Abstract
The term “C-value paradox” was coined by C. A. Thomas, Jr. in 1971 [Thomas CA (1971) Ann Rev Genetics 5:237–256] to describe the initially puzzling lack of correlation between an organism's genome size and its morphological complexity. Polyploidy and the expansion of repetitive DNA, primarily transposable elements, are two mechanisms that have since been found to account for this differential. While the inactivation of retrotransposons by methylation and their removal from the genome by illegitimate recombination have been well documented, the cause of the apparently periodic bursts of retrotranposon expansion is as yet unknown. We show that the expansion of the CRM1 retrotransposon subfamily in the ancient allotetraploid crop plant corn is linked to the repeated formation of novel recombinant elements derived from two parental retrotransposon genotypes, which may have been brought together during the hybridization of two sympatric species that make up the present day corn genome, thus revealing a unique mechanism linking polyploidy and retrotransposition.
Keywords: centromere, corn, genome expansion, chromodomain, polyploidy
Many of the world's crop plants are polyploids. Corn, the most widely grown food crop in the United States, is known to have an allotetraploid origin not later than 4.8 million years ago (MYA) (1, 2) from two parental genomes that had diverged from each other ≈11.9 MYA (2). This tetraploidization event was followed by a major genome expansion mediated by retrotransposons (3), the cause of which is unknown.
Lineage-specific retrotransposon amplification has been documented in several plant genomes (4–6), and appears to result from temporary relief of otherwise tight suppression of transcription. Retrotransposon transcription has been shown to be induced by tissue culture (7), microbial elicitors of plant defense responses (8), and polyploidization (9), but this temporary increase in retrotransposon transcription has not been shown to effect genome expansion of the magnitude observed in many crop plants.
In contrast, the forces counteracting genome expansion are fairly well understood and documented, and are known to include recombination between long terminal repeats (LTRs) of a single element (10, 11) or adjacent elements (10, 12), leading to solo LTRs or complex hybrid retrotransposon arrangements, and illegitimate recombinations, which result in deletions between very short regions of sequence homology on the same DNA strand (10). These factors contribute to element inactivation and removal and limit the estimated half-life of rice retrotransposons to <6 million years (13). Genome expansion requires that retrotransposition rates be sustained at levels higher than element removal rates. How retrotransposons accomplish this has thus far been completely unknown (14).
Centromeric retrotransposons (CR) comprise a family of elements that show strong preference for integration at active centromeres (15, 16). Two CR subfamilies have been recognized in maize (CRM1 and CRM2) and rice (CRR1 and CRR2) for some time (17–19). Four additional subfamilies were recently discovered (CRM3, CRM4, CRR3 and CRR4), and orthologous relationships between the maize and rice subfamilies were established (CRM1-CRR3, CRM2-CRR2, CRM3-CRR1, CRM4-CRR4) (20). Furthermore, a remarkable expansion of the CRM1 subfamily of elements during the past 3 to 4 million years was documented, but no plausible mechanism was provided (20). Notably, although the CRM1 subfamily accounts for >70% of the full-length CRM elements characterized thus far in the corn inbred B73, its rice ortholog (CRR3) is absent from the Oryza sativa ssp. japonica genome (20). Here we show, through detailed sequence analysis of all full-length CRM1 elements found thus far in the B73 genome, that CRM1 deletion derivatives and recombinant elements are active at different evolutionary times, deduce their evolutionary relationships, and propose a mechanism by which these recombinants are formed.
Results and Discussion
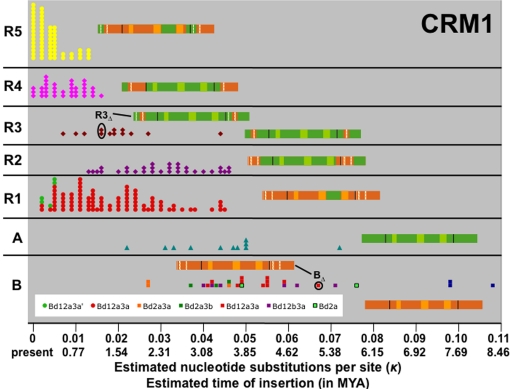
Analysis of 264 full-length CRM1 elements with target-site duplications (TSDs) that were identified in 13,918 sequenced BACs representing 2,256,428,478 nucleotides (nt) of the maize inbred B73 genome, revealed two parental variants (CRM1A and CRM1B) that have given rise to a number of deletion and recombination derivatives in the past 3 to 4 million years. The recombinant nature of all five major derivatives (R1 to R5) is illustrated by SimPlot graphs [supporting information (SI) Fig. S1]. The Maximum Chi Squared Test was used to map the precise recombination breakpoints, which in all cases mapped to the region in which SimPlots show a change in parental allele (Fig. S2 a–d). Fig. 1 provides an overview of all full-length CRM1 elements arranged by insertion time estimated for each element using the method of SanMiguel et al. (3) and measured in κ, the estimated number of nucleotide substitutions per site, as well as years since insertion calculated with the conversion factor used by SanMiguel et al. (3). The oldest recognizable full-length elements, which are of the parental types (A and B), as well as the two deletion derivatives that formed the five recombinants (discussed below), are represented schematically and account for 250 full-length CRM1 elements.
Fig. 1.
Evolution of the CRM1 subfamily. Recombinant and deletion derivatives of the CRM1 subfamily have different periods of activity. Events are listed in chronological order from right (oldest) to left (youngest). Each data point corresponds to a single full-length CRM1 element: 28 B, 11 A, 89 R1, 31 R2, 13 R3, 35 R4 and 57 R5. The oldest full-length elements of BΔ and R3Δ are circled, and a graphical representation is provided, in which green and orange represent sequence fragments derived from the A and B parent, respectively. The three lightly shaded boxes represent the gag, RT, and integrase domains. LTRs are separated from the rest of the element by black bars, and deleted regions (relative to parent A or B) are indicated by white spaces. Deletion derivatives of the B and R1 elements are indicated by the letter “d” and represented in different colors, as noted in the legend. For details on these deletions, see Fig. S3.
The recombination event with the apparently largest impact on the fitness of the element, resulted in the replacement of the CRM1B element RNase H sequence (RHB) with its counterpart from CRM1A (RHA). While this recombinant element family (R1) and its derived recombinant (R5) have proliferated extensively, the last documented insertion of a full-length element containing RHB occurred at κ = 0.027 or ≈2 MYA (Fig. 1), and 236 (89%) of the 264 elements shown in Fig. 1 contain RHA.
The reason for this obvious evolutionary advantage of RHA over RHB remains to be determined. Acquisition of the RH gene from a non-LTR retrotransposon has previously been recognized as a pivotal event in the evolution of the vertebrate retroviral lineage from retrotransposons (21), but the selective advantage of the new RH in that lineage is also as yet unknown. A region of the Ty1 RH that had been shown in mutagenesis experiments to have a large effect on retrotransposition rate because the defective RH is unable to remove the polypurine tract primer from the cDNA during retrotransposition (22), is identical in RHA and RHB.
The other recombinants that feature prominently in the evolution of the CRM1 subfamily all involve portions of the LTR (see Fig. 2 for a detailed schematic of a CRM1 element). R2 represents an A element in which the 3′ end of the integrase, as well as the U3 of the LTR, have been replaced by the B equivalent (Fig. 1). A curious feature of CR elements is that the polyprotein extends into the 3′ LTR, so that the 3′ end of the integrase is encoded by the first ≈172 nt of the LTR. Thus, the region replaced in R2 contains the chromodomain and any promoter sequences contained in the U3. In R3, the 3′ portion of U3 (but not the chromodomain) of an A element has been replaced by the B allele. R4 is similar to R3, but in this subgroup the U5 has also been replaced by the B allele. Finally, the currently most active CRM1 element (R5), which also represents the most complex recombinant, consists of a B polyprotein containing RHA and the 3′ end of the A integrase, with the remainder of the LTR derived from B (Fig. 2).
Fig. 2.
Major linkage blocks of inbred B73 CRM1 elements. The abundance of the A and B alleles for each linkage block in the population of the 246 elements belonging to the nine major subgroups represented schematically in Fig. 1 is indicated below the schematic of an R5 element. Domains are highlighted (g, gag; RT, reverse transcriptase; RH, RNase H; I, integrase).
Creation of these recombinants allows independent selection of specific retroelement regions as summarized in Fig. 2. The regions experiencing the strongest positive selection include the U3B 3′ region, favored by 23:1, and the RHA region, favored by 8.4:1. At first glance there appears to be no overall selection for the chromodomain (C) region of the integrase, and the main benefit of the first recombination in the integrase gene appears to have been incorporation of the U3B 3′ region into the A element. However, closer examination reveals a temporal trend: CA has been favored in recent insertions (κ ≤ 0.003, or ≤0.23 MYA) at a ratio of ≈10:1 (41:4).
In addition to interelement recombinations, deletions in the LTRs appear to play a major role in the evolution of CRM1 elements. For example, CRM1BΔ12a3a (Fig. S3) is the most successful of the six CRM1B LTR deletion derivatives detectable in the B73 genome (Fig. 1), and deletion 3a (Fig. S3) is present in 245 (93%) of the 264 CRM1 LTR pairs represented in Fig. 1. A 21 nt deletion in the U3 of the R3 element has created R3Δ, which subsequently recombined to form the currently most active R5 subgroup. Similarly, a different deletion in the R1 LTR has created a recently (≤0.385 MYA) active derivative in that lineage (BΔ12a3a′).
Shared polymorphisms of consensus sequences derived for each element group suggest the following series of events, which is in agreement with the chronology of element formation based on insertion time in Fig. 1:
Generation of CRM1B deletion derivatives including CRM1BΔ12a3a (BΔ).
Recombination between BΔ and A to yield R1.
Recombination between BΔ and A to yield R2.
Recombination between R1 or BΔ or CRM1BΔ12b3a and A to yield R3.
Replacement of U5A in R3 with U5 of R1 to yield R4.
Deletion in LTR of R3 to yield R3Δ.
Recombination between R3Δ and R1 to yield R5.
While elements that do not recombine or form deletion derivatives appear to have extremely low retrotransposition rates, each of the major recombination or deletion events described above was followed by a period of increased activity for the recombinant element. This is most easily observed for the more recently active elements, as many of the older elements have presumably suffered deletions and are excluded from our tally of full-length elements. The overall effect is a steady or increasing CRM1 population composed of different subgroups at different times.
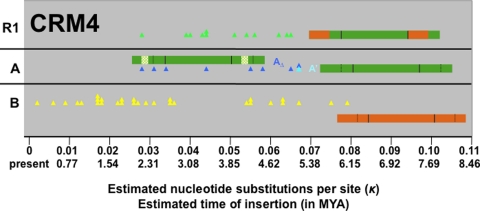
Several of the events documented in the CRM1 subfamily of elements can also be observed in the CRM4 subfamily (Fig. 3): (i) B elements represent the oldest subgroup; (ii) the oldest extant full-length CRM4A element inserted at approximately the same time (κ = 0.067 ≈ 5.15 MYA) as the oldest full-length CRM1A element (κ = 0.072 ≈ 5.54 MYA); (iii) an LTR deletion derivative of CRM4A formed; and (iv) recombined with CRM4B to create R1, a CRM4A element in which the U3 is replaced by the B allele (see Fig. 3). It is noteworthy that, unlike CRM1, CRM4 elements are not localized to active corn centromeres (20), even though they contain the chromodomain characteristic of CR elements and recognizable sequence homology to centromeric CRM subfamilies. The fact that CRM4 elements are more dispersed in the maize genome may have played a role in limiting this subfamily to only one recombinant subgroup, as opposed to the five found in the CRM1 subfamily.
Fig. 3.
Evolution of the CRM4 subfamily. CRM4AΔ LTRs contain a 180 nt deletion relative to CRM4A. The periods of activity of CRM4AΔ and recombinant R1 overlap. Only CRM4B has retrotransposed recently.
We cannot be certain whether the recombination mechanism at work here operates on genomic DNA or during the retrotransposition process. In retroviruses, recombination has been demonstrated to occur frequently during cDNA synthesis (23) which, like yeast Ty1 retrotransposon replication (24), occurs in particles containing two viral or retrotransposon transcripts. Canonical reverse transcription involves two programmed strand-switching events. First, the U5 portion of the reverse transcribed 5′ LTR switches from the 5′ end to the 3′ end, where it serves as a template for reconstruction of the 3′LTR, then an exact copy of the entire 3′ LTR is transferred to the 5′ end of the element (reviewed in ref. 23). These template-switching events would appear to provide an obvious mechanism to create recombinant CRM elements. Furthermore, template switching has been documented for other stages of reverse transcription, specifically because of minus-strand recombination, where a defective RNA template causes the growing minus strand cDNA to switch templates, and plus-strand recombination that is initiated by internally displaced DNA primers (23).
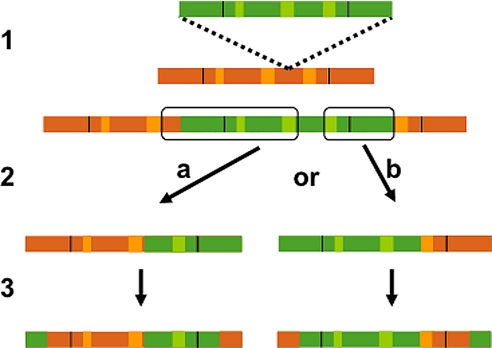
Alternatively, formation of the recombinant elements may be attributable to genomic rearrangements caused by the same DNA recombination machinery that causes solo LTR formation (10, 11) and deletion of genomic DNA between the LTRs of two proximal elements (10, 12). This homologous recombination machinery does act on CRM elements, as evidenced by the presence of CRM solo LTRs (20). If such recombinations occurred between the polyprotein regions of adjacent or nested elements, a chimeric element lacking TSDs would form (Fig. 4). During the retrotransposition process, the upstream U5A would effectively replace the downstream U5B, and this newly formed chimeric downstream LTR would serve as a template to regenerate a chimeric upstream LTR to create the kinds of molecules represented by R2, R3, and R5. Creation of R1 could be accomplished by two sequential recombinations between two nested or three tandem elements (Fig. 4).
Fig. 4.
Generation of recombinant retrotransposons by intrastrand homologous recombination in the polyprotein region: 1, Insertion of a CRM1A into a CRM1B element; 2, Recombination between homologous regions illustrated in “a” and “b” causes deletion of the outlined region and yields primary recombinants; 3, During retrotransposition, the U5 region of the upstream element is transferred to the 3′ terminus of the transcript, generating a chimeric LTR, which is then used as a template to reconstitute the 5′ LTR, resulting in the full-length elements shown. Single recombinations are proposed to have generated R2, R3, and R5. Sequential recombination of “a” and “b” result in the formation of recombinant R1 (see Fig. 1).
Of these two recombination mechanisms, the one operating at the genomic DNA level is more parsimonious for several reasons. First, it requires colocalization of the elements in the genome, but not their coexpression or copackaging. In fact, the highly recombinogenic CRM1 elements are localized to currently active centromeres, while the less recombinogenic CRM4 elements are more widely distributed in the genome. Second, several of the recombinants that are expected to be created during reverse transcription are absent from the population of recombinants observed here. For example, strand switching of the 5′U5A region to a copackaged B template should result in a B element with U5A, but we observe only a single (much more complex) recombinant (R4) that may have been generated in this manner (i.e., strong stop template switching). Note, however, that we cannot exclude the possibility of strong selection against such recombinants. Finally, recombination during retrotransposition should yield elements with identical LTRs and TSDs, whereas recombination of genomic DNA should result in chimeric elements (e.g., with different 5′ and 3′ LTRs) and lacking TSDs. In fact, 75 full-length CRM1 elements (excluded from Fig. 1) lack TSDs and, therefore, presumably represent genomic recombinants between adjacent elements (Fig. S4). Twenty-nine of these are obviously chimeric (three have unique recombination points within the polyprotein, the others have different 5′ and 3′ LTR types), while the remainder are presumably due to recombination between elements of the same subtype. (However, because these BAC sequences are not yet finished, some of these 75 chimeric CRM1 elements may be caused by misassembly.) In contrast, 51 of 52 full-length CRM4 elements (which have formed only one recombinant subfamily) contain TSDs, again suggesting much lower levels of chimeras and recombination in these more genomically dispersed elements.
The existence of solo LTRs, which are thought to be formed as a result of recombination between LTRs of the same element, indicates that the recombination mechanism functions for highly homologous regions separated by the length of an element minus the length of its LTR (5.5–7 kb for CRMs). Thus, nested insertions involving two types of elements should represent suitable substrates for interelement recombination, provided that they meet as yet unknown minimum length and homology criteria. These recombinations are expected to occur in regions of relatively high sequence similarity between the CRM1A and B elements. Indeed, all five regions for which we document recombinations (see Fig. S2 a–d) are flanked by regions of high sequence similarity in CRM1A and CRM1B: the recombined RH region is flanked by 27 and 30 identical nt, the two recombinations within the integrase gene occur within regions of 11 and 12 nt identity, and the LTR recombination that yielded R3 occurred in a region of 11 identical nt. Similarly, the recombination that yielded CRM4 R1 occurred in a region of 22 nt that are identical in CRM4AΔ and CRM4B (Fig. S2 e). Although we cannot exclude the possibility that centromeric chromatin is somehow more recombinogenic than other genomic regions, it is likely that the higher level of recombination activity in centromeric (CRM1) versus noncentromeric (CRM4) elements is at least partly attributable to the high density of CRM1 elements in centromeres.
It is ironic that the very mechanism that eliminates retrotransposons by reducing them to solo LTRs may generate highly promiscuous recombinant elements, conceivably even from two elements with complementary defects. Discovery of recombinant CRM elements was made possible by the availability of large amounts of high quality genomic sequence from a retrotransposon-rich plant genome, and by the detailed phylogenetic analysis performed for each domain of the CRM polyproteins (20). To investigate whether recombination between retrotransposons extends beyond the CRM family, we performed a preliminary analysis of full-length Opie elements that revealed recombination sites in both the LTR and polyprotein regions (Figs. S5 and S6), indicating that retrotransposon recombination is not limited to the centromere-specific CRM family, which belongs to the Ty3/gypsy group of retrotransposons, but also occurs in this Ty1/copia element that is broadly distributed along Zea chromosome arms (6). However, the high frequency of recombinations among Opie elements, combined with the efficient removal of full-length elements from their chromosomal arm locations, provides a very spotty record of Opie element evolution and makes it difficult to reconstruct the history of these elements, let alone measure the effects on retrotransposition rate for individual recombination events. Analysis of other large plant genomes may well prove interelement recombination of the type observed here to play a major role in the evolution and activity of retrotransposons. The tantalizing observations by Vicient et al. (12) of regions with higher sequence similarity between the wheat WIS-2-a1 and the barley BARE-2, than between two BARE elements, further strengthens our assertion that interelement recombinations may be a general feature in the evolution of plant retrotransposons (WIS-2-a1 and BARE elements are noncentromeric and belong to the Ty1-copia family). If so, methods that take into account retrotransposon recombination need to be developed to properly model their evolution.
Genetic recombination offers little advantage in the absence of genetic variation. The success of retrotransposons in plants may be a result, in part, of the propensity of plants to form allopolyploids and wide hybridizations (particularly in wind-pollinated outcrossers), which provide an opportunity to unite retrotransposon alleles in one genome following a period of divergence. Using a conversion factor of 6.5 × 10−9 substitutions per synonymous site per year (3), the estimated nucleotide substitutions per site of the oldest CRM1 (κ = 0.045) and CRM4 (κ = 0.065) recombinants correspond to insertion times of 3.46 MYA and 5 MYA, respectively. Both are near to or more recent than the calculated time of the maize allotetraploidization event at >4.8 MYA (2), suggesting that the A and B alleles were joined as a result of the hybridization event that yielded the allotetraploid progenitor of present day maize. Recent work (25) suggests that the substitution rate for repetitive regions may be approximately fivefold higher than that used here (3.3 × 10−8), which would place all recombinant element insertion times well within the period following allotetraploidization. A survey of the available Oryza and sorghum genome sequences revealed no subgroups of the CRM1 orthologs in those species, again pointing to a wide allopolyploidization event as the basis of the creation of recombinant retrotransposons. Based on Ks estimates of 0.4343 for the polyprotein regions of the most closely related CRM1A and CRM1B element, we estimate the divergence time of CRM1A/CRM1B at between 13 and 34 MYA, suggesting that the two CRM1 subgroups found in present-day corn may have diverged with their respective host genomes 11.9 MYA to generate the genetic diversity that has been exploited since the polyploidization event, the benefits of which are described in this work.
Methods
Identification of TSDs.
Full-length CRM elements and solo LTRs were extracted as in ref. 20 and analyzed for the presence of a 5 nt flanking direct repeats (TSDs). A single mismatch in an otherwise perfect 5 nt direct repeat was assumed to be a point mutation and counted as a TSD.
Estimation of CR Element Insertion Time.
Separate alignments were generated for each CRM1 subgroup of all 5′ and 3′ LTRs from full-length elements with TSDs using ClustalX (26). Apparent misalignments were manually corrected using BioEdit (27). The shape parameter α for each CRM1 (B, BΔ, A, R1, R2, R3, R4 and R5) and CRM4 (A′, AΔ, B, and R1) subgroup was estimated using the program PAML version 3.0 (28) and used to estimate the evolutionary distances between each LTR pairs (k = estimated number of nucleotide substitutions per site) using the γ-K2P model in MEGA version 3.1 (29).
Recombinant Sequence Analysis.
SimPlots: BioEdit was used to generate 80% consensus sequences from full-length sequence alignments for each CRM1 subgroup generated with ClustalX. The consensus sequence of each recombinant subgroup was aligned to the parent groups (A and B) using ClustalX, and the similarity of the recombinant group to the parents was plotted using SimPlot (30) using a sliding window of 200 nt and step size of 20.
Maximum χ2 Test: BioEdit was used to manually edit and subsequently generate 70% consensus sequences from separate multiple sequence alignments generated for each CRM1 LTR, polyprotein and CRM4 LTR subgroup using ClustalX. All single base unique insertions were deleted during editing. Visual inspection of the consensus sequences was used to identify likely parental and derived sequences, which were aligned using ClustalX. The recombination breakpoints for the recombinant sequences were identified using the “2 parental, 1 derived” Maximum χ2 test (31) used in the program START2 (http://pubmlst.org/software/analysis/start2) (32).
Supplementary Material
Acknowledgments.
We thank Kelly Dawe for thoughtful comments on this manuscript. Analysis of CR elements would not have been possible in this detail without the high-quality genome sequence resulting from the ongoing clone-by-clone Maize Genome Sequencing Project. We thank the Washington University Genome Sequencing Center for making the maize genome sequence available to the public. This work was supported by National Science Foundation Grant 0421671.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805694105/DCSupplemental.
References
- 1.Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swigonova Z, et al. Close split of sorghum and maize genome progenitors. Genome Res. 2004;14:1916–1923. doi: 10.1101/gr.2332504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.SanMiguel P, Gaut B, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 4.Piegu B, et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006;16:1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamb JC, Birchler JA. Retroelement genome painting: cytological visualization of retroelement expansions in the genera Zea and Tripsacum. Genetics. 2006;173:1007–1021. doi: 10.1534/genetics.105.053165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pouteau S, Grandbastien MA, Boccara M. Microbial elicitors of plant defense responses activate transcription of a retrotransposon. Plant J. 1994;5:535–542. [Google Scholar]
- 9.Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devos KM, Brown JKM, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vitte C, Panaud O. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotranpsosons in rice Oryza sativa L. Mol Biol Evol. 2003;20:528–540. doi: 10.1093/molbev/msg055. [DOI] [PubMed] [Google Scholar]
- 12.Vicient CM, Kalendar R, Schulman AH. Variability, recombination, and mosaic evolution of the barley BARE-1 retrotransposon. J Mol Evol. 2005;61(3):275–291. doi: 10.1007/s00239-004-0168-7. [DOI] [PubMed] [Google Scholar]
- 13.Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennetzen JL. Patterns in grass genome evolution. Current Opinion in Plant Biology. 2007;10:1–6. doi: 10.1016/j.pbi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Miller JT, Dong F, Jackson SA, Song J, Jiang J. Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics. 1998;150:1615–1623. doi: 10.1093/genetics/150.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Presting GG, Malysheva L, Fuchs J, Schubert I. A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 1998;16:721–728. doi: 10.1046/j.1365-313x.1998.00341.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhong CX, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant cell. 2002;14:2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagaki K, et al. Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics. 2003;163:759–770. doi: 10.1093/genetics/163.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagaki K, et al. Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol Biol Evol. 2005;22:845–855. doi: 10.1093/molbev/msi069. [DOI] [PubMed] [Google Scholar]
- 20.Sharma A, Presting GG. Centromeric retrotransposon lineages predate maize/rice divergence and differ in abundance and activity. Mol Genet Genomics. 2008;279:133–147. doi: 10.1007/s00438-007-0302-5. [DOI] [PubMed] [Google Scholar]
- 21.Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 22.Atwood-Moore A, Ejebe K, Levin HL. Specific recognition and cleavage of the plus-strand primer by reverse transcriptase. J Virol. 2005;79:14863–14875. doi: 10.1128/JVI.79.23.14863-14875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 24.Feng Y-X, Moore SP, Garfinkel DJ, Rein A. The genomic RNA in Ty1 virus-like particles is dimeric. J Virol. 2000;74:10819–10821. doi: 10.1128/jvi.74.22.10819-10821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark RM, Tavaré S, Doebley J. Estimating a nucleotide substitution rate for maize from polymorphism at a major domestication locus. Mol Biol Evol. 2005;22:2304–2312. doi: 10.1093/molbev/msi228. [DOI] [PubMed] [Google Scholar]
- 26.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 28.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 30.Lole KS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34(2):126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 32.Jolley KA, Feil EJ, Chan MS, Maiden MC. Sequence type analysis and recombinational tests (START) Bioinformatics. 2001;17:1230–1231. doi: 10.1093/bioinformatics/17.12.1230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.