Abstract
The nucleotide sequence of the parvovirus H-1 has been determined by the chain-terminating method of Sanger. The sequence is 5,176 nucleotides long. Two large open reading frames (1 and 2) and two smaller open reading frames (3 and 4) of potential importance were identified in the plus-strand sequence. Promoter sequences are located at map positions 4 and 38 when map positions are expressed as percent of genome length from the 3' end of the virion minus strand. The locations for the genes for the parvovirus capsid proteins and a 76,000-dalton noncapsid protein (NCVP1) were mapped by hybrid-arrested translation. The gene for the capsid proteins VP1 and VP2' is located in the 5' half of the virus genome. The gene for NCVP1 is located in the 3' half of the viral DNA.
Full text
PDF






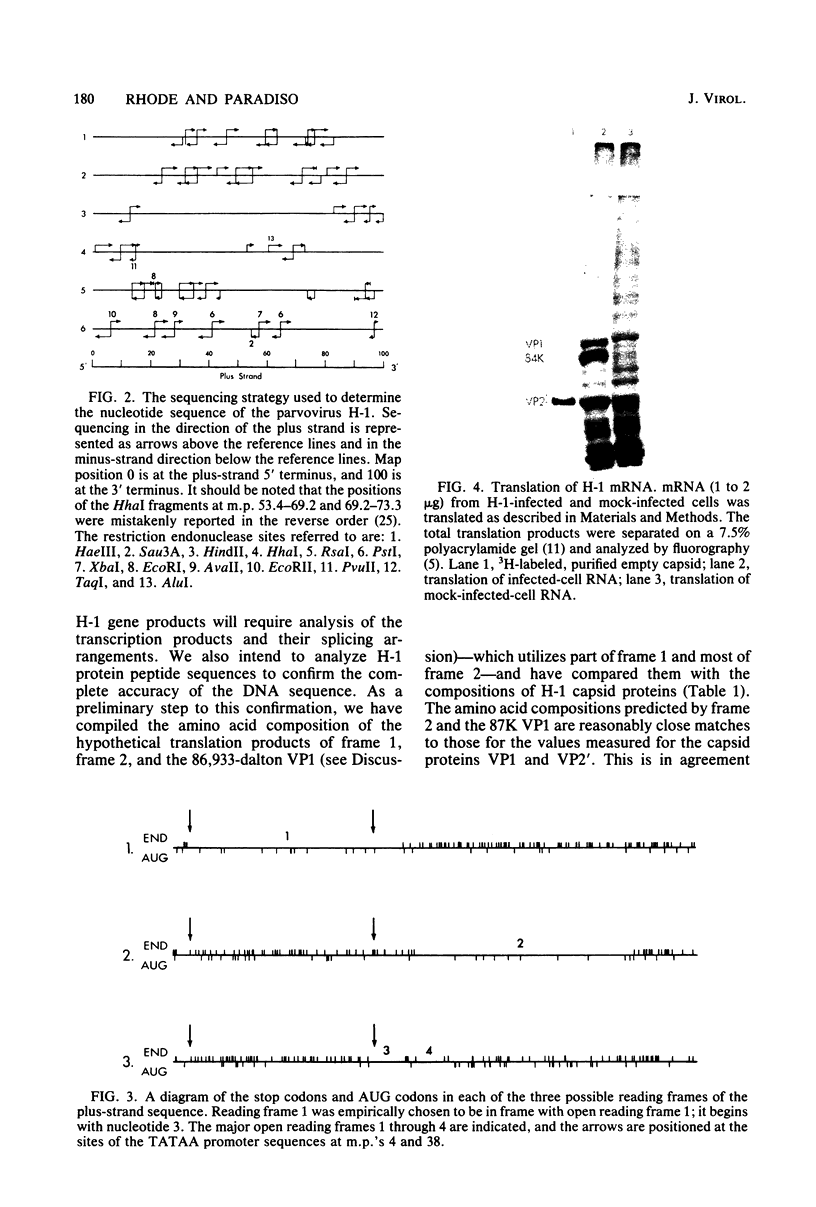

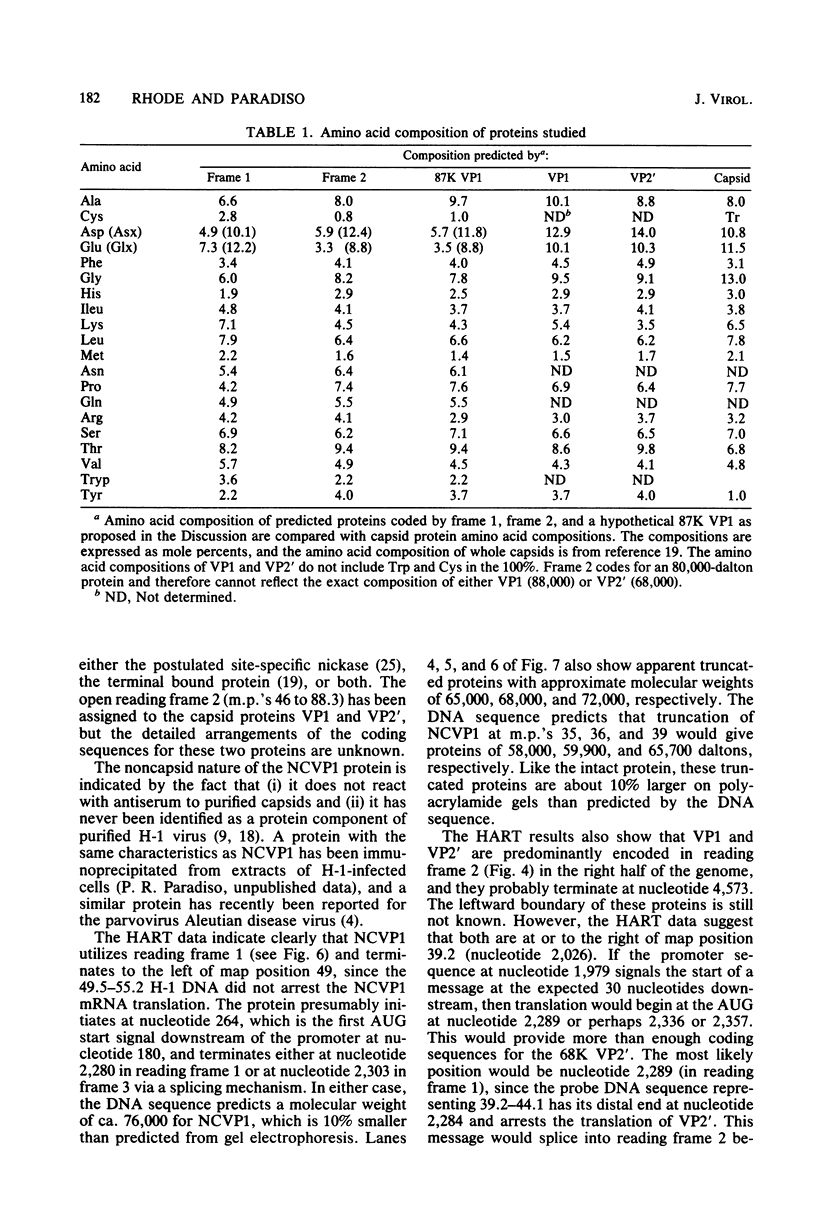


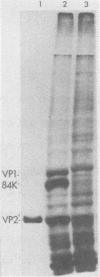
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams S. L., Sobel M. E., Howard B. H., Olden K., Yamada K. M., de Crombrugghe B., Pastan I. Levels of translatable mRNAs for cell surface protein, collagen precursors, and two membrane proteins are altered in Rous sarcoma virus-transformed chick embryo fibroblasts. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3399–3403. doi: 10.1073/pnas.74.8.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astell C. R., Smith M., Chow M. B., Ward D. C. Structure of the 3' hairpin termini of four rodent parvovirus genomes: nucleotide sequence homology at origins of DNA replication. Cell. 1979 Jul;17(3):691–703. doi: 10.1016/0092-8674(79)90276-9. [DOI] [PubMed] [Google Scholar]
- Bloom M. E., Race R. E., Wolfinbarger J. B. Identification of a nonvirion protein of Aleutian disease virus: mink with Aleutian disease have antibody to both virion and nonvirion proteins. J Virol. 1982 Aug;43(2):608–616. doi: 10.1128/jvi.43.2.608-616.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Green M. R., Lebovitz R. M., Roeder R. G. Expression of the autonomous parvovirus H1 genome: evidence for a single transcriptional unit and multiple spliced polyadenylated transcripts. Cell. 1979 Aug;17(4):967–977. doi: 10.1016/0092-8674(79)90336-2. [DOI] [PubMed] [Google Scholar]
- Kongsvik J. R., Gierthy J. F., Rhode S. L., 3rd Replication process of the parvovirus H-1. IV. H-1-specific proteins synthesized in synchronized human NB kidney cells. J Virol. 1974 Dec;14(6):1600–1603. doi: 10.1128/jvi.14.6.1600-1603.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsvik J. R., Toolan H. W. Capsid components of the parvovirus H-1. Proc Soc Exp Biol Med. 1972 Apr;139(4):1202–1205. doi: 10.3181/00379727-139-36329. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Crawford L. V., Follett E. A. The DNAs of three parvoviruses. J Gen Virol. 1970 Jan;6(1):33–40. doi: 10.1099/0022-1317-6-1-33. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Beard P., Engers H. D., Hirt B. Characterization of an immunosuppressive parvovirus related to the minute virus of mice. J Virol. 1981 Apr;38(1):317–326. doi: 10.1128/jvi.38.1.317-326.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison J. M., Keir H. M., Subak-Sharpe H., Crawford L. V. Nearest neighbour base sequence analysis of the deoxyribonucleic acids of a further three mammalian viruses: Simian virus 40, human papilloma virus and adenovirus type 2. J Gen Virol. 1967 Jan;1(1):101–108. doi: 10.1099/0022-1317-1-1-101. [DOI] [PubMed] [Google Scholar]
- Paradiso P. R. Infectious process of the parvovirus H-1: correlation of protein content, particle density, and viral infectivity. J Virol. 1981 Sep;39(3):800–807. doi: 10.1128/jvi.39.3.800-807.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson B. M., Roberts B. E., Kuff E. L. Structural gene identification and mapping by DNA-mRNA hybrid-arrested cell-free translation. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4370–4374. doi: 10.1073/pnas.74.10.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revie D., Tseng B. Y., Grafstrom R. H., Goulian M. Covalent association of protein with replicative form DNA of parvovirus H-1. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5539–5543. doi: 10.1073/pnas.76.11.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Complementation for replicative form DNA replication of a deletion mutant of H-1 by various parvoviruses. J Virol. 1982 Jun;42(3):1118–1122. doi: 10.1128/jvi.42.3.1118-1122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Defective interfering particles of parvovirus H-1. J Virol. 1978 Aug;27(2):347–356. doi: 10.1128/jvi.27.2.347-356.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd, Klaassen B. DNA sequence of the 5' terminus containing the replication origin of parvovirus replicative form DNA. J Virol. 1982 Mar;41(3):990–999. doi: 10.1128/jvi.41.3.990-999.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Replication process of the parvovirus H-1 V. Isolation and characterization of temperature-sensitive H-1 mutants. J Virol. 1976 Feb;17(2):659–667. doi: 10.1128/jvi.17.2.659-667.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode S. L., 3rd Replication process of the parvovirus H-1. X. Isolation of a mutant defective in replicative-form DNA replication. J Virol. 1978 Jan;25(1):215–223. doi: 10.1128/jvi.25.1.215-223.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Tal J., Ron D., Tattersall P., Bratosin S., Aloni Y. About 30% of minute virus of mice RNA is spliced out following polyadenylation. Nature. 1979 Jun 14;279(5714):649–651. doi: 10.1038/279649a0. [DOI] [PubMed] [Google Scholar]
- Tattersall P., Shatkin A. J., Ward D. C. Sequence homology between the structural polypeptides of minute virus of mice. J Mol Biol. 1977 Apr 25;111(4):375–394. doi: 10.1016/s0022-2836(77)80060-0. [DOI] [PubMed] [Google Scholar]