Abstract
All inbred strains of mice transmit one or more copies of mouse mammary tumor virus (MMTV) DNA integrated as proviral sequences. This complicates efforts to define viral-induced mammary carcinogenesis. Here we report the use of surgical nonlethal splenectomy in tissue typing mice and the development of an MMTV-negative mouse strain. The MMTV-negative strain allows study of the involvement of non-MMTV genes in mammary carcinogenesis. In addition, it can be used as a sterile background into which MMTV variants can be introduced. Through the techniques described here, mice containing single MMTV loci or specific combinations can be specially chosen and rapidly developed. In this manner, the oncogenecity of particular MMTV variants may be assessed.
Full text
PDF

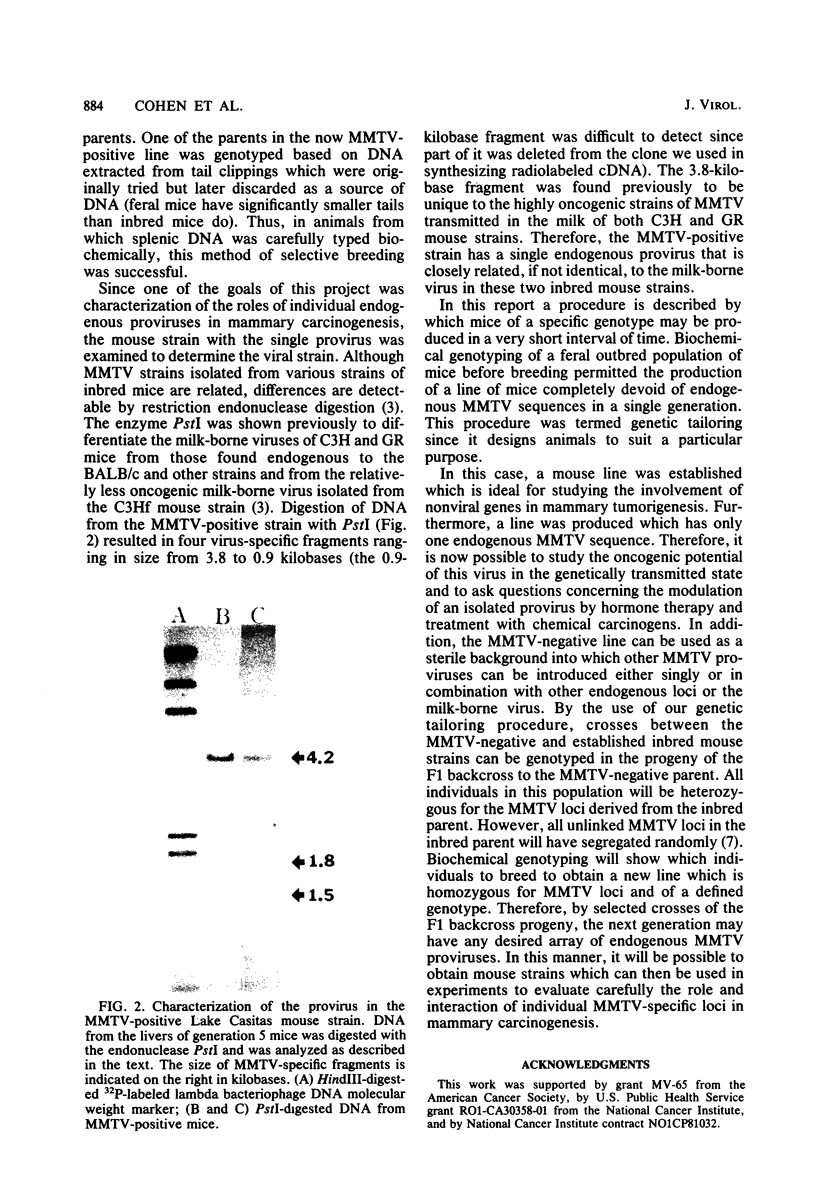

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bentvelzen P. Host-virus interactions in murine mammary carcinogenesis. Biochim Biophys Acta. 1974 Dec 31;355(3-4):236–259. doi: 10.1016/0304-419x(74)90012-2. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Majors J. E., Varmus H. E. Organization of mouse mammary tumor virus-specific DNA endogenous to BALB/c mice. J Virol. 1979 Nov;32(2):483–496. doi: 10.1128/jvi.32.2.483-496.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C., Shank P. R., Morris V. L., Cardiff R., Varmus H. E. Integration of the DNA of mouse mammary tumor virus in virus-infected normal and neoplastic tissue of the mouse. Cell. 1979 Feb;16(2):333–345. doi: 10.1016/0092-8674(79)90010-2. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Morris V. L., Medeiros E., Ringold G. M., Bishop J. M., Varmus H. E. Comparison of mouse mammary tumor virus-specific DNA in inbred, wild and Asian mice, and in tumors and normal organs from inbred mice. J Mol Biol. 1977 Jul;114(1):73–91. doi: 10.1016/0022-2836(77)90284-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., Shank P. R. X-Ray Intensifying Screens Greatly Enhance the Detection by Autoradiography of the Radioactive Isotopes 32P and 125I. Anal Biochem. 1978 May;86(1):184–192. doi: 10.1016/0003-2697(78)90333-0. [DOI] [PubMed] [Google Scholar]
- Traina V. L., Taylor B. A., Cohen J. C. Genetic mapping of endogenous mouse mammary tumor viruses: locus characterization, segregation, and chromosomal distribution. J Virol. 1981 Dec;40(3):735–744. doi: 10.1128/jvi.40.3.735-744.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]