Abstract
Accurate chromosome segregation during mitotic division of budding yeast depends on the multiprotein kinetochore complex, Dam1 (also known as DASH). Purified Dam1 heterodecamers encircle microtubules (MTs) to form rings that can function as “couplers,” molecular devices that transduce energy from MT disassembly into the motion of a cargo. Here we show that MT depolymerization develops a force against a Dam1 ring that is sixfold larger than the force exerted on a coupler that binds only one side of an MT. Wild-type rings slow depolymerization fourfold, but rings that include a mutant Dam1p with truncated C terminus slow depolymerization less, consistent with the idea that this tail is part of a strong bond between rings and MTs. A molecular-mechanical model for Dam1-MT interaction predicts that binding between this flexible tail and the MT wall should cause a Dam1 ring to wobble, and Fourier analysis of moving, ring-attached beads corroborates this prediction. Comparison of the forces generated against wild-type and mutant complexes confirms the importance of tight Dam1-MT association for processive cargo movement under load.
Keywords: depolymerization, kinetochore, laser trapping, mitosis, tubulin
Depolymerizing microtubules (MTs) can generate enough force to move mitotic chromosomes in the absence of MT-dependent, minus-end-directed motors (1–6). MT shortening can do work, thanks to the disassembly pathway of tubulin-containing protofilaments (PFs), the 13 linear polymers that comprise the MT wall. During depolymerization, PFs lose their lateral attachments and bend out from the MT axis (7, 8). As PFs curl toward their minimum energy shape, they can do mechanical work, e.g., pushing on a microbead statically attached to the MT wall via biotin-streptavidin links (9). Such beads experience a “single-shot” power stroke from the bending PFs, after which they detach, together with dissociating tubulins.
Similar power strokes might move mitotic chromosomes. Chromosomes could be attached to depolymerizing MTs via encircling rings (10), so the MT depolymerization force is collected from all 13 bending PFs (11). It is imperative, however, that a chromosome–MT coupler does not detach, because this would lead to chromosome loss. A successful ring-shaped coupler should therefore be efficient in taking advantage of the energy from MT depolymerization and have stable attachment, so the chromosome motion is processive.
The ring hypothesis received a boost with the discovery that the Dam1 kinetochore complex from budding yeast can form MT-encircling rings under physiological conditions in vitro (12, 13). Structural, biochemical, and kinetic properties of these rings have suggested that they might indeed function as chromosome couplers in yeasts (12–18). The inner diameter of a Dam1 ring is ≈10 nm wider than the outer diameter of an MT, but the ring binds directly to the MT wall thanks to inward-directed protrusions from Dam1 complexes (16, 17). Several polypeptides contribute to these protein arms, but deletion of 138 amino acids from the C terminus of only one of them, Dam1p, noticeably reduces the mass of the protrusions and the strength of Dam1-MT binding (12, 16, 17).
These discoveries have prompted theoretical work on the biomechanical design of the Dam1 ring (19, 20). Modeling has suggested that at least two features of Dam1 rings facilitate the transduction of a large fraction of the MT depolymerization energy: the ring's large diameter and the flexibility of its connections with the MT wall (11, 16, 19). If these Dam1 protrusions bind specific sites on tubulin dimers, their flexibility would provide an additional benefit by allowing more ring–MT bonds, thereby increasing the ring's affinity for an MT.
Once ring diameter and linker flexibility are defined, the most critical remaining parameter of ring coupling in our model is the strength of the ring–MT bonds (19). Rings that are otherwise identical but vary in the strength of this bond can all follow a shortening MT end, but the “conformational wave” of PF bending promotes motility of weakly vs. strongly bound rings by different mechanisms. Rings that bind weakly diffuse fast on the MT wall, so bending PFs serve mostly as ratchets to bias these thermal motions. Intuitively, this situation seems excellent, allowing rings to slide without much resistance and making biased diffusion efficient. However, such couplers are vulnerable to variations in the rate of tubulin depolymerization; if PF flaring is lost or decreased, e.g., through a pause in shortening or thermal fluctuations, a weakly bound ring can readily detach from the MT end, even under a very small load (19). Thus, other factors, such as additional protein complexes or some not yet specified features of the Dam1 complex itself, would have to help a weakly bound ring to hang on to the shortening MT end.
This problem is elegantly solved if the Dam1 complex binds strongly to the MT lattice. Binding, even as strong as 15–17 kBT per bond, does not preclude ring motion at the shortening MT end; PF bending can still move a ring that is this tightly bound, provided it has a relatively large diameter and flexible connecting structures [a “forced walk” mechanism (19)]. Unfortunately, the existing biochemical data on Dam1-MT affinity provide values that differ by two orders of magnitude (12, 21). Other measurements of binding strength have also led to a broad range of estimates, so these data do not help to distinguish between possible mechanisms for Dam1 ring motility.
Here we test the idea that the mechanisms for coupler motility on shortening MTs can be clarified by certain properties of Dam1 motion. We use an in vitro system based on purified Dam1 complexes and tubulin polymers to assess both the force that shortening MTs can exert on two alleles of Dam1 oligomers and the effects of these complexes on the rates of MT shortening. The data support the notion that wild type Dam1 binds strongly to the MT lattice through the inward-pointing projection. Our results explain how a mobile ring coupler can both harness a large force from MT disassembly and ensure processive chromosome motion.
Results
Dam1 Rings Capture a Significant Force from Disassembling MTs.
A coupler that encircles an MT but does not bind its wall could in theory experience a force of ≈75 pN from a depolymerizing MT, given the energy associated with GTP hydrolysis (11). A coupler modeled on the Dam1 ring is expected to harness a significant fraction of this force, although rings that are bound more tightly to the MT will stall under a smaller load, so their energy efficiency should be smaller (19). To measure the force that MT depolymerization exerts on a Dam1 ring in vitro, we have used an established method (9) for tethering the minus end of an MT grown from purified bovine tubulin to a coverslip, leaving its plus end free but stabilized by a photodissociable cap of tubulin, assembled in the presence of guanosine-5′-[(α,β)-methyleno]triphosphate (GMPCPP) (Fig. 1A). A bead bound to the wall of such an MT can be clasped in a laser trap and pulled gently toward the MT plus end to generate small tension, and its position can be followed with nanometer precision by a quadrant photodetector (QPD). This method, unlike others (22, 23), allows quantitative comparisons between depolymerization forces transmitted by couplers with different geometries. Here we compare force transients measured with a Dam1 ring with those obtained with previously characterized, nonencircling attachments via biotin-streptavidin links (9).
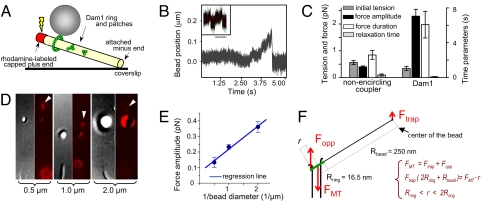
Fig. 1.
Quantitative analysis of the force-transducing attachments. (A) A schematic of the experimental system. Position of beads bound to the GDP-MT walls was followed with QPD before and after induction of MT disassembly. (B) Unprocessed QPD records of representative signals from a Dam1-coated bead in the presence of soluble Dam1 and a streptavidin-coated bead (Inset) relative to the center of the laser trap. (C) Mean values for the four parameters that describe force signals (see ref. 9 for details). (Left) 0.5-μm streptavidin-coated beads assayed with biotinylated MTs (n = 35). (Right) Wild type Dam1-coated beads with soluble Alexa488–Dam1 (n = 26). (D and E) Force measurements carried out with streptavidin-coated beads: 0.5 μm (n = 45), 1 μm (n = 81; includes data from ref. 9), 2 μm (n = 35). Photos show DIC images of representative beads attached to MT walls and their fluorescence images (red) taken during dissolution of the rhodaminated GMPCPP caps (arrowheads). Graph shows median values for force amplitudes for these beads. Trap stiffness, ≈0.008 pN/nm. (F) Drawing (not to scale) of the forces from MT depolymerization and the laser trap. The fulcrum is at ring's edge on the bead-distal side of the MT; because of the ring's tilt, FMT is likely to act slightly off the MT axis, so its lever arm r is defined by a range. The bead (not shown) is 15-times larger than the ring, so a relatively small trapping force can stall the MT-disassembly-driven movement of the ring.
Shortly after a pulse of light removes the stable cap, allowing the MT to shorten, the MT-associated, 0.5-μm bead displays a brief movement toward the MT minus end as depolymerization passes by [supporting information (SI) Movie S1]. The displacement observed, and thus the force exerted on the bead, differed depending on the coupling. If the binding was to only one side of the wall (probably to two PFs), the average force was ≈0.4 pN (Fig. 1B Inset). We then coated beads with bacterially expressed Dam1 complexes labeled with Alexa488 fluorophores (12, 18). When the bead was MT bound under conditions previously identified to promote the formation of MT-encircling oligomers of Dam1 (18), the measured force was 5.7-fold larger. This value is close to the 6.5-fold expected from the action of 13 PFs, rather than 2 PFs (Fig. 1 B and C). The Dam1-coupled forces were slow to develop and lasted significantly longer than those obtained with nonencircling couplers. Because a longer-lasting force signal corresponds to a slower rate of MT depolymerization (9), Dam1 rings appear to retard MT disassembly. The termination of force development is also different in these two systems. In ≈50% of our previous measurements with nonencircling couplers, depolymerization-dependent forces decreased gradually (0.8 ± 0.2 s), presumably because of the randomness of which PFs depolymerized first, bead-associated or not (9). With Dam1 couplers, the relaxation was always <30 ms (Fig. 1C), as one would expect if the PFs were held together by a ring that finally fell off the MT end.
These forces are significantly less than those predicted by our model of a Dam1 ring where the load is attached uniformly (19). The forces recorded here were, however, applied to the Dam1 complex asymmetrically, via a laterally attached bead (Fig. 1A). We have previously suggested that, in this arrangement, the trapping force creates a torque, so the force measured at the center of the bead is smaller than the force actually exerted by the bending PFs; the ratio of these forces should depend on the diameter of the bead (9). We have tested this supposition by using streptavidin-coated beads with three diameters, all bound to biotinylated MTs and measured under the same conditions (Fig. 1D). The force signals for these beads were inversely proportional to their radii (Fig. 1E), strongly supporting the validity of our mechanical interpretations of this experimental system.
We can apply this interpretation to a ring-based coupler with the following logic. When a bead is attached to an MT wall via a Dam1-ring, the torque from the trapping force should increase the ring's tilt (Fig. 1F). In this configuration, the bead's movement will stall when the total MT-parallel force and net torque are zero. The trapping force, Ftrap, at the center of the bead, and the opposing force that is exerted on the ring by the MT, Fopp, are compensated by the sum of forces exerted on the ring by the bending PFs. The component of this sum along the MT axis, FMT, acts in a direction opposite to the trapping force. Thus, bead movement will stall when FMT = Ftrap(Rbead + 2Rring) /r, where Rring and Rbead are the radius of the ring and bead, respectively; r is a distance between the fulcrum and the site of action of FMT. Because
the trapping force that can compensate the MT-disassembly force is roughly 13-fold smaller. Following this logic, the maximum force with which PFs can push [≈80 pN (11)] in our setup should be stalled by a trapping force of ≈6 pN. The average observed trapping force with Dam1 beads was 2.3 pN (range 0.5–5 pN), a little less than half the maximum predicted. This estimate implies that the ring experienced on average ≈30 pN from bending PFs.
With this geometrical correction, the observed force suggests 40% efficiency in energy transduction by a Dam1 ring. One reason for this low value is that we have not truly stalled MT depolymerization; the bead followed the depolymerizing MT end for a short distance in the trap (<250 nm), experiencing an ever stronger force, and then detached abruptly (Fig. 1B). This behavior suggests that the ring detached from the MT or fell apart before or immediately after the stalling force had been reached. Such behavior may be more common when a load is applied asymmetrically, but the result emphasizes the importance of a coupler design that maximizes processivity, not just efficiency of energy transduction. In our model, processivity is provided by comparatively tight ring–MT binding, so we sought ways to examine this idea directly.
Dam1–19 Complexes with Partially Truncated Protrusions Are Poor Force-Transducing Couplers.
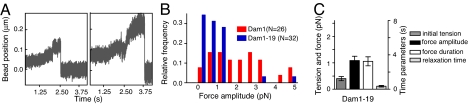
Dam1 heterodecamers associate with the MT wall through inward-directed protrusions (16, 17). The Dam1–19 allele, a deletion of the C terminus of Dam1p, can still oligomerize into rings around MTs but lacks much of the protrusion from each Dam1 heterodecamer, so these complexes bind MT walls less strongly than wild type (12, 17). In the presence of soluble mutant protein, the Dam1–19-coated beads formed stable attachments with the GDP-containing parts of MT walls, just as beads did with wild type Dam1. The Dam1–19 beads moved readily with shortening MTs, showing little effect on disassembly rate (24 ± 5 μm/min, n = 12). We then followed their motions with a stationary laser trap. The overall features of the observed signals and the range of force amplitudes from beads coated with Dam1–19 and wild type Dam1 were quite similar (Fig. 2 A and B). As with wild-type protein, Dam1–19 beads detached from the MT ends before we could see a clear stalling of depolymerization. Some of the Dam1–19 signals reached 5 pN, the largest force we detected with wild type Dam1. The frequency of large signals was low, however, so the average amplitude of the forces with Dam1–19 was one-half that of wild type Dam1, although still significantly higher than that seen with nonencircling couplers (Figs. 1C and 2C). Thus, although Dam1–19 rings can transduce a large force, they frequently fail to do so because they lose their attachment to the shortening MT ends, under comparatively small loads. We concluded that the C-terminal tail of Dam1 protein plays an important role in coupling the Dam1 ring to MTs by contributing to stronger bonding.
Fig. 2.
Force-coupling via Dam1–19 mutant complexes. (A) Typical (Left) and one of the largest (Right) signals obtained with Dam1–19 coated beads in the presence of soluble Dam1–19. Overall features of these signals are similar to those seen with wild-type protein, but the quantitative characteristics are different. (B) Histogram of force amplitudes shows the same range for wild-type and mutant proteins, but most of the Dam1–19 signals were smaller. (C) Average characteristics of Dam1–19 signals. The experiments with different beads were done under similar conditions, including initial tension that was applied to the beads.
Tracking Dam1 Rings Slow the Rate of MT Shortening, Even in the Absence of an External Load.
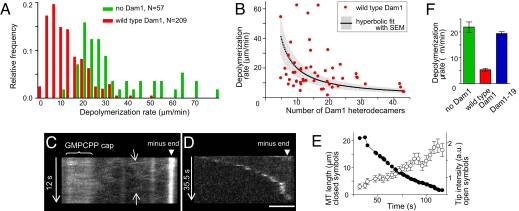
If wild type Dam1 subunits bind tightly to tubulin in the MT wall, then the ring should not slide freely and polymer shortening should be slowed, even with no load attached (19). To study the effect of Dam1 rings on MT dynamics, we added Alexa488–Dam1 complexes to our segmented MTs. We have previously shown that the majority of Alexa488–Dam1 dots that form on MTs in this system comprise single rings or stacks of two rings, but only single rings will track shortening MT ends, moving steadily over the segments of MTs that are free from other complexes (18). We measured the rate of these motions on 117 MTs. The resulting distribution had noticeable asymmetry, with a major peak at 5.4 ± 0.5 μm/min (Fig. 3A). This rate is four times slower than MT shortening in the absence of Dam1 complexes (22 μm/min). This significant retardation of MT disassembly by the end-tracking Dam1 rings suggests that the rings adhere strongly to the MT wall and must be pushed along by the depolymerization process.
Fig. 3.
MT end-tracking by wild-type and mutant Dam1 complexes. (A) Histograms of MT depolymerization rates. (B) Brightness of the tip-tracking Alexa488–Dam1 complexes (soluble concentration 1–2 nM) was normalized with the intensity of a single Alexa488 fluorophore (18) to plot the number of Dam1 subunits vs. their tracking rates. Thermal motion of MTs is the major contributor to this system's noise. (C and D) Lateral diffusion and end-tracking by Alexa488–Dam1–19. The oblique lines on the kymograph in C (arrows) indicate thermal diffusion. (Scale bar, 2 μm.) (E) Typical kinetics of the end tracking and changes in brightness of the tip-associated Dam1–19 complexes. (F) Peak values for the histograms in A and for Dam1–19 rates were determined with Rayleigh fitting (18).
The observed extent of depolymerization slowing is predicted by our model when the strength of MT-Dam1 interaction is 13–15 kBT (19), a value that is consistent with the biochemical affinity determined in ref. 12 but much lower than that measured in ref. 21. The latter estimate of 19 kBT would cause such strong ring–MT adhesion that bending PFs could not displace such rings; because the depolymerization motor is fueled by GTP hydrolysis, there is an upper limit to the force that MT shortening can produce (11, 19).
Dam1 Complexes with Fewer Subunits Slow MT Depolymerization Less.
Approximately 25% of the wild type Dam1 complexes in our experiments moved faster than the bulk of the distribution, as shown by the shoulder in Fig. 3A. This finding could be partially caused by the known variability in the rates with which individual MTs disassemble (24) (Fig. 3A). Consistent with this supposition, the rate of Dam1 movement on any one MT was remarkably constant: The ratio of speeds for two segments of Dam1 tracking on the same MT [e.g., the intervals before and after a moving Dam1 ring encountered another ring (18)] was 1.1 ± 0.1 (n = 67). Quantitative analysis showed, however, that the variability of MT disassembly rates could not account for the distribution seen for Dam1, so we sought additional factors.
The faster rates of MT end-tracking could correspond to Dam1 patches, i.e., oligomers with fewer subunits than in a full ring. Dam1 patches can bind the MT walls and track shortening MT ends (18, 21), but these smaller oligomers might cause less or no slowing of MT shortening. We tested this idea by seeking correlations between the brightness of tracking complexes and the rate with which they track shortening MTs. Complexes with fewer subunits slowed MT disassembly less (Fig. 3B). Thus, Dam1 patches and incomplete rings can account for the faster tracking rates in the asymmetric distribution in Fig. 3A. Importantly, the fact that smaller Dam1 complexes slow MT depolymerization less than bigger ones supports the notion of strong Dam1–tubulin bonds; if Dam1 rings could slide freely on the MT surface, end-tracking rates should be insensitive to the number of tracking subunits.
Retardation of MT Depolymerization by a Dam1 Ring Depends on the C Terminus of the Dam1 Protein.
The role of protein arms, which connect Dam1 heterodecamers with the MT, in ring motility is controversial. In the forced walk mechanism, they ensure strong and specific Dam1-MT bonding that slows MT disassembly (19). In another model, rings are thought to bind the MT wall with no “specific footprint,” and these protrusions facilitate ring “free gliding” (14, 17, 25). We tested these predictions by using the Dam1–19 complexes.
The decoration of MTs walls by Alexa488-labeled Dam1–19 was similar to that by wild-type protein, although more mutant than wild-type complexes showed rapid diffusion on the MT wall (Fig. 3C and Movie S2). Like wild type Dam1, the mutant complexes tracked the ends of shortening MTs (Fig. 3D), but the details of their motilities were distinct. Wild type Dam1 tracked ends with constant speed and brightness (18), but Dam1–19 complexes accumulated fluorescence as the MT shortened, and depolymerization slowed as fluorescence brightened (Fig. 3E). These results are consistent with the notion that Dam1–19 rings adhere less strongly to the MT wall than do wild-type rings (12). Furthermore, the rate of Dam1–19 MT end-tracking was 19.4 ± 0.7 μm/min, considerably faster than that of wild type Dam1 (Fig. 3F). Thus, the protrusions from wild type Dam1 inhibit tracking rather than facilitate it.
Beads Transported by the Dam1 Rings Oscillate Irregularly.
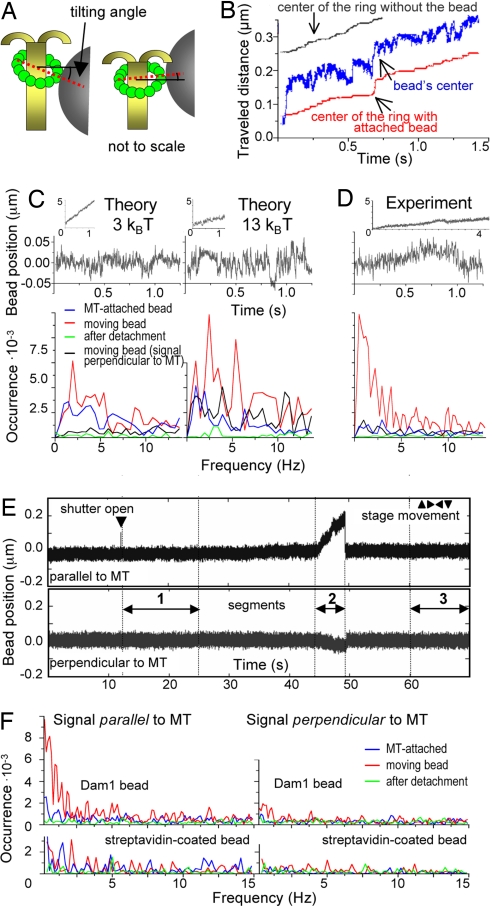
Previous calculations have shown that if a ring's protrusions are flexible and bind to specific sites on tubulins in the wall of helical MT, the minimum energy binding occurs when the plane of the ring is tilted relative the MT axis (19). When a ring follows the end of a depolymerizing MT, its orientation must change as it forms these preferred configurations (Fig. 4A and Fig. S1). We have tested these predictions by looking for oscillations in the motions of ring-associated beads. As the plane of the ring changes, the center of the bead should move with the same average speed as the ring's center, but the amplitude of the bead's oscillations will be significantly larger, because it is 15 times bigger than the ring (Movie S3). The instantaneous rate of bead movement can exceed the rate of ring motion, and the bead can even occasionally move backwards. Thus, the motion of ring-associated beads with shortening MT ends should be jagged (Fig. 4B).
Fig. 4.
Oscillatory motions of the Dam1-ring associated beads. (A) The drawing illustrates the wobbling of the ring and the resulting changes in bead position. (B) Model results for Dam1–tubulin energy 13kBT. (C and D) Comparisons of theoretical and experimental results for 0.5-μm beads coupled to a Dam1 ring in the presence of soluble Dam1. The rising parts of the force signals (Insets, see also Fig. 1B) are not smooth, as beads pause and even reverse their motions. The spectral characteristics of these irregular oscillations were analyzed after fitting these raw QPD signals with lines whose slopes describe the linear rates of bead movement and then subtracting this component to obtain the variable parts (Upper) and their Fourier transformations (Lower). The exact positions of the peaks and their amplitudes were different for repetitions of the experiments and theoretical calculations, because of the stochasticity of the Dam1 ring–MT system (e.g., compare D, F, and Fig. S2). As a control, similar transformations were done for beads attached to MTs or after their detachment. (E) Full, unprocessed QPD signal (for bead in Movie S1), which shows typical positions of the segments used for spectral analysis. (F) Experimental spectra of movements of another bead attached to a Dam1 ring and a streptavidin-coated bead. Dam1 beads display discrete but highly variable peak frequencies during forced movement parallel to MT, because the attached ring wobbles as it transits stochastically between energetically preferred configurations on MT lattice (19).
When these tracings are analyzed with Fourier transformation, the line spectrum should show discrete peaks. The distribution of frequencies, however, is expected to be disordered, because the bead's oscillations do not mirror those of the ring exactly and are further blurred by thermal noise (SI Text). Such features will also depend on the strength of the Dam1 ring binding to MTs, because weaker Dam1–tubulin bonds should, on average, create smoother motions. Fig. 4C compares predictions by our model for beads moving under a depolymerization force with the help of rings that bind to tubulin with 3 or 13 kBT per bond. The 3-kBT ring with a bead moves faster (Fig. 4C Upper Insets), and the amplitude of the bead's oscillations in the low frequency range is, on average, one-half that of the 13-kBT ring–bead pair.
To compare these results with the behavior of a real ring–bead system, we followed the motions of wild type Dam1-coated beads in a weak stationary laser trap (stiffness 0.010–0.025 pN/nm). MT depolymerization did not drive the beads smoothly; there were visible irregularities 20–30 nm in amplitude and with occasional excursions of as much as 50 nm (Fig. 4 D and E). We detected complex oscillations in 17 of 26 beads (signals parallel to MT axis that gave Fourier peaks in a low frequency range with amplitudes >4 × 10−3). General features of the observed Fourier spectra are highly similar to the predictions. For example, oscillations are both predicted and observed in a 2- to 20-Hz range (Fig. S2). They are seen mostly with beads that are moving under the depolymerization force; thermal oscillations of beads attached to the stable MT or of those that are simply trapped with a laser beam (times before and after MT-induced bead movement in Fig. 4E) are smoother, and the peak amplitudes are less than one-tenth as big. In both prediction and experiment, oscillations along the MT axis are significantly greater than in a perpendicular direction.
Furthermore, these nonthermal motions are seen only under conditions where Dam1 is an encircling coupler, not when beads are coupled to depolymerizing MTs via other couplers (Fig. 4F). Only ≈26% of Dam1–19 signals showed characteristic oscillations, significantly less frequently than in experiments with wild-type protein, as one would expect based on their MT affinities. Together, these findings strongly support the conclusion that most beads coupled to MTs in the presence of soluble Dam1 were bound by ring-shaped assemblies that moved in a fashion that resembled our theoretical description.
Discussion
Understanding the mechanisms of the force coupling between kinetochores and the ends of spindle MTs is one of the long-standing goals in cell biology. The recent discoveries that Dam1 heterodecamers can oligomerize into rings and couple MT shortening to cargo motility have sparked significant interest in the biomechanical design of this coupler. There are at least two features of its function that await explanation: how it converts the energy of tubulin depolymerization into chromosome motions and how it maintains stable attachment during processive, MT-driven motions.
Toward a Comprehensive Understanding of Dam1 Ring-Dependent Coupling.
Several properties of Dam1 complexes have so far been studied: (i) their affinity for MTs, (ii) diffusion on the MT wall, (iii) MT-end tracking and other kinetic properties, such as the collection and shedding of moving complexes, and (iv) tracking and force transduction by Dam1-coated beads. All these phenomena reflect some aspects of the interactions between Dam1 heterodecamers and the MT wall, so their quantitative characteristics should correspond. For example, if Dam1 heterodecamers bind strongly to the MT wall, it is highly unlikely that the rings can exhibit fast diffusion. Likewise, rings that diffuse freely on the MT wall should not significantly affect dynamics of the MT end, unless there are justified assumptions about the ring's special behavior near flared PFs. More accurate statements, however, require a mathematic model with which to analyze the quantitative relationships among these various parameters. Even though the data now available are likely to be incomplete, such an approach should help to identify correlations and inconsistencies, as well as to focus future studies on the most critical issues.
There are competing views on various aspects of Dam1-dependent motility (12, 14, 16, 17, 19–21, 25, 26), but the systematic analysis of all experimental data are just starting. We have previously developed a theoretical model of the Dam1 ring coupling that was founded on a single key assumption: The Dam1 heterodecamer has only one specific binding site with tubulin (19). We make no special assumptions about the directionality of Dam1-tubulin interactions: A Dam1 heterodecamer must expend the same energy to break its bond with tubulin, regardless of whether Dam1 will detach from the MT completely or move to the next binding site. By examining ring behavior with varying model parameters and comparing these results with experimental data, we have arrived at a cohesive and consistent description of the biomechanical design of this coupler, summarized below.
We propose that a Dam1 ring moves during MT depolymerization through a forced walk mechanism, a mode of motility exhibited specifically by an encircling coupler with a wide diameter and flexible extensions that bind strongly to the tubulin lattice. We presume that Dam1-tubulin affinity is high (most likely 13–15 kBT), based on biochemical measures (12), the absence of ring accumulation at a shortening MT end (18), the retardation of MT disassembly by Dam1 rings (Fig. 3A), and reduced retardation of MT shortening by Dam1 patches (Fig. 3B), as well as by a mutant Dam1 in which the formation of rings is reduced (18) and by a mutant with partially truncated protrusions (Fig. 3F).
Although small, nonencircling Dam1 oligomers can diffuse fast on the MT wall (18, 21), Dam1 rings show negligible diffusion (D ≤ 10−13 cm2/s) (18). This coefficient implies that the energy of Dam1-tubulin binding is >7kBT (19), which is consistent with the estimate based on the rate of Dam1 tracking. Furthermore, our model describes well the amplitudes of forces experienced by beads associated with wild-type and mutant Dam1–19 rings (Figs. 1 and 2) and the jagged character of bead motions (Fig. 4). Model predictions of a ring's tilting on a stable MT and the lack of ring rotation during processive motion are supported by structural data (12, 13, 19) and by the translational motions of ring-attached beads (Fig. S1) (18). The consistencies between model and experiment indicate that these views of the biomechanical design of the Dam1 ring are not far from reality.
Why Rings?
Since their discovery, this question has been attracting significant attention (12, 27). It became even more interesting with the finding that ringless Dam1 assemblies can also track shortening MTs. Although much remains to be learned about the coupling properties of Dam1 patches, we see three reasons to favor the view of Dam1–ring coupling as more efficient and reliable than smaller oligomeric forms: (i) Couplers that bind fewer than 13 PFs should collect less force (Fig. 1); (ii) PFs that are not held by the coupler may disassemble faster than the coupler-bound PFs, thereby increasing MT flaccidity and cargo detachment (9, 23, 28); and (iii) Couplers with fewer MT-binding subunits are expected to have weaker attachment to the MT (19). Model calculations depicted in Movie S3 provide visual demonstrations for some of these points. When the tracking ring stalls in this sequence, the flared PFs continue their shortening and their splitting creeps into the MT wall downstream from the ring. Although the MT then begins to loose its integrity, the strongly attached ring hangs on and impedes further PF splaying. When opportunity arises, the ring swiftly moves forward and the bead continues its motion. A coupler like this would be particularly useful in budding yeast, where each kinetochore is stably attached to only one MT.
Future Directions.
There are several issues that must be addressed before the above views can be firmly established. First, it is necessary to determine the energy potentials of Dam1–tubulin and Dam1–Dam1 interactions. This analysis should also include various Dam1 mutants, such as Dam1–19. We found that Dam1–19 complexes show faster diffusion and tracking and more abundant collection of complexes but smaller forces and oscillations than wild type. All these findings are consistent with expectations based on our model, but some of these effects are stronger than predicted. Indeed, the fourfold reduction in Dam1–tubulin affinity in Dam1–19 complexes (12) formally corresponds to <2-kBT change in binding energy, so the rate of Dam1–19 tracking was expected to rise to 12 μm/min, whereas we measured 19 μm/min. This increased effect might be a result of a reduced ring oligomerization of Dam1–19, relative to wild type Dam1. This supposition is supported by structural data (17) and by our observation that Dam1–19 beads exhibited longer dissociation times from MT ends (Fig. 2C and Fig. S3), a feature that we attribute to the presence of nonencircling complexes. These findings highlight the importance of creating tests to accurately compare the efficiency of ring formation for different Dam1 complexes.
It will also be necessary to determine accurately the diffusion coefficient for the Dam1 ring. Measuring Dam1 diffusion on MTs that project into solution is technically challenging, because their thermal motions impede the visualization of dim fluorescent dots. This analysis, however, is one of the most direct ways to determine the contribution of a ring's thermal motions to MT-end tracking. Other important unresolved issues include the analysis of interaction site(s) between Dam1 and tubulin and examination of the rigidity of both the ring's extensions and its core. Close comparison of the results of these approaches with rigorous modeling that is based on explicit assumptions will undoubtedly help us learn how the Dam1 ring works.
Methods
All reagents and experimental conditions were as described in ref. 18. Laser trapping experiments were carried out as in ref. 9 with instruments described in SI Text and Fig. S4. In all of our experiments, the QPD was sampled at 4 kHz without additional filtering or processing. Our model for MT depolymerization is based on ref. 29 with modifications and model parameters as in ref. 19. Theoretical description of the motion of ring-associated beads is provided in the SI Text.
Supplementary Material
Acknowledgments.
We thank M. Molodtsov, J. Welburn, P. Grissom, J. Fang, S. Gustafson, S. Karamzin, E. Salova, and A. Zheleznjakov for technical assistance; M. Porter for the kind gift of Chlamydomonas axonemes; and A.I. Vorobjev for support. This work was supported by National Institutes of Health Grant GM033787 (to J.R.M.), Civilian Research and Development Foundation Grant CGP2006B#2863 (to J.R.M. and F.I.A.), and Molecular and Cellular Biology grant from the Russian Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807859105/DCSupplemental.
References
- 1.Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franco A, Meadows JC, Millar JB. The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J Cell Sci. 2007;120:3345–3351. doi: 10.1242/jcs.013698. [DOI] [PubMed] [Google Scholar]
- 7.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: A time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller-Reichert T, Chretien D, Severin F, Hyman AA. Structural changes at microtubule ends accompanying GTP hydrolysis: Information from a slowly hydrolyzable analogue of GTP, guanylyl (α,β)methylenediphosphonate. Proc Natl Acad Sci USA. 1998;95:3661–3666. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- 10.Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- 11.Molodtsov MI, Grishchuk EL, Efremov AK, McIntosh JR, Ataullakhanov FI. Force production by depolymerizing microtubules: A theoretical study. Proc Natl Acad Sci USA. 2005;102:4353–4358. doi: 10.1073/pnas.0501142102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westermann S, et al. Formation of a dynamic kinetochore–microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 13.Miranda JJ, De WP, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 14.Westermann S, et al. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 15.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore–microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miranda JJ, King DS, Harrison SC. Protein arms in the kinetochore–microtubule interface of the yeast DASH complex. Mol Biol Cell. 2007;18:2503–2510. doi: 10.1091/mbc.E07-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang HW, et al. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- 18.Grishchuk EL, et al. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Efremov A, Grishchuk EL, McIntosh JR, Ataullakhanov FI. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proc Natl Acad Sci USA. 2007;104:19017–19022. doi: 10.1073/pnas.0709524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Onuchic JN. A driving and coupling “Pac-Man” mechanism for chromosome poleward translocation in anaphase A. Proc Natl Acad Sci USA. 2006;103:18432–18437. doi: 10.1073/pnas.0608962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gestaut DR, et al. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci USA. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franck AD, et al. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gildersleeve RF, Cross AR, Cullen KE, Fagen AP, Williams RC., Jr Microtubules grow and shorten at intrinsically variable rates. J Biol Chem. 1992;267:7995–8006. [PubMed] [Google Scholar]
- 25.Sandall S, Desai A. When it comes to couple(r)s, do opposites attract? Nat Struct Mol Biol. 2007;14:790–792. doi: 10.1038/nsmb0907-790. [DOI] [PubMed] [Google Scholar]
- 26.Gardner MK, Odde DJ. Dam1 complexes go it alone on disassembling microtubules. Nat Cell Biol. 2008;10:379–381. doi: 10.1038/ncb0408-379. [DOI] [PubMed] [Google Scholar]
- 27.Salmon ED. Microtubules: A ring for the depolymerization motor. Curr Biol. 2005;15:R299–R302. doi: 10.1016/j.cub.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 28.McIntosh JR, Grishchuk EL, West RR. Chromosome-microtubule interactions during mitosis. Annu Rev Cell Dev Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- 29.Molodtsov MI, et al. A molecular-mechanical model of the microtubule. Biophys J. 2005;88:3167–3179. doi: 10.1529/biophysj.104.051789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.