Olson et al. (32). Data were obtained on vertebrates by overlaying range maps of extant species compiled from numerous scientific works, field guides, or directly from experts (32), and on vascular plants from published and unpublished richness data and from a variety of additional information (39). Following Lamoreux et al. (13), we excluded Mangrove ecoregions and large uninhabited parts of Greenland and Antarctica because of lack of data reliability or availability. The resulting database contains 78 datasets (combinations across 13 biomes and 6 taxonomic groups) and covers 792 ecoregions that represent 96.3% of Earth's inhabited land, making our analysis a good descriptor of global distribution patterns (Table S1).
Statistical Analyses.
All statistical analyses conducted in this study were implemented within the R statistical programming environment (R 2.7, ref. 48).
Toward Consensual Inference.
We discriminated the different SAR models in the so-called model selection framework (42, 49), which is now widely used across biological fields (44, 50–52). Through the use of information theoretic criteria such as the Akaike Information Criterion (AIC, ref. 42), it provides a rigorous way in which to evaluate and compare the relative support of nonnested differently parameterized models of a given dataset. In this study, we use Akaike weights derived from the AIC to evaluate the relative likelihood of each SAR model given the data and the set of models. Akaike weights (normalized by construction across the set of candidate models to sum to one) are directly interpreted in terms of probabilities of a given model being the best of a defined set of alternative models in explaining the data (42, 50).
In the model selection framework, model selection uncertainty arises when the data at hand support several models with a similar strength. In such a case, relying on only the best model is inadequate, and multimodel inference is recommended as a way to construct a robust final inference (42). As advocated for differently parameterized models, we use model averaging and consider the weighted average of model predictions with respect to model weights.
One of the most important challenges in information theoretic analyses is the construction of a consistent set of models (42, 52). Here, we propose a set (Table S2), including four convex models (power, exponential, negative exponential, and Monod) and four sigmoidal models (rational function, logistic, Lomolino, and cumulative Weibull). This includes convex, sigmoid, asymptotic, and nonasymptotic functions, thus encompassing the various shapes attributed to SARs in the literature. The linearized forms (via logarithmic transformations) of the power and exponential models were not included in the set because of nonequivalence in the study of the variation in a variable and in its transformation (23, 53) and bias of back-transformed results obtained on a logarithmic scale (54). Furthermore, the nonlinear form of the power equation leads to a more realistic detection of biodiversity hotspots than does the log-linearized power equation (54).
AIC and other model selection criteria that estimate Kullback–Leibler information (see SI Materials and Methods) are used widely in the ecological literature, but other criteria such as the Bayesian Information Criterion (BIC) are also commonly used to carry out model selection (42, 50). AIC and BIC were not derived in similar contexts [AIC is based on the Kullback–Leibler information theory, whereas BIC was derived in a Bayesian context (42, 50)] and have different properties: AIC aims to select the best model approximating reality given the sample size and the set of models, whereas BIC was devised to select the true model that generates the data independently of sample size and given that this true model is one of the candidate models. Although AIC and BIC do not share the same conceptual bases and penalize differently for the dimension of the models (BIC tends to select models with fewer parameters than AIC), the results of our analyses were robust to the criterion used for model selection and averaging. Using the BIC, the model ranks were globally maintained across the datasets, and the substantial uncertainty revealed by the AIC analysis persists (Fig. S1).
Fitting the Models.
Nonlinear regression models were fitted by minimizing the residual sum of squares (RSS) using the unconstrained Nelder–Mead optimization algorithm (55). Assuming normality of the observations, this approach produces optimal maximum likelihood estimates of model parameters (56). Regressions were evaluated by statistical examination of normality and homoscedasticity of residuals: a model was excluded from final averaging if the Lilliefors extension of the Kolmogorov normality test or the Pearson's product moment correlation coefficient with areas was significant at the 5% level. To avoid numerical problems, such as local minima, and speed up the convergence process, we paid particular attention to the starting values that were used to run the optimization algorithm. We obtained initial values for those parameters that were directly interpretable (e.g., an asymptote) by taking corresponding values in the datasets (e.g., the observed maximum of species richness in the case of an asymptote) and calculated initial values for the remaining parameters using the standard procedures of Ratkowsky (57, 58). Although the selection of nonlinear regression models through the use of the coefficient of determination (R2) is not advocated (53, 57), these indices were useful indicators of the proportion of variation in intrabiome species richness explained by area.
Confidence Intervals and Ecoregion Ranking.
By synthesizing and extending recent advances and solving major concerns about the methodology of hotspot detection (6–9, 11, 12, 54), ecoregions were ranked with respect to their positions in the confidence interval of the model-averaged SAR (Fig. 3, SI Materials and Methods). To fully incorporate uncertainty in this process, confidence intervals were calculated by using a nonparametric bootstrapping procedure (59, 60). As advocated for regression (59, 61), we generated bootstrap resamples from the modified residuals (in the sense of ref. 60), and we applied the model selection and averaging procedure to each of these resamples. In so doing, we generated robust confidence intervals explicitly incorporating uncertainty regarding both model selection and parameter estimation.
Comparison of Hotspot Detection Methods.
To investigate the effect of accounting for uncertainty in richness comparison among places of varying area, we assessed the similarity between the ranking obtained from our approach and that obtained from usual methods (e.g., ref. 13). Classical methods rank regions according to their residuals in a log-linear power regression: the higher the residual, the higher the region in the ranking. The percentage similarity was defined as the number of ecoregions identified as hotspots by the two methods, divided by the total number of ecoregions in a set of hotspots (6). For all higher taxa studied and for a varying proportion of ecoregions identified as hotspots, the percentage similarity between the two methods was averaged across the fitted biomes.
Supplementary Material
Acknowledgments.
We thank S. Buckland, K. L. Evans, L. Marini, and three anonymous reviewers for helpful comments and/or discussions. K.J.G. holds a Royal Society-Wolfson Research Merit Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.P.H.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803610105/DCSupplemental.
References
- 1.Cowlishaw G. Predicting the pattern of decline of African primate diversity, an extinction debt from historical deforestation. Conserv Biol. 1999;13:1183–1193. [Google Scholar]
- 2.Brooks TM, et al. Habitat loss and extinction in the hotspots of biodiversity. Conserv Biol. 2002;16:909–923. [Google Scholar]
- 3.Thomas CD, et al. Extinction risk from climate change. Nature. 2004;427:145–148. doi: 10.1038/nature02121. [DOI] [PubMed] [Google Scholar]
- 4.Malcolm JR, Liu CR, Neilson RP, Hansen L, Hannah L. Global warming and extinctions of endemic species from biodiversity hotspots. Conserv Biol. 2006;20:538–548. doi: 10.1111/j.1523-1739.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- 5.Báldi A, Vörös J. Extinction debt of Hungarian reserves: A historical perspective. Basic Appl Ecol. 2006;7:289–295. [Google Scholar]
- 6.Veech JA. Choice of species-area function affects identification of hotspots. Conserv Biol. 2000;14:140–147. [Google Scholar]
- 7.Brummitt N, Lughadha E. Biodiversity: Where's hot and where's not. Conserv Biol. 2003;17:1448. [Google Scholar]
- 8.Hobohm C. Characterization and ranking of biodiversity hotspots: Centres of species richness and endemism. Biodivers Conserv. 2003;12:279–287. [Google Scholar]
- 9.Ovadia O. Ranking hotspots of varying sizes : A lesson from the nonlinearity of the species-area relationship. Conserv Biol. 2003;17:1441. [Google Scholar]
- 10.Mutke J, Barthlott W. Patterns of vascular plant diversity at continental to global scales. Biol Skr. 2005;55:521–531. [Google Scholar]
- 11.Werner U, Buszko J. Detecting biodiversity hotspots using species-area and endemics-area relationships: The case of butterflies. Biodivers Conserv. 2005;14:1977–1988. [Google Scholar]
- 12.Fattorini S. Detecting biodiversity hotspots by species-area relationships: A case study of Mediterranean beetles. Conserv Biol. 2006;20:1169–1180. doi: 10.1111/j.1523-1739.2006.00418.x. [DOI] [PubMed] [Google Scholar]
- 13.Lamoreux JF, et al. Global tests of biodiversity concordance and the importance of endemism. Nature. 2006;440:212–214. doi: 10.1038/nature04291. [DOI] [PubMed] [Google Scholar]
- 14.Arhennius O. Species and area. J Ecol. 1921;9:95–99. [Google Scholar]
- 15.May RM. In: Ecology and Evolution of Communities. Cody ML, Diamond J, editors. Cambridge, MA: Harvard Univ Press; 1975. pp. 81–120. [Google Scholar]
- 16.Sugihara G. Minimal community structure: An explanation of species abundance patterns. Am Nat. 1980;116:770–787. doi: 10.1086/283669. [DOI] [PubMed] [Google Scholar]
- 17.Martín HG, Goldenfeld N. On the origin and robustness of power-law species−area relationships in ecology. Proc Natl Acad Sci USA. 2006;103:10310–10315. doi: 10.1073/pnas.0510605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Southwood TRE, May RM, Sugihara G. Observations on related ecological exponents. Proc Natl Acad Sci USA. 2006;103:6931–6933. doi: 10.1073/pnas.0600988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connor EF, McCoy ED. The statistics and biology of the species-area relationship. Am Nat. 1979;113:791–833. [Google Scholar]
- 20.Williamson M. In: Analytical Biogeography. Myers AA, Giller PS, editors. London: Chapman & Hall; 1988. pp. 91–115. [Google Scholar]
- 21.Drakare S, Lennon JJ, Hillebrand H. The imprint of the geographical, evolutionary and ecological context on species-area relationships. Ecol Lett. 2006;9:215–227. doi: 10.1111/j.1461-0248.2005.00848.x. [DOI] [PubMed] [Google Scholar]
- 22.Tittensor DP, Micheli F, Nyström M, Worm B. Human impacts on the species-area relationship in reef fish assemblages. Ecol Lett. 2007;10:760–772. doi: 10.1111/j.1461-0248.2007.01076.x. [DOI] [PubMed] [Google Scholar]
- 23.He F, Legendre P. On species-area relations. Am Nat. 1996;148:719–737. [Google Scholar]
- 24.Tjørve E. Shapes and functions of species-area curves: a review of possible models. J Biogeogr. 2003;30:827–835. [Google Scholar]
- 25.Palmer MW, White PS. Scale dependence and the species-area relationship. Am Nat. 1994;144:717–740. [Google Scholar]
- 26.Crawley MJ, Harral JE. Scale dependence in plant biodiversity. Science. 2001;291:864–868. doi: 10.1126/science.291.5505.864. [DOI] [PubMed] [Google Scholar]
- 27.Rosenzweig ML. Species Diversity in Space and Time. Cambridge, UK: Cambridge Univ Press; 1995. [Google Scholar]
- 28.He F, Legendre P. Species diversity patterns derived from species-area models. Ecology. 2002;83:1185–1198. [Google Scholar]
- 29.Lennon JJ, Koleff P, Greenwood JJD, Gaston KJ. The geographical structure of British bird distributions: Diversity, spatial turnover and scale. J Anim Ecol. 2001;70:966–979. [Google Scholar]
- 30.Lennon JJ, Kunin WE, Hartley S, Gaston KJ. In: Scaling Biodiversity. Storch D, Marquet PA, Brown JH, editors. Cambridge, UK: Cambridge Univ Press; 2007. pp. 51–76. [Google Scholar]
- 31.Evans KL, Lennon JJ, Gaston KJ. Slopes of avian species-area relationships, human population density, and environmental factors. Avian Conserv Ecol. 2007;2:7. [Google Scholar]
- 32.Olson DM, et al. Terrestrial ecoregions of the world: A new map of life on Earth. Bioscience. 2001;51:933–938. [Google Scholar]
- 33.Margules CR, Pressey RL. Systematic conservation planning. Nature. 2000;405:243–253. doi: 10.1038/35012251. [DOI] [PubMed] [Google Scholar]
- 34.Qian H, Ricklefs RE. Global concordance in diversity patterns of vascular plants and terrestrial vertebrates. Ecol Lett. 2008;11:547–553. doi: 10.1111/j.1461-0248.2008.01168.x. [DOI] [PubMed] [Google Scholar]
- 35.Loucks C, Ricketts TH, Naidoo R, Lamoreux JF, Hoekstra J. Explaining the global pattern of protected area coverage: Relative importance of vertebrate biodiversity, human activities and agricultural suitability. J. Biogeogr. 2008;35:1337–1348. [Google Scholar]
- 36.McDonald RI, Kareiva P, Forman RTT. The implications of current and future urbanization for global protected areas and biodiversity conservation. Biol Conserv. 2008;141:1695–1703. [Google Scholar]
- 37.Wilson KA, et al. Conserving biodiversity efficiently: what to do, where, and when. PloS Biol. 2007;5:e223. doi: 10.1371/journal.pbio.0050223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Underwood EC, et al. Protecting Biodiversity when Money Matters: Maximizing Return on Investment. PLoS ONE. 2008;3:e1515. doi: 10.1371/journal.pone.0001515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kier G, et al. Global patterns of plant diversity and floristic knowledge. J Biogeogr. 2005;32:1107–1116. [Google Scholar]
- 40.Hoekstra JM, Boucher TM, Ricketts TH, Roberts C. Confronting a biome crisis: Global disparities of habitat loss and protection. Ecol Lett. 2005;8:23–29. [Google Scholar]
- 41.McKnight MW, et al. Putting beta-diversity on the map: broad-scale congruence and coincidence in the extremes. PloS Biol. 2007;5:e272. doi: 10.1371/journal.pbio.0050272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. 2nd Ed. New York: Springer; 2002. [Google Scholar]
- 43.Stiles A, Scheiner SM. Evaluation of species-area functions using Sonoran Desert plant data: Not all species-area curves are power functions. Oikos. 2007;116:1930–1940. [Google Scholar]
- 44.Hobbs NT, Hilborn R. Alternatives to statistical testing in ecology: a guide to self teaching. Ecol Appl. 2006;16:5–19. doi: 10.1890/04-0645. [DOI] [PubMed] [Google Scholar]
- 45.Flather CH. Fitting species−accumulation functions and assessing regional land use impacts on avian diversity. J Biogeogr. 1996;23:155–168. [Google Scholar]
- 46.Gaston KJ. Biodiversity and extinction: Macroecological patterns and people. Prog Phys Geogr. 2006;30:258–269. [Google Scholar]
- 47.Williamson M, Gaston KJ, Lonsdale WM. The species-area relationship does not have an asymptote! J Biogeogr. 2001;28:827–830. [Google Scholar]
- 48.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 49.Kadane JB, Lazar NA. Methods and criteria for model selection. J Am Stat Assoc. 2004;99:279–290. [Google Scholar]
- 50.Johnson JB, Omland KS. Model selection in ecology and evolution. Trends Ecol Evol. 2004;19:101–108. doi: 10.1016/j.tree.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 51.Fidler F, Burgman MA, Cumming G, Buttrose R, Thomason N. Impact of criticism of null-hypothesis significance testing on statistical reporting practices in conservation biology. Conserv Biol. 2006;20:1539–1544. doi: 10.1111/j.1523-1739.2006.00525.x. [DOI] [PubMed] [Google Scholar]
- 52.Stephens PA, Buskirk SW, del Rio CM. Inference in ecology and evolution. Trends Ecol Evol. 2007;22:192–197. doi: 10.1016/j.tree.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Gitay H, Roxburgh SH, Wilson JB. Species-area relations in a New Zealand tussock grassland, with implications for nature reserve design and for community structure. J Veg Sci. 1991;2:113–118. [Google Scholar]
- 54.Fattorini S. To fit or not to fit? A poorly fitting procedure produces inconsistent results when the species-area relationship is used to locate hotspots. Biodivers Conserv. 2007;16:2531–2538. [Google Scholar]
- 55.Dennis JE, Schnabel RB. Classics Appl Math. Vol. 16. Englewood Cliffs, NJ: Prentice–Hall; 1983. Numerical methods for unconstrained optimization an nonlinear equations. [Google Scholar]
- 56.Rao CR. Linear Statistical Inference and Its Applications. New York: Wiley; 1973. [Google Scholar]
- 57.Ratkowsky DA. Nonlinear Regression Modelling. A Unified Practical Approach. New York: Dekker; 1983. [Google Scholar]
- 58.Ratkowsky DA. Handbook of Nonlinear Regression Models. New York: Dekker; 1990. [Google Scholar]
- 59.Buckland ST, Burnham KP, Augustin NH. Model selection: An integral part of inference. Biometrics. 1997;53:603–618. [Google Scholar]
- 60.Davison AC, Hinkley DV. Bootstrap Methods and Their Application. Cambridge, UK: Cambridge Univ Press; 1997. [Google Scholar]
- 61.Efron B. Bootstrap methods: Another look at the jackknife. Ann Stat. 1979;7:1–26. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.