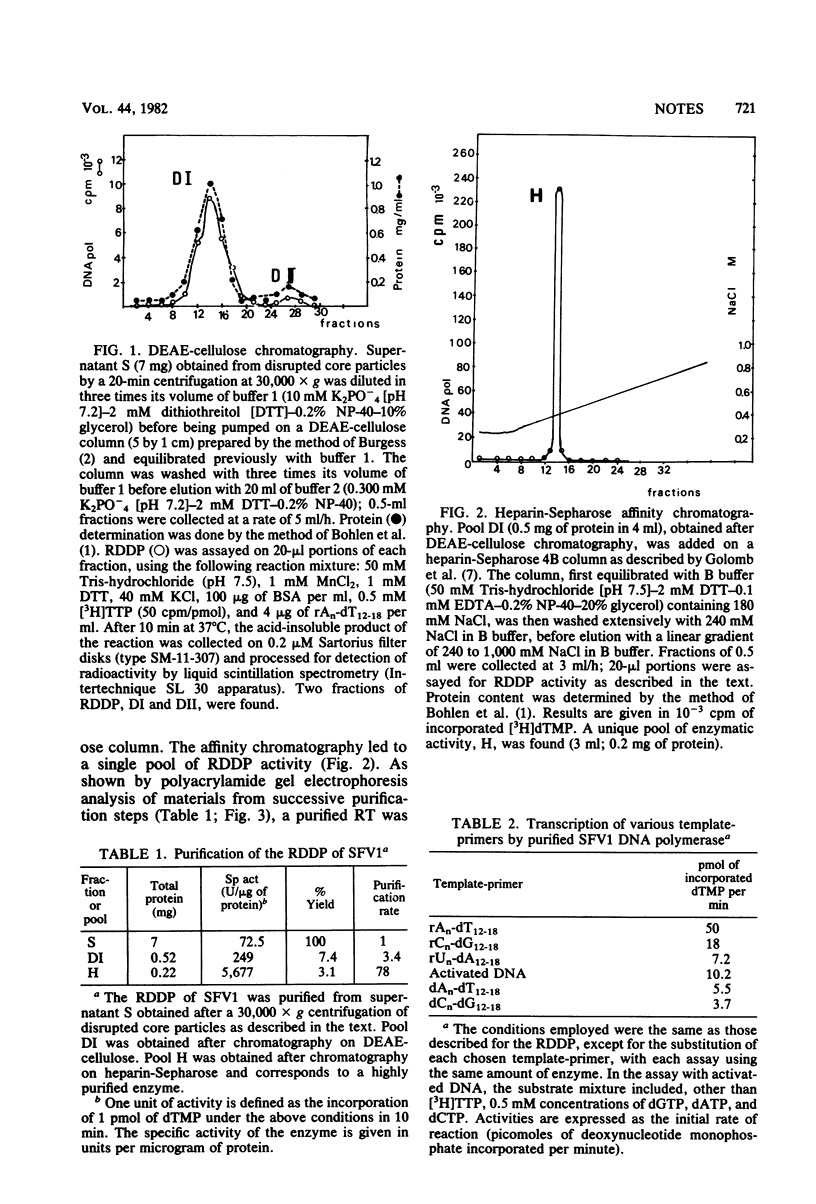
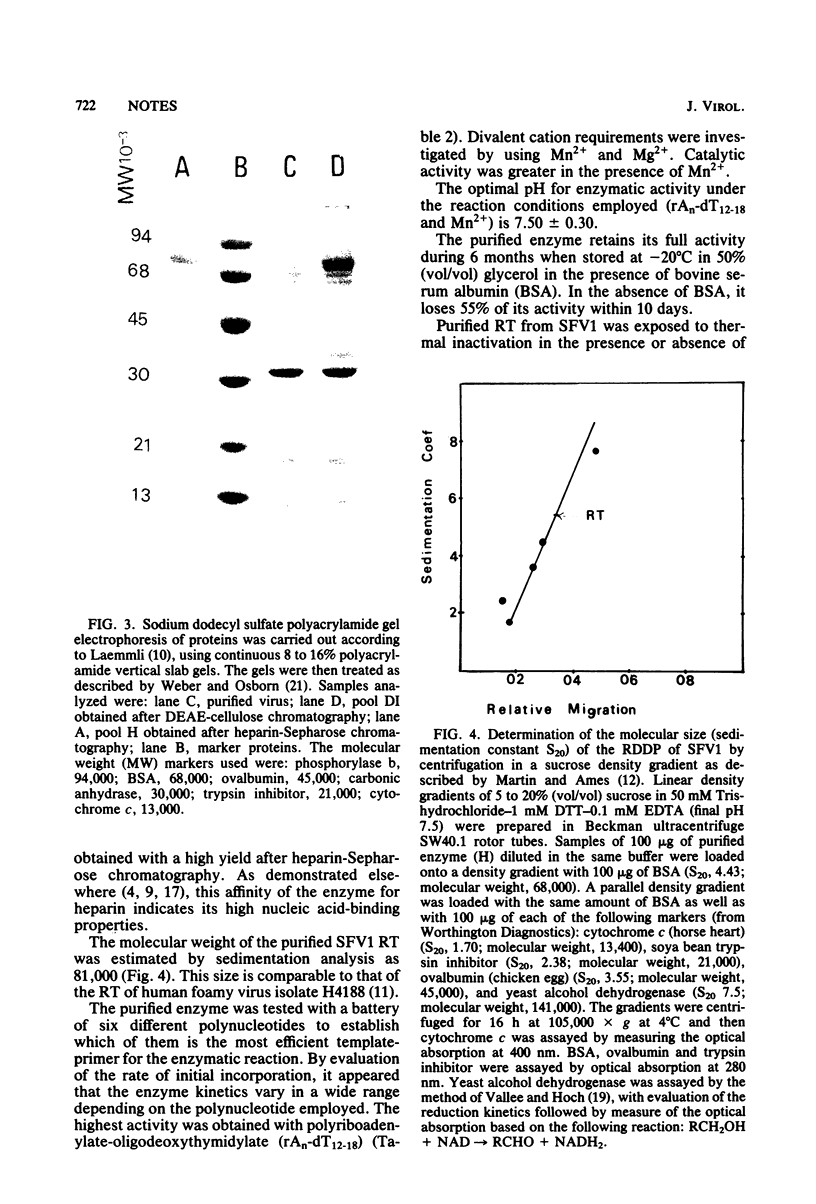
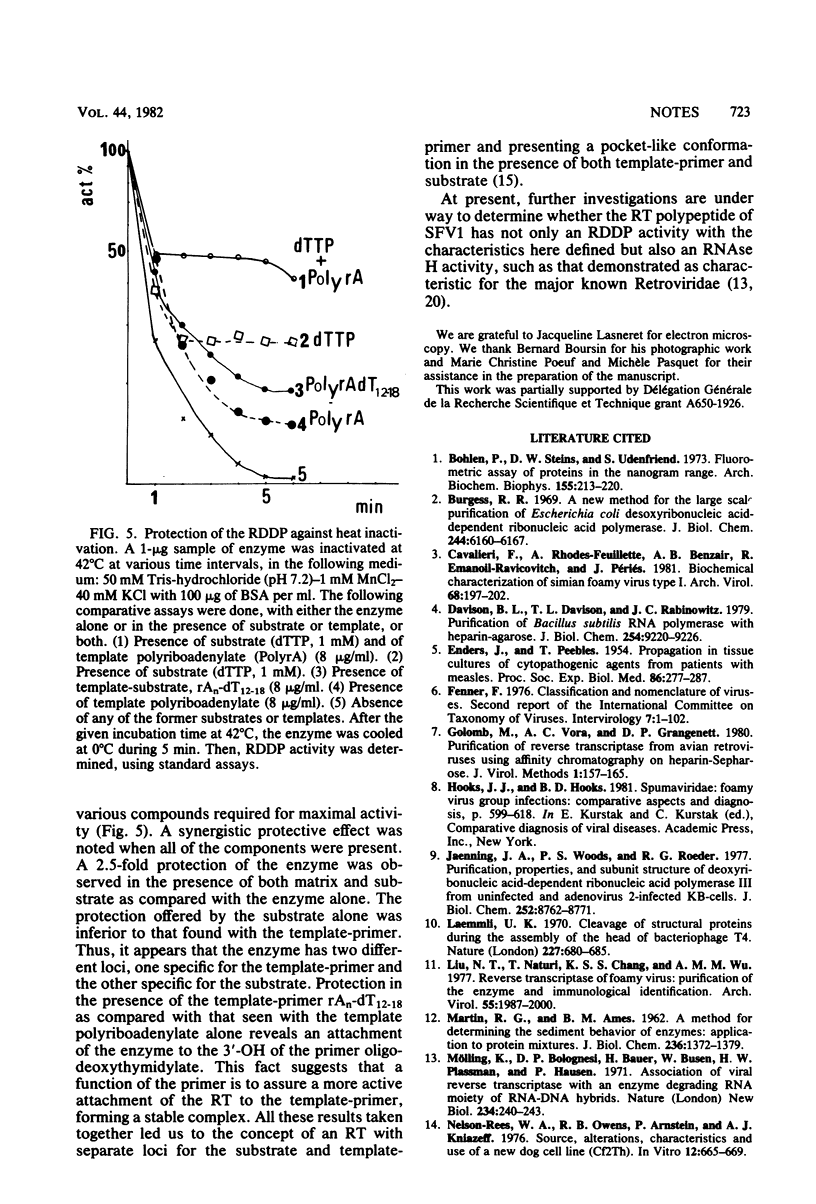
Abstract
Chromatography on heparin-Sepharose, known for its affinity for nucleotide-binding polypeptides, was used to purify the viral RNA-dependent DNA polymerase (reverse transcriptase) from the core polypeptides of simian foamy virus type 1. This procedure allowed the recovery of highly purified enzyme with a high specific activity. The average molecular weight of this monomeric enzyme is 81,000 and is thus comparable to that found for other known primate retroviruses. Reverse transcriptase activity of simian foamy virus type 1 requires a ribonucleotide template as a primer or otherwise a DNA with 3'-OH ends. Other optimal conditions of activity are reviewed. Heat inactivation studies led to the concept of an enzyme with two loci, one specific for the substrate and the other for the template-primer.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burgess R. R. A new method for the large scale purification of Escherichia coli deoxyribonucleic acid-dependent ribonucleic acid polymerase. J Biol Chem. 1969 Nov 25;244(22):6160–6167. [PubMed] [Google Scholar]
- Böhlen P., Stein S., Dairman W., Udenfriend S. Fluorometric assay of proteins in the nanogram range. Arch Biochem Biophys. 1973 Mar;155(1):213–220. doi: 10.1016/s0003-9861(73)80023-2. [DOI] [PubMed] [Google Scholar]
- Cavalieri F., Rhodes-Feuillette A., Benzair A. B., Emanoïl-Ravicovitch R., Peries J. Biochemical characterization of simian foamy virus type i. Arch Virol. 1981;68(3-4):197–202. doi: 10.1007/BF01314572. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Leighton T., Rabinowitz J. C. Purification of Bacillus subtilis RNA polymerase with heparin-agarose. In vitro transcription of phi 29 DNA. J Biol Chem. 1979 Sep 25;254(18):9220–9226. [PubMed] [Google Scholar]
- ENDERS J. F., PEEBLES T. C. Propagation in tissue cultures of cytopathogenic agents from patients with measles. Proc Soc Exp Biol Med. 1954 Jun;86(2):277–286. doi: 10.3181/00379727-86-21073. [DOI] [PubMed] [Google Scholar]
- Fenner F. Classification and nomenclature of viruses. Second report of the International Committee on Taxonomy of Viruses. Intervirology. 1976;7(1-2):1–115. doi: 10.1159/000149938. [DOI] [PubMed] [Google Scholar]
- Golomb M., Vora A. C., Grandgenett D. P. Purification of reverse transcriptase from avian retroviruses using affinity chromatography on heparin-sepharose. J Virol Methods. 1980;1(3):157–165. doi: 10.1016/0166-0934(80)90012-9. [DOI] [PubMed] [Google Scholar]
- Jaehning J. A., Woods P. S., Roeder R. G. Purification, properties, and subunit structure of deoxyribonucleic acid-dependent ribonucleic acid polymerase III from uninfected and adenovirus 2-infected KB cells. J Biol Chem. 1977 Dec 10;252(23):8762–8771. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Mölling K., Bolognesi D. P., Bauer H., Büsen W., Plassmann H. W., Hausen P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat New Biol. 1971 Dec 22;234(51):240–243. doi: 10.1038/newbio234240a0. [DOI] [PubMed] [Google Scholar]
- Nelson-Rees W. A., Owens R. B., Arnstein P., Kniazeff A. J. Source, alterations, characteristics and use of a new dog cell line (Cf2Th). In Vitro. 1976 Oct;12(10):665–669. doi: 10.1007/BF02797468. [DOI] [PubMed] [Google Scholar]
- Papas T. S., Marciani D. J., Samuel K., Chirikjian J. G. Mechanism of release of active alpha subunit from dimeric alpha beta avian myeloblastosis virus DNA polymerase. J Virol. 1976 Jun;18(3):904–910. doi: 10.1128/jvi.18.3.904-910.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks W. P., Todaro G. J., Scolnick E. M., Aaronson S. A. RNA dependent DNA polymerase in primate syncytium-forming (foamy) viruses. Nature. 1971 Jan 22;229(5282):258–260. doi: 10.1038/229258a0. [DOI] [PubMed] [Google Scholar]
- Sternbach H., Engelhardt R., Lezius A. G. Rapid isolation of highly active RNA polymerase from Escherichia coli and its subunits by matrix-bound heparin. Eur J Biochem. 1975 Dec 1;60(1):51–55. doi: 10.1111/j.1432-1033.1975.tb20974.x. [DOI] [PubMed] [Google Scholar]
- Strand M., August J. T. Structural proteins of ribonucleic acid tumor viruses. Purification of envelope, core, and internal components. J Biol Chem. 1976 Jan 25;251(2):559–564. [PubMed] [Google Scholar]
- Vallee B. L., Hoch F. L. ZINC, A COMPONENT OF YEAST ALCOHOL DEHYDROGENASE. Proc Natl Acad Sci U S A. 1955 Jun 15;41(6):327–338. doi: 10.1073/pnas.41.6.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma I. M. Studies on reverse transcriptase of RNA tumor viruses III. Properties of purified Moloney murine leukemia virus DNA polymerase and associated RNase H. J Virol. 1975 Apr;15(4):843–854. doi: 10.1128/jvi.15.4.843-854.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]