Abstract
Background
Germline mutations in the Chek2 kinase gene (CHEK2) have been associated with a range of cancer types. Recently, a large deletion of exons 9 and 10 of CHEK2 was identified in several unrelated patients with breast cancer of Czech or Slovak origin. The geographical and ethnic extent of this founder allele has not yet been determined.
Participants and methods
We assayed for the presence of this deletion, and of three other CHEK2 founder mutations, in 1864 patients with prostate cancer and 5496 controls from Poland.
Results
The deletion was detected in 24 of 5496 (0.4%) controls from the general population, and is the most common CHEK2 truncating founder allele in Polish patients. The deletion was identified in 15 of 1864 (0.8%) men with unselected prostate cancer (OR 1.9; 95% CI 0.97 to 3.5; p = 0.09) and in 4 of 249 men with familial prostate cancer (OR 3.7; 95% CI 1.3 to 10.8; p = 0.03). These ORs were similar to those associated with the other truncating mutations (IVS2+1G→A, 1100delC).
Conclusion
A large deletion of exons 9 and 10 of CHEK2 confers an increased risk of prostate cancer in Polish men. The del5395 founder deletion might be present in other Slavic populations, including Ukraine, Belarus, Russia, Baltic and Balkan countries. It will be of interest to see to what extent this deletion is responsible for the burden of prostate cancer in other populations.
The Chek2 kinase gene (CHEK2) is a key component of the DNA damage signalling pathway. Activation of this protein in response to DNA damage prevents cellular entry into mitosis.1,2 Germline mutations in CHEK2 have been associated with a range of cancer types, in particular cancer of the breast and the prostate.3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20 Most predisposing CHEK2 alleles are protein truncating, but a missense variant (I157T) has also been associated with cancer. Recently, a large deletion in CHEK2 was identified in several unrelated patients with breast cancer of Czech or Slovak origin.21 Haplotype analysis confirmed that the mutation had a single source.21 The geographical and ethnic extent of this founder allele has not yet been determined. We estimated the frequency of this deletion in the Polish population and asked whether it is associated with an increased risk of prostate cancer. Poland is well suited for association studies because of the relative genetic homogeneity of the population.
Previously, we genotyped 690 Polish men with prostate cancer for three common CHEK2 mutations.9 We found that the risk of prostate cancer was increased for carriers of both the truncating mutations (IVS2+1G→A and 1100delC) and the missense mutation I157T. We have now extended our patient series to include 1864 incident cases of prostate cancer and have genotyped these for the presence of all four CHEK2 founder mutations, including the large deletion.
Patients and methods
Patients
We studied prostate cancer cases diagnosed between 1999 and 2005 in 13 centres situated throughout Poland. This study was initiated in Szczecin, Poland, in 1999 and was extended to include Białystok and Olsztyn in 2002, and Opole in 2003. Nine other centres began recruiting patients in 2005 (Koszalin, Gdańsk, Lublin, Łódź, Warszawa, Wrocław, Poznań, Rzeszów and Sucha Beskidzka). Patients were recruited from the urology services of the contributing hospitals and were unselected for age or family history. All men with invasive prostate cancer were invited to participate. In all, 1864 unselected patients with prostate cancer were enrolled, representing 78.3% of all patients with prostate cancer who were invited to participate in the study. The mean age at diagnosis was 67.3 (range 43–92 years). A family history of prostate cancer (and of other cancers) in relatives was obtained from each participant. Of them, 249 (13.4%) patients had ⩾1 first‐degree or second‐degree relatives diagnosed with prostate cancer (familial cases). The ethics committee of Pomeranian Medical University, Szczecin, Poland, approved the study.
Controls
To estimate the frequency of the Polish founder mutations in the general population, three control groups were combined. The first group consisted of 2183 newborn children from 10 cities in Poland (Szczecin, Białystok, Gorzów, Katowice, Wrocław, Poznań, Opole, Łódź and Rzeszów) between 2003 and 2006. Samples of cord blood from unselected infants were forwarded to the study centre in Szczecin. The second control group included healthy adult patients (1079 women and 817 men) of three family doctors practising in the Szczecin region. These people were selected randomly from the patient lists of family doctors. The third control group consisted of 1417 young adults (705 women and 712 men) from Szczecin who submitted blood for paternity testing.
Search for CHEK2 large deletion in the Polish population
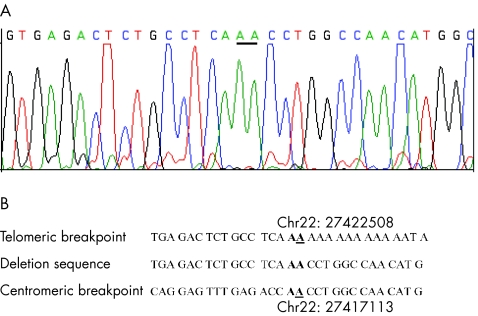
Three samples of pooled DNA, each including pooled DNA from about 500 people from Poland, were amplified with primers described previously for the detection of the deletion of exons 9 and 10 in Czechs and Slovaks.21 Ninety unpooled DNA samples representing individual controls were also included. Short extension times (2 min) were applied during polymerase chain reaction (PCR) to amplify only a short allele containing the large deletion. A single PCR product of about 1.3 kb was amplified from all samples with the pooled DNA. The short product was seen in 1 of 90 DNA samples from single patients. The PCR products from all positive cases were sequenced. The short PCR products included the deletion of exons 9 and 10, and the deletion breakpoints were characterised at a nucleotide level (fig 1). This deletion removes 5395 bp (del5395), incuding exons 9 and 10 of CHEK2.
Figure 1 A 5395 bp deletion of exons 9 and 10 of CHEK2 detected in the Polish population. A. Sequencing chromatogram of PCR product containing the deletion. B. Localization of deletion breakpoints at chromosome 22 in Alu‐repeats.
Genotyping
Two primer pairs were designed specifically for genotyping of a large deletion of exons 9 and 10 in a multiplex PCR. The first pair (CHLdel2F 5′‐TGT AAT GAG CTG AGA TTG TGC‐3′; CHLc2R 5′‐CAG AAA TGA GAC AGG AAG TT‐3′) flanked breakpoint site in intron 8. The second pair (CHLdelR 5′‐GTC TCA AAC TTG GCT GCG‐3′; CHLcF 5′‐CTC TGT TGT GTA CAA GTG AC‐3′) flanked breakpoint site in intron 10. In mutation‐negative cases, only two PCR fragments of 379 and 522 bp were amplified from the wild‐type allele. The forward primer of the first pair and the reverse primer of the second pair amplified an additional PCR product of 450 bp in mutation‐positive cases. In all multiplex PCR‐positive samples, the presence of the deletion was confirmed by sequencing. Experimental conditions of the multiplex PCR are available on request.
The other three mutations in CHEK2 (IVS2+1G→A, 1100delC and I157T) were genotyped as described previously.9 These variants are detected by allele‐specific oligonucleotide‐PCR or restriction fragment length polymorphism‐PCR analyses. In all reaction sets, positive and negative controls (without DNA) were used. All PCRs or enzymatic digestions were carried out under a layer of mineral oil.
Statistical analysis
The prevalence of each of the three CHEK2 alleles in patients and in controls was compared. Odds ratios (ORs) were generated from 2×2 tables and statistical significance was assessed using Fisher's exact test. ORs were used as estimates of relative risk.
Results
A large deletion of exons 9 and 10 of CHEK2 was detected in 24 of 5496 (0.4%) controls from the general population in Poland. This was the single most common protein‐truncating variant observed in the population (the IVS2+1G→A variant was seen 22 times and the 1100delC variant was seen 12 times). The breakpoints of the deletion were verified at a nucleotide level. All patients with a deletion carried the same deletion of 5395 bp of exons 9 and 10. Sequencing also confirmed that the deletion was in the functional copy of CHEK2 at chromosome 22 (not in pseudogenes located elsewhere).
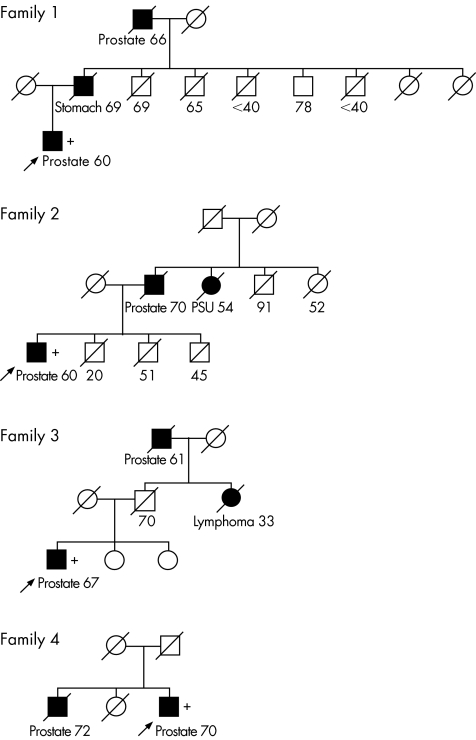
The 5395 deletion was present in 15 of 1864 (0.8%) patients (OR 1.9, p = 0.09). Of these 1864 patients, 249 (13%) men had a family history of prostate cancer. The deletion was found in four familial cases (OR 3.7, p = 0.03). Each of the deletion‐positive families contained two men affected with prostate cancer (fig 2).
Figure 2 Pedigrees of four familial prostate cancer cases with genomic CHEK2 deletion from a series of 1864 unselected prostate cancer cases. Arrows indicate probands. Squares denote males; circles denote females. The cancer type and age at diagnosis are shown near all blackened symbols, which denote patients affected with cancer. Current age (or the age of death) is shown below the blank symbols, which denote family members unaffected with cancer. All symbols with a diagonal denote deceased individuals. A (+) near the symbol indicates the presence of the deletion. PSU, primary cancer site unknown.
One of four founder CHEK2 mutations was identified in 184 of 1864 (9.9%) men with prostate cancer, including I157T (142 times), the del5395 deletion (15 times), IVS2+1G→A (15 times) and 1100delC (14 times). Overall, OR for prostate cancer given a CHEK2 mutation was 1.8 (95% confidence interval (CI) 1.5 to 2.1; p<0.001; table 1). OR was higher for men with a truncating mutation (OR 2.3; 95% CI 1.5 to 3.4) than for men with a missense mutation (OR 1.6; 95% CI 1.3 to 2.0), but the difference was not significant. A CHEK2 mutation was present in 17% of men with familial prostate cancer (OR 3.3; 95% CI 2.3 to 4.6; p<0.001). Again, OR was higher for carriers of a truncating mutation (OR 4.7; 95% CI 2.5 to 9.0) than for carriers of a missense variant (OR 2.7; 95% CI 1.8 to 4.1).
Table 1 Prevalence of mutations in the Chek2 kinase gene in patients with prostate cancer and controls, with corresponding odds ratios.
| Mutation case | Frequency (%) | OR (95% CI) | p Value |
|---|---|---|---|
| CHEK2 mutation | |||
| Controls | 321/5496 (5.8) | 1.0 | |
| Unselected cases | 184/1864 (9.9) | 1.8 (1.5 to 2.1) | <0.001 |
| Familial cases | 42/249 (16.9) | 3.3 (2.3 to 4.6) | <0.001 |
| Large deletion | |||
| Controls | 24/5496 (0.4) | 1.0 | |
| Unselected cases | 15/1864 (0.8) | 1.9 (0.97 to 3.5) | 0.09 |
| Familial cases | 4/249 (1.6) | 3.7 (1.3 to 10.8) | 0.03 |
| 1100delC | |||
| Controls | 12/5496 (0.2) | 1.0 | |
| Unselected cases | 14/1864 (0.8) | 3.5 (1.6 to 7.5) | 0.002 |
| Familial cases | 3/249 (1.2) | 5.6 (1.6 to 19.9) | 0.02 |
| IVS2+1G→A | |||
| Controls | 22/5496 (0.4) | 1.0 | |
| Unselected cases | 15/1864 (0.8) | 2.0 (1.05 to 3.9) | 0.052 |
| Familial cases | 5/249 (2.0) | 5.1 (1.9 to 13.6) | 0.002 |
| Any truncating mutation | |||
| Controls | 58/5496 (1.1) | 1.0 | |
| Unselected cases | 44/1864 (2.4) | 2.3 (1.5 to 3.4) | <0.001 |
| Familial cases | 12/249 (4.8) | 4.7 (2.5 to 9.0) | <0.001 |
| I157T | |||
| Controls | 264/5496 (4.8) | 1.0 | |
| Unselected cases | 142/1864 (7.6) | 1.6 (1.3 to 2.0) | <0.001 |
| Familial cases | 30/249 (12.0) | 2.7 (1.8 to 4.1) | <0.001 |
Frequency is calculated as the ratio of number of carriers to total number.
Two patients and one control had both truncating and missense mutations.
Two patients and two controls were homozygotic for the I157T variant.
The frequencies of CHEK2 alleles (including the large deletion) in the control group were similar in newborns and adults, and in men and women. Also, we found no difference in CHEK2 mutation frequency between newborns from different counties of Poland.
Discussion
A predisposing CHEK2 mutation was present in 10% of Polish patients with prostate cancer. On average, men who carry a mutation have a 1.8‐fold increased risk of prostate cancer, and we estimate that mutations in CHEK2 are responsible for about 7% of all prostate cancer cases in the country. The large deletion of exons 9 and 10 in CHEK2, first described in Czechs and Slovaks, is also a founder mutation in Poland. By our estimate, the length of this deletion is 5395 bp, and not 5567 bp as described in the original report.21
The deletion of exons 9 and 10 was the single most commonly observed protein‐truncating mutation in the Polish population. We observed the deletion in 0.4% of the Polish controls. The deletion was originally reported to be associated with breast cancer risk in the Czech Republic and in Slovakia, but prostate cancer was not studied there. In that study, the deletion was not observed among controls, but only 367 healthy people were studied.21 Poles, Czechs and Slovaks are of Slavic origin. On the basis of our results, we expect the del5395 deletion to be present in other Slavic populations, including Ukraine, Belarus, Russia and the Baltic and Balkan countries.
Our results suggest that this deletion confers an approximately twofold increase in the risk of prostate cancer (p = 0.09) and an approximately fourfold increase in the risk of familial prostate cancer (OR 3.7; p = 0.03). The effects of the three truncating mutations (1100delC, IVS2+1G→A and del5395) on prostate cancer risk seem to be similar, but our subgroups were small and the results should be confirmed on other large series. The large CHEK2 deletion leads to premature protein truncation at codon 381. The 1100delC also leads to protein truncation at codon 381, but exons 9 and 10 are retained. The splice‐site mutation (IVS2+1G→A) results in a 4‐bp insertion as a result of an abnormal splicing, and creates a termination codon in exon 3. The detection of mRNAs of abnormal lengths suggests that these mutations do not lead to complete transcript loss due to nonsense‐mediated decay.13,21 Therefore, in theory, the effect of these truncating mutations on cancer risk might differ. It is uncertain whether the difference in disease risk between truncating and missense variants is due to a quantitative difference in kinase activity or whether a dominant negative effect of I157T is present.10
The strengths of our study include the large number of patients studied and the sampling of incident cases unselected for family history. Incident cases are preferred to prevalent cases if there is concern that the hereditary subgroup might have a prognosis different from patients with cancer in general. Our patients were not selected for family history and unselected familial cases were drawn from the same database. Our control group was large (5496 controls)—this permitted us to estimate the frequency of the deletion in the Polish population at large.
In conclusion, there are (at minimum) four founder alleles of the CHEK2 gene, which predispose the Polish population to prostate cancer. These founder alleles account for 7% of patients with prostate cancer in Poland. The most common of these is a missense mutation (I157T). Among the truncating variants is a 5395‐bp deletion. It will be of interest to see to what extent this deletion is responsible for cancer burden in other populations.
Abbreviations
CHEK2 - Chek2 kinase gene
PCR - polymerase chain reaction
Footnotes
Competing interests: None.
Ethical approval: This study was approved by the Ethics Committee of Pomeranian Medical University, Szczecin, Poland.
References
- 1.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge S J. Ataxia telangiectasia–mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA 20009710389–10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaturvedi P, Eng W K, Zhu Y, Mattern M R, Mishra R, Hurle M R, Zhang X, Annan R S, Lu Q, Faucette L F, Scott G F, Li X, Carr S A, Johnson R K, Winkler J D, Zhou B B. Mammalian Chk2 is a downstream effector of the ATM‐dependent DNA damage checkpoint pathway. Oncogene 1999184047–4054. [DOI] [PubMed] [Google Scholar]
- 3.Bell D W, Varley J M, Szydlo T E, Kang D H, Wahrer D C R, Shannon K E, Lubratovich M, Verselis S J, Isselbacher K J, Fraumeni J F, Birch J. Li FP, Garber JE, Haber DA. Heterozygous germ line hCHK2 mutations in Li‐Fraumeni syndrome. Science 19992862528–2531. [DOI] [PubMed] [Google Scholar]
- 4.Vahteristo P, Tamminen A, Karvinen P, Eerola H, Eklund C, Aaltonen L A, Blomqvist C, Aittomaki K, Nevanlinna H. p53, CHK2, and CHK1 genes in Finnish families with Li‐Fraumeni syndrome: further evidence of CHK2 in inherited cancer predisposition. Cancer Res 2001615718–5722. [PubMed] [Google Scholar]
- 5. CHEK2 Breast Cancer Consortium. Low‐penetrance susceptibility to breast cancer due to CHEK2*1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 20023155–59. [DOI] [PubMed] [Google Scholar]
- 6.Oldenburg R A, Kroeze‐Jansema K, Kraan J, Morreau H, Klijn J G, Hoogerbrugge N, Ligtenberg M J, van Asperen C J, Vasen H F, Meijers C, Meijers‐Heijboer H, de Bock T H, Cornelisse C J, Devilee P. The CHEK2*1100delC variant acts as a breast cancer risk modifier in non‐BRCA1/BRCA2 multiple‐case families. Cancer Res 2003638153–8157. [PubMed] [Google Scholar]
- 7.Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomaki K, Heikkila P, Holli K, Blomqvist C, Bartek J, Kallioniemi O P, Nevanlinna H. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet 200271432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. CHEK2 Breast Cancer Case‐Control Consortium. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies, Am J Hum Genet 2004741175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cybulski C, Gorski B, Huzarski T, Masojc B, Mierzejewski M, Debniak T, Teodorczyk U, Byrski T, Gronwald J, Matyjasik J, Zlowocka E, Lenner M, Grabowska E, Nej K, Castaneda J, Medrek K, Szymanska A, Szymanska J, Kurzawski G, Suchy J, Oszurek O, Witek A, Narod S A, Lubinski J. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet 2004751131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilpivaara O, Vahteristo P, Falck J, Syrjakoski K, Eerola H, Easton D, Bartkova J, Lukas J, Heikkila P, Aittomaki K, Holli K, Blomqvist C, Kallioniemi O P, Bartek J, Nevanlinna H. CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer 2004111543–547. [DOI] [PubMed] [Google Scholar]
- 11.Shaag A, Walsh T, Renbaum P, Kirchhoff T, Nafa K, Shiovitz S, Mandell J B, Welcsh P, Lee M K, Ellis N, Offit K, Levy‐Lahad E, King M C. Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum Mol Genet 200514555–563. [DOI] [PubMed] [Google Scholar]
- 12.Cybulski C, Huzarski T, Górski B, Masojc B, Mierzejewski M, Debniak T, Gliniewicz B, Matyjasik J, Zlowocka E, Kurzawski G, Sikorski A, Posmyk M, Szwiec M, Czajka R, Narod S A, Lubinski J. A novel founder CHEK2 mutation is associated with increased prostate cancer risk. Cancer Res 2004642677–2679. [DOI] [PubMed] [Google Scholar]
- 13.Dong X, Wang L, Taniguchi K, Wang X, Cunningham J M, McDonnell S K, Qian C, Marks A F, Slager S L, Peterson B J, Smith D I, Cheville J C, Blute M L, Jacobsen S J, Schaid D J, Tindall D J, Thibodeau S N, Liu W. Mutations in CHEK2 associated with prostate cancer risk. Am J Hum Genet 200372270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meijers‐Heijboer H, Wijnen J, Vasen H, Wasielewski M, Wagner A, Hollestelle A, Elstrodt F, van den Bos R, de Snoo A, Fat G T, Brekelmans C, Jagmohan S, Franken P, Verkuijlen P, van den Ouweland A, Chapman P, Tops C, Moslein G, Burn J, Lynch H, Klijn J, Fodde R, Schutte M. The CHEK2 1100delC mutation identifies families with a hereditary breast and colorectal cancer phenotype. Am J Hum Genet 2003721308–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allinen M, Huusko P, Mantyniemi S, Launonen V, Winqvist R. Mutation analysis of the CHK2 gene in families with hereditary breast cancer. Br J Cancer 200185209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schutte M, Seal S, Barfoot R, Meijers‐Heijboer H, Wasielewski M, Evans D G, Eccles D, Meijers C, Lohman F, Klijn J, van den Ouweland A, Futreal P A, Nathanson K L, Weber B L, Easton D F, Stratton M R, Rahman N. Breast Cancer Linkage Consortium. Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am J Hum Genet 2003721023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogdanova N, Enssen‐Dubrowinskaja N, Feshchenko S, Lazjuk G I, Rogov Y I, Dammann O, Bremer M, Karstens J H, Sohn C, Dork T. Association of two mutations in the CHEK2 gene with breast cancer. Int J Cancer 2005116263–266. [DOI] [PubMed] [Google Scholar]
- 18.Seppala E H, Ikonen T, Mononen N, Autio V, Rokman A, Matikainen M P, Tammela T L, Schleutker J. CHEK2 variants associate with hereditary prostate cancer. Br J Cancer 2003891966–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilpivaara O, Alhopuro P, Vahteristo P, Aaltonen L A, Nevanlinna H. CHEK2 I157T associates with familial and sporadic colorectal cancer. J Med Genet 200643e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudd M F, Sellick G S, Webb E L, Catovsky D, Houlston R S. Variants in the ATM‐BRCA2‐CHEK2 axis predispose to chronic lymphocytic leukaemia. Blood 2006108638–644. [DOI] [PubMed] [Google Scholar]
- 21.Walsh T, Casadei S, Coats K H, Swisher E, Stray S M, Higgins J, Roach K C, Mandell J, Lee M K, Ciernikova S, Foretova L, Soucek P, King M C. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 20062951379–1388. [DOI] [PubMed] [Google Scholar]