Abstract
Agrobacterium represents the only natural example of transkingdom transfer of genetic information, from bacteria to plants. Before the bacterial transferred DNA (T- DNA) can integrate into the plant genome, it should be targeted to and bind the host chromatin. However, the T-DNA association with the host chromatin has not been demonstrated. Here, we study T-DNA binding to plant nucleosomes in vitro and show that it is mediated by bacterial and host proteins associated with the T-DNA. The main factor that determines nucleosomal binding of the T-DNA is the cellular VirE2-interacting protein 1 (VIP1), which functions as a molecular link between the T-DNA-associated bacterial virulence protein VirE2 and core histones. The presence of both VIP1 and VirE2 is required for association of the T-DNA with mononucleosomes in which the DNA molecule exists as a tripartite complex DNA–VirE2–VIP1. Furthermore, this nucleosome-associated ternary complex can bind another bacterial virulence factor, VirF, which is an F-box protein known to target both VirE2 and VIP1 for proteasomal degradation and uncoat the T-DNA.
Keywords: histones, VirE2-interacting protein 1, VirE2, chromatin targeting, T-complex
In nature, Agrobacterium tumefaciens genetically transforms plant cells, causing neoplastic growths in many plant species (1, 2). Under laboratory conditions, however, Agrobacterium can be used as a gene transfer agent for a wide variety of eukaryotic organisms, from fungi to humans (3, 4). Thus, the mechanism by which Agrobacterium introduces its transferred DNA (T-DNA) into the eukaryotic genome most likely is conserved between most eukaryotes. This mechanism includes three major types of DNA traffic: export into the eukaryotic cell, import into the cell nucleus, and targeting to and association with the eukaryotic chromatin. Whereas the first two events are relatively well studied (e.g., refs. 1 and 5), the last process has not been studied, or even demonstrated, at all.
Agrobacterium T-DNA is exported into the eukaryotic cell via the type IV secretion system as a ssDNA molecule (6), the T-strand, and its transport is mediated by bacterial virulence (Vir) proteins, some of which accompany the T-strand into the host cell. For example, the VirD2 protein is covalently attached to the 5′ end of the T-strand (7), whereas VirE2, an ssDNA binding protein, is transported separately from the T-strand, and is thought to associate with the T-strand in the host cell cytoplasm, producing a core T-complex (8–10). Two additional virulence proteins, VirE3 and VirF, are exported into the host cell to facilitate the nuclear import and proteasomal uncoating of the T-complex, respectively (11–16). The T-complex nuclear import is thought to occur via the importin α-dependent pathway, in which VirD2 (17) directly interacts with the importin α and VirE2 is recognized by the importin α via a molecular adaptor (18, 19), VirE2-interacting protein 1 (VIP1), which is encoded by the plant cell and is able to bind both VirE2 and importin α. VIP1 is not an abundant protein, but its function in the T-complex nuclear import is augmented by the bacterial effector, VirE3 (14, 15). Once in the cell nucleus, the T-complex is expected to recognize and bind the host chromatin by an as-yet-unknown mechanism that may involve VIP1, known to interact with individual core histones (20, 21). Consistent with this idea, core histones have been shown to play a role in Agrobacterium infection (22–24). Finally, the chromatin-bound T-complex is thought to uncoat its proteins via proteasomal degradation mediated by the F-box protein VirF (25) that recognizes VIP1 and destabilizes both VIP1 and its associated VirE2 (16).
Here, we focused on the least-studied event of the T-complex association with the host chromatin. To this end, we developed an in vitro system for detection and characterization of the association of the synthetic T-DNA with plant mononucleosomes. Using this approach, we demonstrated that the reconstituted core T-complex, i.e., ssDNA coated with VirE2 molecules, is able to bind to plant nucleosomes and that this binding requires the presence of VIP1.
Results
VIP1 Associates with Nucleosomes.
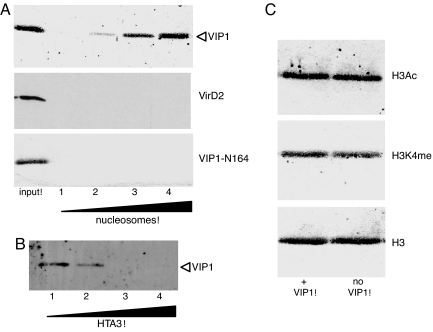
Among all known proteins, i.e., VirD2, VirE2, and VIP1, that are thought to associate with the T-strand after its nuclear import, VIP1 has been shown to bind individual core histones (20, 21), thus representing the best candidate for a factor that may mediate interaction with nucleosomes. Indeed, VIP1 efficiently bound purified mononucleosomes in a concentration-dependent manner (Fig. 1A). This binding was specific because no nucleosomal association was observed for VirD2; furthermore, no binding was detected with the VIP1-N164 mutant (Fig. 1A), known to retain most functions of VIP1, but not its capacity to bind the core histones (20). The specificity of the VIP1–nucleosome interaction was also supported by the ability of the purified Arabidopsis histone H2A, HTA3, to inhibit this binding competitively (Fig. 1B).
Fig. 1.
Specific binding of VIP1 to mononucleosomes. (A) VIP1, but not VirD2 or VIP1-N164, shows a dose-response binding to increasing amounts of immobilized nucleosomes. Lanes 1–4: 0, 5, 10, and 20 μg protein of mononucleosomal preparation, respectively; Input: 1 μg of the indicated ligand. (B) Competitive inhibition of VIP1 (1 μg) binding to immobilized mononucleosomes by increasing concentrations of the plant histone H2A, HTA3, premixed with VIP1 before binding. Lanes 1–4: 0, 1, 2, and 3 μg HTA3, respectively. (C) VIP1 binding to immobilized histones is independent of histone H3 acetylation and histone H3 K4 methylation. The amount of the total histone H3 preparation loaded on gel (no VIP1) was normalized to the total amount of VIP1-bound H3 histone (+VIP1) based on quantification using anti-H3 antibodies; this normalization allows us to compare directly what fraction of the H3 population is represented by the H3Ac and H3K4Me subpopulations.
Next, we examined whether major histone modifications indicative of the active chromatin state affect VIP1 binding. To this end, commercial preparations of total histones were incubated with immobilized VIP1, and the bound histones were analyzed by Western blotting using antibodies specific to dimethyl-histone H3 K4 and acetyl-histone H3. Under our binding conditions where VIP1 does not saturate the entire histone preparation (data not shown), preferential binding of VIP1 to the modified histones would result in enrichment of the corresponding immunoblot signal as compared with that detected in the original histone preparation with antibody against the same modified histone. Fig. 1C shows that no such enrichment was observed with either of the two antibodies, indicating that VIP1 binding to nucleosomes is independent of H3 acetylation and H3K4 methylation. Western blotting using anti-H3 antibodies indicated that all samples contained equal amounts of total H3 histone (Fig. 1C).
VIP1 Links Between Nucleosomes and VirE2.
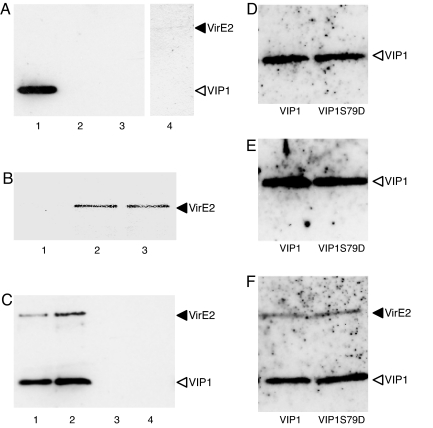
Unlike VIP1, VirE2 was unable to bind nucleosomes under our experimental conditions, although, at low stringency, some residual association was detected (Fig. 2A). Thus, we examined whether VIP1 can mediate association of VirE2 with nucleosomes. As expected, VIP1 as well as VIP1-N164 bound VirE2 in our experimental system (Fig. 2B). Addition of VIP1 to VirE2 resulted in the association of both proteins with nucleosomes (Fig. 1C). Importantly, VIP1-N164, which retains its VirE2 binding activity, but is unable to bind nucleosomes, did not promote nucleosomal association of VirE2 (Fig. 1C). These results indicate formation of ternary VirE2–VIP1–nucleosome complexes in which VIP1 most likely functions as a molecular adapter between VirE2 and the nucleosome.
Fig. 2.
VIP1 mediates association of VirE2 with mononucleosomes. (A) VirE2 does not bind to immobilized mononucleosomes directly. Lane 1, nucleosomes + VIP1 (1.0 μg); lane 2, nucleosomes + VirE2 (0.5 μg); lane 3, nucleosomes + VirE2 (1.0 μg); lane 4, nucleosomes + VirE2 (1.0 μg) under low-stringency binding conditions. (B) VirE2 binds to immobilized VIP1 and VIP1-N164. Lane 1, VirE2 alone (0.5 μg); lane 2, VirE2 (0.5 μg) + VIP1 (1.0 μg); lane 3, VirE2 (0.5 μg) + VIP1-N164 (1.0 μg). (C) VIP1-mediated association of VirE2 with immobilized mononucleosomes. Lane 1, nucleosomes + VIP1 (1. 0 μg) + VirE2 (0.5 μg); lane 2, nucleosomes + VIP1 (1. 0 μg) + VirE2 (1.0 μg); lane 3, nucleosomes + VIP1–164N (1. 0 μg) + VirE2 (1.0 μg); lane 4, VIP1 (1. 0 μg) + VirE2 (1.0 μg). (D–F) VIP1 and VIP1S79D (1 μg each) exhibit similar abilities to bind immobilized mononucleosomes (D) and immobilized VirE2 (1 μg) (E) or to promote association of VirE2 (1 μg) association with immobilized mononucleosomes (F).
Next, we examined the effect of mimicking VIP1 phosphorylation at Ser-79, known to enhance its nuclear import (26), on its ability to bind VirE2 and associate with nucleosomes. To this end, we compared nucleosome binding of the recombinant VIP1, which is not phosphorylated, with a VIP1 mutant, VIP1S79D, containing a negatively charged aspartate residue known to reproduce the biological effects of VIP1 phosphorylation (26). We observed no detectible differences in nucleosomal association between VIP1 and its phosphorylation-mimicking mutant (Fig. 2D). We also detected no differences between VIP1 and VIP1S79D in their capacity to bind VirE2 (Fig. 2E) or promote the association of VirE2 with nucleosomes (Fig. 2F). Thus, VIP1 phosphorylation is not involved in chromatin targeting of this protein.
Nucleosomal Association of Synthetic T-Complexes.
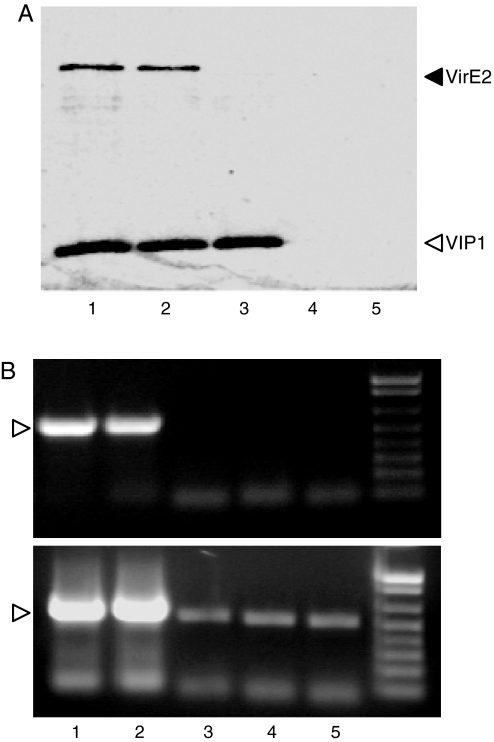
Can VIP1 mediate nucleosomal binding of not only VirE2, but also the T-complexes? Because VirE2 is the major structural and functional protein component of the T-complex, and because T-DNA is sequence nonspecific, a simple T-complex can be reconstituted from ssDNA and VirE2 in vitro. When such synthetic T-complexes were incubated with nucleosomes in the presence of VIP1, both proteins (Fig. 3A) and ssDNA (Fig. 3B Upper) showed nucleosomal association. In the absence of VirE2, VIP1 still bound the nucleosomes, but no significant nucleosomal association of ssDNA was observed. Also, no nucleosomal binding of the ssDNA–VirE2 complexes occurred in the absence of VIP1 or the presence of VIP1-N164 (Fig. 3). Thus, synthetic T-complexes can bind eukaryotic chromatin, and this binding is VIP1-dependent.
Fig. 3.
VIP1 mediates association of ssDNA–VirE2 complexes with mononucleosomes. (A) Detection of protein. (B) Detection of ssDNA. Lane 1, nucleosomes + VIP1 (1. 0 μg) + VirE2 (1.0 μg) + ssDNA (0.1 μg); lane 2, nucleosomes + VIP1 (1.0 μg) + VirE2 (0.5 μg) + ssDNA (0.1 μg); lane 3, nucleosomes + VIP1 (1. 0 μg) + ssDNA (0.1 μg); lane 4, nucleosomes + VirE2 (1.0 μg) + ssDNA (0.1 μg); lane 5, nucleosomes + VIP1-N164 (1. 0 μg) + VirE2 (1.0 μg) + ssDNA (0.1 μg). (Upper) Twenty PCR cycles. (Lower) Thirty-five PCR cycles. Arrowhead indicates the ssDNA-specific 500-bp PCR fragment.
Interestingly, at much more sensitive detection conditions, ssDNA was also found associated with the nucleosomes irrespective of the presence of VirE2 and functional VIP1 in the amounts dramatically lower than those achieved with T-complexes and VIP1 (Fig. 3B Lower). These results are consistent with the long-standing observation that, although plant-stable transformation can be achieved by using free DNA, delivered, for example, by electroporation or microbombardment, the transformation efficiency is much higher with Agrobacterium as gene vector.
Effect of VIP1-Specific F-Box Protein, VirF, on VIP1 Binding to Nucleosomes.
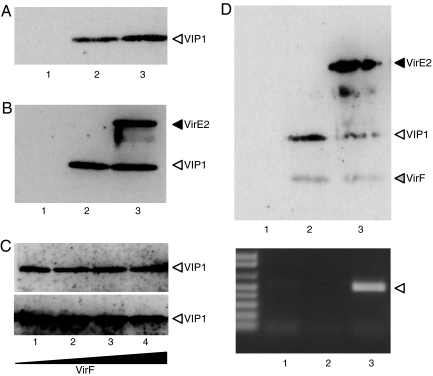
Once at the host chromatin, VirF is thought to mediate uncoating of the T-complex via proteasomal degradation of VIP1 and VirE2 (16). In this scenario, VirF should interact with the T-complex bound to nucleosomes, and/or it should not inhibit this nucleosomal binding. Because VirF interacts directly with VIP1 (Fig. 4A), we first tested whether this interaction impairs the ability of VIP1 to bind VirE2 and/or nucleosomes. Fig. 4B shows that VIP1 was able to form ternary complexes with VirF and VirE2, whereas Fig. 4C demonstrates that increasing concentrations of VirF had no effect on the VIP1 ability to bind nucleosomes. Furthermore, in the presence of VIP1, VirF itself became associated with nucleosomes (Fig. 4D Upper), presumably via its interaction with VIP1. Similarly, the VIP1–VirF association did not interfere with VIP1–VirE2 binding, allowing formation of ternary VirF–VIP1–VirE2 complexes (Fig. 4D Upper). These complexes also bound to the nucleosomes, and the binding depended on VIP1 because neither VirF nor VirE2 were found associated with the nucleosomes in the absence of VIP1 (Fig. 4D Upper). Importantly, ssDNA bound to VirE2 also was detected in the nucleosome-associated VirF–VIP1–VirE2 complexes (Fig. 4D Lower). Thus, T-complexes can be recognized by VirF while they are bound to nucleosomes.
Fig. 4.
Nucleosomal binding of VirF–VIP1–VirE2-ssDNA complexes. (A) VIP1 binds immobilized VirF. Lane 1. VIP1 alone (1 μg); lane 2, VIP1 (1 μg) + VirF (3 μg); lane 3, VIP1 (1 μg) + VirF (5 μg). (B) Both VIP1 and VirE2 bind to immobilized VirF. Lane 1, VIP1 (1.0 μg) + VirE2 (0.5 μg); lane 2, VIP1 (1.0 μg) + VirF (5.0 μg); lane 3, VIP1 (1.0 μg) + VirE2 (0.5 μg) + VirF (5.0 μg). (C) VirF does not affect VIP1 binding to immobilized mononucleosomes. Lane 1, nucleosomes + VIP1; lane 2, nucleosomes + VIP1 + VirF (3 μg); lane 3, nucleosomes + VIP1 + VirF (5 μg); lane 4, nucleosomes + VIP1 + VirF (7 μg). (Upper) One microgram of VIP1. (Lower) Two micrograms of VIP1. (D) VIP1, VirE2, ssDNA, and VirF together associate with immobilized mononucleosomes. Lane 1, nucleosomes + VirE2 (0.5 μg) + ssDNA (0.1 μg) + VirF (2.0 μg); lane 2, nucleosomes + VIP1 (1.0 μg) + ssDNA (0.1 μg) + VirF (2 μg); lane 3, nucleosomes + VIP1 (1 μg) + VirE2 (0.5 μg) + ssDNA (0.1 μg) + VirF (2.0 μg). (Upper) Detection of proteins. (Lower) Detection of ssDNA. Arrowhead indicates the ssDNA-specific 500-bp PCR fragment produced after 20 PCR cycles.
Discussion
A long-standing question in the field of genetic engineering of eukaryotic cells is the mechanism by which the transgene associates with the target chromatin before integration. Because of its ability to transfer genes to most eukaryotic species, Agrobacterium represents a useful model system to study this question. The bacterial T-DNA is packaged into a deoxyribonucleoprotein T-complex composed primarily of ssDNA and the VirE2 and VirD2 proteins (2). The ssDNA is sequence-nonspecific as any DNA placed between the T-DNA borders on the bacterial tumor-inducing (Ti) plasmid is transferred to plants and, therefore, acts as a T-DNA. Thus, the biological functions of the T-complex are fulfilled by its VirE2 and VirD2 components. Of those, VirE2 is by far predominant, accounting for >99.9% of the protein content of the T-complex (27). Based on these considerations, a synthetic ssDNA–VirE2 complex represents a simple and well defined paradigm of the Agrobacterium T-complex.
Because the number of the T-complexes that ultimately become integrated into the genome is low, on average 1.5 T-DNA inserts per genome (28), and the T-complex is most likely very short-lived because of its proteasomal degradation (16), it was necessary to develop an in vitro system to study the T-complex–chromatin interactions. To this end, we used synthetic T-complexes and purified plant mononucleosomes. Using this approach, we demonstrated nucleosomal association of the T-complexes which occurs by a VIP1-dependent mechanism. VIP1 is a plant protein that Agrobacterium has evolved to take advantage of during several critical steps of the infection process. First, the bacterial VirE2 protein binds VIP1, most likely in the host cell cytoplasm, and uses it for piggy-back nuclear import via an importin α-dependent pathway (18); this process is up-regulated by a host MAP kinase that phosphorylates VIP1 (26). Because VirE2 is associated with the T-DNA, VIP1 effectively mediates nuclear import of the entire T-complex, facilitating the genetic transformation. Next, as our present results demonstrate, VIP1 mediates chromatin association of the T-complex by acting as a molecular link between VirE2 and nucleosomes via interactions with the core histones. Interestingly the VIP1-nucleosome binding does not depend on the VIP1 phosphorylation, effectively uncoupling the nuclear import and nucleosomal binding activities of this protein.
Whether or not the T-DNA integration occurs preferentially within the active chromatin has been a subject of debate. Whereas several early studies suggested integration bias toward transcriptionally active regions (29–31), a more recent study challenged this view, suggesting that the integration occurs equally in the heterochromatin and the euchromatin of the host cell (32). Our studies of the VIP1–nucleosome interaction showed that it does not depend on acetylation and K4 methylation of the histone H3, which represent the hallmarks of euchromatin (33). This observation supports and provides a molecular basis for the view that T-DNA integration in the plant genome occurs irrespective of its transcriptional state.
VIP1 is also thought to play a role in proteasomal uncoating of the T-complex before integration. This process is mediated by the bacterial F-box protein, VirF, which recognizes VIP1 and targets it for degradation (16). Although VirF does not recognize VirE2, it would still promote its degradation when VirE2 is bound to VIP1. This model is based on a hitherto unproven assumption that binding of VirF to the VIP1-associated T-complex can occur at the site of integration, i.e., at the host chromatin. This notion is now supported by our observations that VirF can exist in complex with VIP1, VirE2, and nucleosomes, in which VIP1 represents the nucleation site for all of the other components. Furthermore, because VirF can promote degradation of VIP1-bound VirE2 (16), it is tempting to speculate that it also can induce degradation of the VIP1-bound histones, effectively perturbing the host chromatin and facilitating integration of the bacterial T-DNA.
Materials and Methods
Plasmids.
Coding sequences of HTA3 (At1g54690) and VIP1-N164 (the gene segment corresponding to the 164 N-terminal amino acids of VIP1) were PCR-amplified from an Arabidopsis thaliana cDNA library, using forward primers 5′-CCGGAATTATGAGTTCCGGCGCCGGCAGT-3′ and 5′-CCGGAATTCATGGAAGGAGGAGGAAGAGGA-3′, and reverse primers 5′-CCGCTCGAGTTAAAACTCTTGAGAAGCAGATCC-3′ and 5′-CCGCTCGAGTTATTCGATATTAAACGACGCCGA-3′, respectively, and subcloned in the EcoRI–XhoI sites of pET28-a (Novagen). The VirF ORF was PCR-amplified from purified Ti-plasmid from Agrobacterium strain 15955, using forward primer 5′-CGCGGATTCCGATGAGAAATTCGAGTTTGCGTG-3′ and reverse primer 5′-CGCGTCGACTAGACCGCGCGTTGATCG-3′, and subcloned into the BamHI–SalI sites of pET28-c (Novagen). The S79D mutant of VIP1 was cloned in two steps; first, the DNA segment encoding the 79 N-terminal amino acids of VIP1 and containing a point mutation replacing serine with aspartate at position 79, was PCR-amplified, using forward and reverse primers 5′-CCGGAATTCATGGAAGGAGGAGGAAGAGGA-3′ and 5′-CATGGGATCAGCTTGCGGTTG-3′, respectively; then a second PCR was performed, using the fragment amplified in the first step as forward primer and the reverse primer 5′-CCGCTCGAGTCAGCCTCTCTTGGTGAAATCCATGTA-3′, and its product was subcloned as an EcoRI–XhoI fragment into pET28a. All PCR products in these reactions were obtained by using a high-fidelity DNA polymerase (Pfu), and all constructs were verified by DNA sequencing. The plasmids for expression of VirE2 and VIP1 have been described (18).
Protein Purification.
Proteins were expressed in BL21(DE3) Escherichia coli strain (Novagen) and extracted by using a 10 mM phosphate buffer, pH 8, supplemented with either 1 M NaCl for purification of HTA3 or 1 M NaCl and 4 M urea for purification of all other proteins, in the presence of 1 mM PMSF and 1 mM β-mercaptoethanol. Protein extracts were adsorbed onto a nickel-agarose resin (Qiagen) and renatured on the resin by sequential washes with the 10 column volumes of 10 mM phosphate buffer, pH 8, containing 1 mM PMSF, 10 mM imidazole, and 1 mM β-mercaptoethanol and supplemented first with 1 M NaCl and decreasing concentrations of urea (4, 3, 2, and 1 M) and then only with decreasing concentrations of NaCl (1, 0.6, and 0.3 M). Proteins were then eluted in 10 mM phosphate buffer, pH 8, containing 0.3 M NaCl, 1 mM PMSF, 10% glycerol, 250 mM imidazole, and 1 mM β-mercaptoethanol, and dialyzed extensively against 10 mM phosphate buffer, pH 8, containing 100 mM NaCl, 1 mM, PMSF, 10% glycerol, and 1 mM β-mercaptoethanol. All proteins were purified to near homogeneity (95%), as verified by SDS/PAGE and silver staining. In our experience, His tag has no detectible nonspecific effects on VirE2, VirF, or VIP1 activities (data not shown). Here, this point is illustrated by specific differences between biological activities of His-tagged VIP1 and VIP1-N164, both of which interacted with VirE2, but only VIP1 interacted with nucleosomes.
Nucleosome Purification.
As source for nucleosomes, we chose cauliflower florets; cauliflower is a close relative of A. thaliana, its florets provide large amounts of tissue devoid of chloroplasts that may interfere with nuclei extraction, and they have low concentrations of oxidative molecules as compared with leaf tissues. Mononucleosomes were purified from cauliflower florets as described (34); briefly, chromatin extracted from purified nuclei was partially digested by micrococcal nuclease and the products of digestion were separated by ultracentrifugation on a 10–40% glycerol gradient for 20 h at 45000 × g and 4°C. The fractions containing a DNA band of ≈150 bp, all four core histones, but lacking the linker histone H1, were pooled and concentrated to ≈1 mg/ml.
Protein Binding to Nucleosomes.
A modified ELISA was used, in which purified mononucleosomes (10 μg protein unless indicated otherwise) were first adsorbed onto Nunc MaxiSorp 96-well plates overnight at 4°C in a 50 mM sodium carbonate buffer, pH 9.5, followed by blocking for 4 h at 4°C with 5% BSA in PBS, pH 7.4. The same adsorption protocol was used when purified proteins were used as binding substrates. Next, the tested proteins in PBS (pH 7.4) supplemented with 1% BSA, 0.05% Tween 20 and 1 mM DTT were added in the wells and incubated for 50 min at room temperature. The wells were rinsed three times for 1 min with PBS (pH 7.4) containing 0.1% Tween 20 for detections of binding at low stringency and 10 times for 3 min with PBS (pH 7.4) containing 1% Tween 20 for all other experiments. For experiments testing binding of multiprotein complexes, the corresponding components were premixed in the same buffer and allowed to form complexes for 20 min at 4°C. For studies of effects of histone modifications, purified calf thymus histones (Sigma) were used. Proteins bound to the immobilized mononucleosomes or other substrates were resuspended in Laemmli buffer, resolved by SDS/PAGE and analyzed by Western blotting using the corresponding primary antibodies [i.e., rabbit polyclonal anti-VIP1 (1:1,000 titer) or anti-VirE2 antibodies (1:1,000 titer), mouse monoclonal anti-T7 tag antibodies (Sigma) (1:5,000 titer), or antibodies against histone H3, acetyl-histone H3, dimethyl-histone H3 lysine 4 (K4) (1:2,000 titer each) (Upstate Biotechnology)], and secondary anti-rabbit or anti-mouse antibodies conjugated to alkaline phosphatase or horseradish peroxidase (1:3,000 titer) (Pierce) detected by using chemiluminescent substrate according to the manufacturer's instructions (Millipore). All experiments were repeated at least three times.
ssDNA-Protein Binding to Nucleosomes.
M13mp18 ssDNA (New England Biolab) and purified VirE2 were allowed to form a complex as described (18), VIP1 or one of its derivatives was then added and the mix was used for binding experiments as described above. After resuspension in Laemmli buffer and DNA extraction using the GE kit (GE Healthcare), the bound ssDNA was detected by PCR using ExTaq polymerase (TaKaRa) and the M13mp18-specific forward primer 5′-AGGCGATGATACAAATCTCC-3′ and reverse primer 5′-CAACAGTTTCAGCGGAGTGA-3′, which amplify a 500-bp fragment. The PCRs were performed for 20 or 35 cycles under the following conditions: denaturation for 30 s at 94°C, annealing for 30 s at 56°C, and elongation for 1 min at 72°C.
Acknowledgments.
This work was supported by grants from the National Institutes of Health, the National Science Foundation, National Research Initiative U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service, the United States–Israel Binational Agricultural Research and Development Fund, and Cooperative Development Research– U.S. Agency for International Development (to V.C.) and a grant from the United Sates–Israel Binational Science Foundation (to V.C. and A.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Gelvin SB. Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev. 2003;67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzfira T, Citovsky V. Agrobacterium-mediated genetic transformation of plants: Biology and biotechnology. Curr Opin Biotechnol. 2006;17:147–154. doi: 10.1016/j.copbio.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet. 2005;48:1–17. doi: 10.1007/s00294-005-0578-0. [DOI] [PubMed] [Google Scholar]
- 4.Lacroix B, Tzfira T, Vainstein A, Citovsky V. A case of promiscuity: Agrobacterium's endless hunt for new partners. Trends Genet. 2006;22:29–37. doi: 10.1016/j.tig.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Citovsky V, et al. Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol. 2007;9:9–20. doi: 10.1111/j.1462-5822.2006.00830.x. [DOI] [PubMed] [Google Scholar]
- 6.Scheiffele P, Pansegrau W, Lanka E. Initiation of Agrobacterium tumefaciens T-DNA processing: Purified proteins VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J Biol Chem. 1995;270:1269–1276. doi: 10.1074/jbc.270.3.1269. [DOI] [PubMed] [Google Scholar]
- 7.Young C, Nester EW. Association of the VirD2 protein with the 5′ end of T-strands in Agrobacterium tumefaciens. J Bacteriol. 1988;170:3367–3374. doi: 10.1128/jb.170.8.3367-3374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie PJ, Ward JE, Winans SC, Nester EW. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J Bacteriol. 1988;170:2659–2667. doi: 10.1128/jb.170.6.2659-2667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Citovsky V, Wong ML, Zambryski PC. Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: Implications for the T-DNA transfer process. Proc Natl Acad Sci USA. 1989;86:1193–1197. doi: 10.1073/pnas.86.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelvin SB. Agrobacterium VirE2 proteins can form a complex with T strands in the plant cytoplasm. J Bacteriol. 1998;180:4300–4302. doi: 10.1128/jb.180.16.4300-4302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vergunst AC, et al. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 12.Schrammeijer B, den Dulk-Ras A, Vergunst AC, Jurado Jácome E, Hooykaas PJJ. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: Evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vergunst AC, et al. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci USA. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacroix B, Vaidya M, Tzfira T, Citovsky V. The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J. 2005;24:428–437. doi: 10.1038/sj.emboj.7600524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Rodríguez FM, Schrammeijer B, Hooykaas PJJ. The Agrobacterium VirE3 effector protein: A potential plant transcriptional activator. Nucleic Acids Res. 2006;34:6496–6504. doi: 10.1093/nar/gkl877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 17.Ballas N, Citovsky V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci USA. 1997;94:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzfira T, Vaidya M, Citovsky V. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzfira T, Vaidya M, Citovsky V. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis VIP1 gene. Proc Natl Acad Sci USA. 2002;99:10435–10440. doi: 10.1073/pnas.162304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci USA. 2005;102:5733–5738. doi: 10.1073/pnas.0404118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loyter A, et al. The plant VirE2 interacting protein 1. A molecular link between the Agrobacterium T-complex and the host cell chromatin? Plant Physiol. 2005;138:1318–1321. doi: 10.1104/pp.105.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mysore KS, Nam J, Gelvin SB. An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc Natl Acad Sci USA. 2000;97:948–953. doi: 10.1073/pnas.97.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anand A, et al. Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol Plant-Microbe Interact. 2007;20:41–52. doi: 10.1094/MPMI-20-0041. [DOI] [PubMed] [Google Scholar]
- 24.Gelvin SB, Kim SI. Effect of chromatin upon Agrobacterium T-DNA integration and transgene expression. Biochim Biophys Acta. 2007;1769:409–420. doi: 10.1016/j.bbaexp.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Schrammeijer B, et al. Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol. 2001;11:258–262. doi: 10.1016/s0960-9822(01)00069-0. [DOI] [PubMed] [Google Scholar]
- 26.Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- 27.Abu-Arish A, et al. Three-dimensional reconstruction of Agrobacterium VirE2 protein with single-stranded DNA. J Biol Chem. 2004;279:25359–25363. doi: 10.1074/jbc.M401804200. [DOI] [PubMed] [Google Scholar]
- 28.Azpiroz-Leehan R, Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: Going back and forth. Trends Genet. 1997;13:152–156. doi: 10.1016/s0168-9525(97)01094-9. [DOI] [PubMed] [Google Scholar]
- 29.Szabados L, et al. Distribution of 1000 sequenced T-DNA tags in the Arabidopsis genome. Plant J. 2002;32:233–242. doi: 10.1046/j.1365-313x.2002.01417.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen S, et al. Distribution and characterization of over 1,000 T-DNA tags in rice genome. Plant J. 2003;36:105–113. doi: 10.1046/j.1365-313x.2003.01860.x. [DOI] [PubMed] [Google Scholar]
- 31.Alonso JM, et al. Genomewide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 32.Kim SI, Veena, Gelvin SB. Genomewide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under nonselective conditions. Plant J. 2007;51:779–791. doi: 10.1111/j.1365-313X.2007.03183.x. [DOI] [PubMed] [Google Scholar]
- 33.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Bowler C, et al. Chromatin techniques for plant cells. Plant J. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]