Abstract
Background
A previous study identified two peaks of allelic association between psoriasis and single nucleotide polymorphisms (SNPs) mapping to distal chromosome 17q, including a disease associated SNP that leads to loss of a RUNX1 transcription factor binding site, and additional SNPs in the third intron of the RAPTOR gene. Another study found an association with SNPs in the RAPTOR gene, but not with the RUNX1 binding site polymorphism.
Methods
In an effort to confirm these observations, we genotyped 579 pedigrees containing 1285 affected individuals for three SNPs immediately flanking and including the RUNX1 binding site, and for three SNPs in the RAPTOR gene.
Results
Here we report further evidence for linkage to distal chromosome 17q, with a linkage peak mapping 1.7 cM distal to the RUNX1 binding site (logarithm of the odds 2.26 to 2.73, depending upon statistic used). However, we found no evidence for association to individual SNPs or haplotypes in either of the previously identified peaks of association. Power analysis demonstrated 80% power to detect significant association at genotype relative risks of 1.2 (additive and multiplicative models) to 1.5 (dominant and recessive models) for the RUNX1 binding site, and 1.3 to 1.4 for the RAPTOR locus under all models except dominant.
Conclusions
Our data provide no support for the previously identified RUNX1 binding site or for the RAPTOR locus as genetic determinants of psoriasis, despite evidence for linkage of psoriasis to distal chromosome 17q.
Keywords: association, linkage, psoriasis, RAPTOR, RUNX1
Psoriasis is a common, immunologically mediated, hyperproliferative skin disease that is influenced by multiple genes, including a major gene in the major histocompatibility complex.1,2,3 Because the penetrance of the disease allele at this locus is only about 10%, and based on recurrence risk4 and linkage5 analysis, it is apparent that additional loci also influence susceptibility to psoriasis. An early candidate for such a locus was identified by linkage analysis on distal human chromosome 17q,6 and designated psoriasis susceptibility 2 (PSORS2). While we and others have provided some confirmatory evidence for genetic linkage to this locus,7,8,9 the PSORS2 gene itself has remained unidentified for a decade.
A recent study by Helms et al of single nucleotide polymorphisms (SNPs) and microsatellite markers in the distal 17q region identified allelic association between psoriasis and SNPs mapping in and between the SLC9A3R1 and NAT9 genes on distal chromosome 17q.10 The disease associated allele of one of these polymorphisms was shown to inactivate a binding site for RUNX1, which is a haematopoietic transcription factor implicated in leukaemogenesis.11 Variant RUNX1 binding sites have also been genetically implicated in systemic lupus erythaematosus12 and rheumatoid arthritis.13,14 Thus, the RUNX1 binding site polymorphism is an attractive candidate for PSORS2. However, only one marker in this peak exceeded the p = 0.05 level of significance after the most stringent level of correction for multiple testing,10 making independent confirmation critical.
The studies of Helms et al also identified a second peak of association 6 Mb distal to the RUNX1 binding site, which mapped to the third intron of the RAPTOR gene. A second study of an independent set of subjects found a weak (p = 0.027) association with one SNP in the RAPTOR gene, but not with the RUNX1 binding site polymorphism.15 A third, independent study found no association with the RUNX1 binding site in any of three independent German cohorts.16
In this study, we genotyped 579 pedigrees of various structures for three SNPs mapping to the 3′ end of SLC9A3R1 and the interval between SLCA3R1 and NAT9, including the implicated RUNX1 binding site. We also typed the three SNPs in the RAPTOR gene that were previously reported to be associated with psoriasis.10,15 Our pedigree sample allowed us to refine and extend our previous linkage analysis of chromosome 17q7 by adding 159 pedigrees informative for linkage to our original cohort of 115 pedigrees. We found further evidence for linkage to distal chromosome 17q, with a linkage peak mapping quite close to the RUNX1 binding site. However, we found no evidence for association to individual SNPs or haplotypes in either of the previously identified peaks of association. Simulations demonstrated excellent power of our 517 informative families to detect significant association at realistic genotype relative risks (GRRs) for both regions, with the exception of the RAPTOR locus under the dominant model. Taken together, our data provide no support for either region as genetic determinants of psoriasis, despite demonstration of evidence for linkage of psoriasis to distal chromosome 17q.
Methods
A detailed description of the methods used, including marker primers and the composition of our pedigree sample, is provided as three supplemental appendices at http://www.jmedgenet.com/supplemental.
Subject recruitment
Psoriasis was defined as previously described,17 and ascertainment was for age at onset of ⩽40 years in the proband.18 After providing informed consent, all participants received a total body skin examination and provided a blood sample. A total of 579 families were recruited, 102 from northern Germany and the remaining 477 from the United States, largely from south eastern Michigan. Enrolment of subjects and genotyping was carried out under protocols approved by the medical ethical committees of the University of Michigan, Henry Ford Hospital, and the University of Kiel. This study was conducted according to Declaration of Helsinki principles at all participating institutions.
Genotyping
Genomic DNA amplification was performed using the same primers employed by Helms et al.10 After amplification, SNPs rs745318, rs734232, and rs895691 mapping to the RUNX1 binding site region (rs734232 representing the RUNX1 polymorphism itself) and rs1564864, rs2019154, and rs869190 mapping to the RAPTOR gene, were genotyped using SnapShot SNP assay reagents and GeneMapper software (Applied Biosystems, Foster City, CA). Microsatellites were typed utilising 32P‐labelled oligonucleotide primers as described.19
Linkage analysis
The sample consisted of 274 families, including 115 families that were used in a prior genome‐wide linkage analysis.7 The 38 marker set for linkage analysis consisted of the six RAPTOR and RUNX1 SNPs and 32 microsatellite markers located across chromosome 17. Marker density was greatest in the vicinity of the PSORS2 locus on distal 17q.6,7
Non‐parametric linkage analysis was performed using Merlin version 0.10.6.20 Logarithm of the odds (LOD) scores and p values were computed using the Kong and Cox linear model,21 with both the NPL‐all and NPL‐pairs scoring functions.22 Because there is strong linkage disequilibrium (LD) among some of the markers, tightly linked markers were first clustered to avoid positive bias of the LOD scores.23
Haplotype reconstruction
Maximum likelihood haplotypes for analysis by the transmission/disequilibrium test (TDT) and the pedigree disequilibrium test (PDT) were created using the “best” option of Merlin (version 0.10.2). Phase ambiguities in the most likely Merlin haplotypes were resolved whenever possible using PHASE version 2.1.1.24,25 Although PHASE is designed for constructing haplotypes of independent individuals, when families are available current methods of haplotype reconstruction in pedigrees (for example, Merlin) can use information about gene flow in the family to infer founder haplotypes at many loci, and PHASE can then use this known phase information to estimate any remaining ambiguous phases.24 Before input into the TDT and PDT, all haplotypes with any inferred or remaining phase uncertain alleles were converted to missing (0.4% of haplotypes of collected family members for the RUNX1 binding site, 0.7% for RAPTOR).
Haplotypes for the family based association test (FBAT) were reconstructed internally by the FBAT program in a probabilistic manner using a conditioning approach that allows use of haplotypes with missing genotype or phase information without introducing bias.26
Family based association analysis
Pedigrees were analysed for the putative disease associated alleles and haplotypes with three different FBATs: the TDT,27 the PDT,28,29 and the FBAT.26,30 For the TDT, a single trio was randomly extracted from each pedigree. Since results vary depending upon the particular random selection, the analysis was repeated 999 times with different random number seeds, and the median result reported. For the PDT, we utilised the PDT‐avg test which gives equal weighting to all families. All trios and discordant sibpairs in a family contributed to the test. We also computed D̄, a standardised measure of LD between the disease and marker loci as assessed by the PDT.31D̄ has a range of [−1,1] and is equal to 0 in the absence of evidence for LD. For the FBAT, version 1.5.5 of the software32 was used with the empirical variance and an offset of 0.
Power tests
All power computations used a type I error rate of 0.05, a range of values for GRR for test allele homozygotes (GRR2) broad enough to encompass 60–99% power, and four genetic models (dominant, additive, multiplicative, and recessive). The marker locus was assumed to be in complete LD (r2 measure of disequilibrium = 1.0) with the true disease locus. Power for the TDT was determined analytically by the first approximation method of Knapp,33 using the observed number of fully typed and independent trios in our family sample and the observed frequency of the test allele or haplotype among founders. Power for the PDT and FBAT was determined by simulation, under the alternative hypothesis of LD between psoriasis and the marker locus. Genotypes for simulated pedigrees identical in structure, founder allele frequencies, and disease phenotypes to the observed sample were generated using a gene drop algorithm with rejection sampling. One thousand pedigree samples were simulated for each combination of locus, GRR2, and genetic model of inheritance and subjected to a Monte Carlo test of power.
Results
Linkage analysis
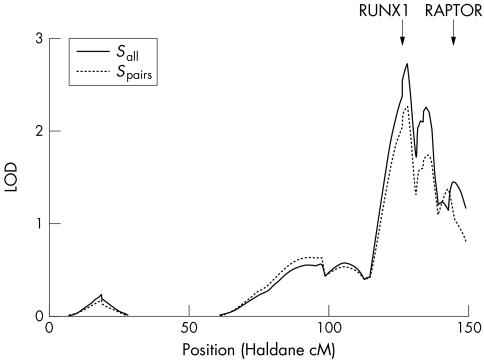
As shown in fig 1, non‐parametric multipoint linkage analysis of the chromosome 17 markers yielded suggestive evidence for linkage by genome‐wide criteria34 for both allele sharing statistics (maximum LOD = 2.73, p = 2.0×10−4 with Sall and maximum LOD = 2.26, p = 6.3×10−4 with Spairs). Evidence for linkage in both cases peaked at 127.9 Haldane cM (marker D17S785) in chromosome region 17q25.1, only 1.7 cM distal to the RUNX1 SNPs.
Figure 1 Non‐parametric linkage results for chromosome 17. Solid and dotted lines are LOD score plots using the linear Kong and Cox model with the Sall or Spairs allele sharing statistic, respectively. The locations of the RUNX1 binding site and RAPTOR gene are shown.
Both the older7 and newer families independently showed evidence for linkage. The original 115 families yielded maximum LOD = 1.32, p = 0.0068 with Sall and maximum LOD = 0.93, p = 0.019 with Spairs, and the subsequent 159 families yielded maximum LOD = 1.63, p = 0.0031 with Sall and maximum LOD = 1.53, p = 0.0040 with Spairs. LOD scores peaked between D17S674 and D17S1847 (134.5–135.5 Haldane cM) for the original sample and at D17S785 (127.9 Haldane cM) for the subsequent sample. The similar linkage peak locations and positive LOD scores obtained in two independent samples increase our confidence that our linkage results for the pooled sample are not a false positive.
Haplotype reconstruction
The two approaches used for reconstructing haplotypes (Merlin combined with PHASE or FBAT), although very different, gave nearly identical results. For the RUNX1 interval, the combined frequency of the two most common haplotypes was estimated as 99.2% by both methods; for the RAPTOR interval it was estimated as 99.8% using Merlin and PHASE and 99.9% using FBAT. The preponderance of only two of the eight possible haplotypes indicates nearly complete LD (r2≈1) among the three biallelic loci of each interval.
Family based association tests
Using the full pedigree sample, no significant association of psoriasis was found with any of the previously reported disease associated alleles for either the RUNX1 or RAPTOR intervals (table 1A). As expected given the nearly complete LD among SNPs in each interval, results for the three‐locus haplotypes were essentially identical to those for individual SNPs. A weak positive but clearly non‐significant (p value⩾0.40) association with psoriasis was measured by all three tests with both the putative RUNX1 disease alleles (51.5–52.0 percent transmission (%T), 0.006 to 0.010 D̄, 0.07 to 0.17 FBAT statistic Z) and the putative RAPTOR disease alleles (50.6–51.2 %T, 0.030 to 0.041 D̄, 0.17 to 0.35 Z). The strength of association originally reported for the RUNX1 SNPs and haplotypes, 55–58 %T by the TDT or PDT, was much higher.10 Neither of the reports of significant association with RAPTOR10,15 provided measures of the strength of association.
Results were not improved by restricting the analysis to those pedigrees with a documented family history of psoriasis, which we defined as two or more affected individuals in a family larger than a trio (table 1B). p values were generally higher (⩾0.6), and the strength of association was diminished for RUNX1 (49.7–50.5 %T, −0.037 to −0.026 D̄, −0.53 to −0.36 Z) and essentially unchanged for RAPTOR (50.9–51.2 %T, 0.021 to 0.037 D̄, 0.11 to 0.23 Z).
Power tests
Power of the full pedigree sample is very good for detecting association with the markers or haplotypes in the vicinity of the RUNX1 binding site under all genetic models (table 2). Power is good for all three tests of association but consistently best for the FBAT (table 2 and fig 2). When carrying one copy of the test allele (two copies for the recessive model), 80% power with the FBAT is attained with GRRs of 1.23 to 1.53 (depending on genetic model; details in table 2) and 95% power with GRRs of 1.31 to 1.75. Power is also very good for detecting association with the RAPTOR markers or haplotypes under all models except the dominant (table 2); 80% power with the FBAT is attainable with GRRs of 1.31 to 1.41 and 95% power is achieved with GRRs of 1.42 to 1.64. The putative disease allele and haplotypes for the RAPTOR locus are very common (frequency 0.77) and carried by 94.7% of individuals in our sample. Therefore, power to detect associations with RAPTOR under a dominant model is poor, being only 24% and 52% for GRR2 of 2 and 5, respectively. As expected, for either interval, association tests of the haplotype had power essentially identical to tests of single SNPs (data not shown).
Table 2 Genotype relative risks needed to achieve power of 0.80 and 0.95 in family based tests of the association of psoriasis with RAPTOR and RUNX1 SNPs*.
| Locus† | Genetic model | GRR2/GRR1 for power of 0.80 | GRR2/GRR1 for power of 0.95 | ||||
|---|---|---|---|---|---|---|---|
| TDT | PDT | FBAT | TDT | PDT | FBAT | ||
| RUNX1 | Dominant | 1.69/1.69 | 1.61/1.61 | 1.52/1.52 | 2.00/2.00 | 1.92/1.92 | 1.75/1.75 |
| Additive | 1.69/1.35 | 1.62/1.31 | 1.53/1.27 | 1.96/1.48 | 1.87/1.44 | 1.73/1.37 | |
| Multiplicative | 1.69/1.30 | 1.62/1.27 | 1.52/1.23 | 1.96/1.40 | 1.86/1.36 | 1.72/1.31 | |
| Recessive | 1.69/– | 1.63/– | 1.53/– | 1.93/– | 1.84/– | 1.70/– | |
| RAPTOR | Dominant | –‡ | –‡ | –‡ | –‡ | –‡ | –‡ |
| Additive | 2.09/1.54 | 1.99/1.50 | 1.81/1.41 | 2.76/1.88 | 2.59/1.80 | 2.28/1.64 | |
| Multiplicative | 1.92/1.38 | 1.84/.1.36 | 1.71/1.31 | 2.33/1.53 | 2.23/1.49 | 2.01/1.42 | |
| Recessive | 1.50/– | 1.47/– | 1.40/– | 1.69/– | 1.65/– | 1.55/– | |
*GRRs needed to achieve power of 0.80 and 0.95 when testing for association of the three‐locus RUNX1 and RAPTOR haplotypes are essentially identical to the GRRs of their constituent SNPs.
†For each locus, power was tested for the allele listed in table 1.
‡Power of 0.80 or 0.95 is not attainable. For both single SNPs and the three‐locus haplotype, power at a GRR2 ( = GRR1) of 2 and 5 is 0.18 and 0.42 for the TDT, 0.22 and 0.47 for the PDT, and 0.24 and 0.52 for the FBAT.
GRR1, genotype relative risk for test allele heterozygotes; GRR2, genotype relative risk for test allele homozygotes
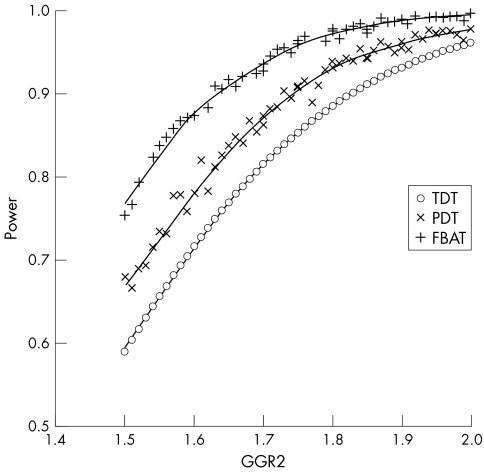
Figure 2 Comparison of power curves for the RUNX1 binding site locus under the additive model, type I error rate = 0.05. The TDT power curve was analytically derived using the method of Knapp,33 whereas the power calculations for the PDT and FBAT were carried out as described in Methods. For purposes of comparison, GRR2 for the MHC linked PSORS1 locus is approximately 10 under the additive model.31
Discussion
The search for genes conferring risk for common diseases with a genetic component continues to be a major challenge.35 Independent replication of findings is a critical component of this process. In this study, we have analysed 517 pedigrees of various structures for association to three SNPs within a 2.5 kb interval including the RUNX1 binding site, and three SNPs within a 26.5 kb interval in the RAPTOR gene, that were previously described by Helms et al.10 However, we failed to demonstrate any significant associations.
Given these disappointing findings, we considered it essential to carefully evaluate the power of our full sample to detect significant association. Our sample contains a diverse mix of families, including many extended pedigrees. The TDT is restricted to a single trio per family when implemented as a test of association, but the PDT and FBAT can more fully use multiplex and multigenerational families, including those with missing parents. Analytical power methods are available for the TDT36 but not for the PDT and FBAT when applied to samples like ours. We therefore developed a gene drop algorithm with rejection sampling to determine power of the PDT and FBAT by simulating family samples with founder allele frequencies, pedigree structures, and disease phenotypes matching our observed sample. As illustrated in fig 2 for an additive model and the RUNX1 locus, the power of our families to detect association is substantially greater with the PDT and FBAT than with the TDT. The PDT and FBAT not only utilise a larger variety of pedigrees (compare number of families in table 1) but also a larger portion of the information contained in each family. Greatest power was achieved with the FBAT, which uses the greatest number of phenotypically informative families and which does the best job of utilising families with missing parental information.
Table 1 Results of family based tests of the association of psoriasis with RAPTOR and RUNX1 SNPs and three‐locus haplotypes.
| Locus | Associated allele* | TDT | PDT | FBAT | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Base | Freq | No. fam† | No. inf fam‡ | T:NT§ | p value¶ | No. fam† | No. inf fam‡ | D̄** | p value¶ | No. fam† | No. inf fam‡ | Z†† | p value¶ | |
| A: All pedigrees | ||||||||||||||
| RUNX1 | ||||||||||||||
| rs745318 | T | 0.442 | 467 | 351 | 233:218 | 0.51 | 503 | 372 | 0.009 | 0.53 | 515 | 307 | 0.13 | 0.89 |
| rs734232 | A | 0.440 | 466 | 350 | 235:219 | 0.48 | 502 | 371 | 0.006 | 0.51 | 514 | 306 | 0.17 | 0.87 |
| rs895691 | A | 0.444 | 466 | 350 | 233:219 | 0.54 | 502 | 371 | 0.010 | 0.55 | 514 | 307 | 0.07 | 0.95 |
| Haplotype | TAA | 0.435 | 466 | 350 | 236:218 | 0.43 | 502 | 371 | 0.009 | 0.48 | 514 | 364 | 0.17 | 0.87 |
| RAPTOR | ||||||||||||||
| rs1564864 | T | 0.772 | 468 | 286 | 163:159 | 0.87 | 504 | 304 | 0.030 | 0.96 | 516 | 271 | 0.17 | 0.86 |
| rs2019154 | C | 0.772 | 467 | 284 | 168:160 | 0.73 | 503 | 302 | 0.037 | 0.83 | 515 | 270 | 0.31 | 0.76 |
| rs869190 | G | 0.773 | 468 | 284 | 165:159 | 0.78 | 505 | 303 | 0.039 | 0.86 | 517 | 269 | 0.31 | 0.76 |
| Haplotype | TCG | 0.771 | 467 | 285 | 164:157 | 0.76 | 503 | 301 | 0.041 | 0.84 | 515 | 315 | 0.35 | 0.79 |
| B: Non‐trio pedigrees | ||||||||||||||
| RUNX1 | ||||||||||||||
| rs745318 | T | 0.452 | 181 | 143 | 90:89 | 1.00 | 217 | 164 | −0.031 | 0.87 | 229 | 129 | −0.40 | 0.69 |
| rs734232 | A | 0.448 | 180 | 142 | 91:90 | 1.00 | 216 | 163 | −0.033 | 0.90 | 228 | 129 | −0.36 | 0.72 |
| rs895691 | A | 0.452 | 181 | 143 | 88:89 | 1.00 | 217 | 164 | −0.037 | 0.74 | 229 | 130 | −0.53 | 0.59 |
| Haplotype | TAA | 0.442 | 181 | 143 | 92:90 | 0.94 | 217 | 164 | −0.026 | 0.98 | 229 | 187 | −0.36 | 0.72 |
| RAPTOR | ||||||||||||||
| rs1564864 | T | 0.767 | 182 | 112 | 65:62 | 0.86 | 218 | 130 | 0.032 | 0.88 | 230 | 110 | 0.11 | 0.91 |
| rs2019154 | C | 0.766 | 182 | 112 | 59:57 | 0.93 | 218 | 130 | 0.031 | 0.86 | 230 | 111 | 0.11 | 0.91 |
| rs869190 | G | 0.768 | 181 | 110 | 59:57 | 0.93 | 218 | 129 | 0.037 | 0.88 | 230 | 108 | 0.17 | 0.87 |
| Haplotype | TCG | 0.765 | 182 | 112 | 62:59 | 0.86 | 218 | 130 | 0.036 | 0.91 | 230 | 149 | 0.23 | 0.82 |
Non‐trio pedigrees: those families larger than a trio with at least two affected members.
*The alleles and haplotypes tested are those reported to be associated with psoriasis (Helms et al10 for RUNX1; Anne Bowcock, personal communication for RAPTOR). Allele frequency is based on all founders in the pedigrees.
†The number of families shown for each association test counts only those families with at least one typed and phenotypically informative unit. For the TDT this unit is a trio (an affected child and both parents), for the PDT it is a trio or a discordant sibpair (an affected and unaffected sib), and for the FBAT with the settings used here it is a trio, discordant sibpair, or a sibship with three or more affected sibs.
‡The number of informative families is the subset of the typed and phenotypically informative families that are genotypically informative for the allele being tested.
§T:NT is the ratio of transmissions to non‐transmissions of the test allele from heterozygous parents to affected children in the TDT.
¶All p values are uncorrected for multiple testing.
**D̄ is a standardised measure of disequilibrium for the PDT (see Methods).
††Large sample test statistic for association, distributed as a standard normal.
Our findings for the RUNX1 binding site constitute the third negative report for this locus.15,16 The power of the recent study by Hüffmeier et al16 is good but substantially less than ours. In order to directly compare the power of the TDT in these two samples according to the method of Knapp,33 we reset our type I error rate to 0.01, the genotype relative risk of test allele heterozygotes (GRR1) to 1.6, and the GRR2 to 2.8, as specified by Hüffmeier et al.16 With these parameters, the power of our sample was 0.999, as compared to 0.93 for the family sample of Hüffmeier et al. Power calculations were not reported by Capon et al,15 but given their sample size, the power of their study should be lower than that of Hüffmeier et al or of the present study (233 v 300 v 467 trios, respectively). Furthermore, the only positive report of RUNX1 binding site association10 found no significant association of psoriasis with any of the three RUNX1 SNPs we tested or their haplotype when a single affected individual was drawn from each family and tested against an independent control sample. This is puzzling, as the TDT and case control tests are known to have nearly identical power for a given genetic model when the numbers of trios and case control pairs are equivalent.37 The difference is not likely to be due to population admixture, as the allele frequencies of the associated SNPs in the independent control sample did not differ from those of non‐transmitted alleles and haplotypes identified by the family based tests.
Our findings stand in contrast to two previous positive reports of association between psoriasis and RAPTOR.10,15 However, the Capon et al study found only weak evidence of association to only one of the RAPTOR SNPs (p = 0.027 for rs2019154) unless analysis was restricted to those trios with a documented family history of psoriasis.15 A similar stratification in the present study failed to increase the significance or strength of association for any of the RAPTOR SNPs or their haplotype (table 1B).
Association analysis is generally more powerful than linkage analysis.38 Capon et al15 cite this fact to explain why they found significant association with the RAPTOR loci when two previous studies39,40 using many of the same families failed to find significant linkage. By the same reasoning, one would expect association analysis of our sample to readily reveal a bona fide PSORS2 locus, given that our pedigree sample provides evidence for linkage to the PSORS2 region of chromosome 17q. However, it did not. Indeed, our current linkage results attain a much higher level of significance than those previously reported by Speckman et al41 for the cohort used to identify associations to the RUNX1 binding site and the RAPTOR gene10 (p = 0.00020 or 0.00063 for this study v p = 0.05 for Speckman et al).
In the original report,10 only one marker each in the RUNX1 binding site and RAPTOR peaks of association remained significant at the p = 0.05 level after correcting for multiple testing with the false discovery rate method under the assumption of correlated data,42 raising the possibility of a false positive result. The appearance of three independent negative attempts to confirm this result for the RUNX1 binding site (Capon et al15, Hüffmeier et al,16 and this study) heightens this concern. Our failure to find any evidence for association to the RAPTOR locus could be due to the low power of our sample to detect a dominant gene. However, if this were the case, it would be very surprising that two prior studies,10,16 both of which reported allele frequencies similar to our sample but were based on fewer families than this one, would be able to detect it. Taken together, the available information suggests that the PSORS2 locus or loci exist, but the corresponding gene(s) remains to be identified. Additional studies, and meta‐analysis of available studies,43 will be necessary to provide a definitive answer to this important question.
Electronic‐database information
Three supplemental appendices describing the methods used are available at http://www.jmedgenet. com/supplemental.
Supplementary Material
Acknowledgements
The authors thank all the psoriasis patients and family members who were willing to participate in this study.
Abbreviations
FBAT - family based association test
GRR - genotype relative risk
GRR1 - genotype relative risk for test allele heterozygotes
GRR2 - genotype relative risk for test allele homozygotes
LD - linkage disequilibrium
LOD - logarithm of the odds
PDT - pedigree disequilibrium test
PSORS2 - psoriasis susceptibility 2
SNP - single nucleotide polymorphism
%T - percent transmission
TDT - transmission/disequilibrium test
Footnotes
This work was supported by the Ann Arbor VA Hospital (JTE), by awards (R01 AR 042742 and R01 AR 050511) from the National Institutes of Arthritis and Musculoskeletal Diseases, National Institutes of Health (JTE, RN, PS), and by an award (Grant 01 GS 0171) from the National Genome Research Network, Germany (SJ, MW)
Competing interests: none declared
Three supplemental appendices describing the methods used are available at http://www.jmedgenet. com/supplemental.
References
- 1.Lowes M A, Lew W, Krueger J G. Current concepts in the immunopathogenesis of psoriasis. Dermatol Clin 200422(4)349–69, vii. [DOI] [PubMed] [Google Scholar]
- 2.Elder J T, Nair R P, Henseler T, Jenisch S, Stuart P, Chia N, Christophers E, Voorhees J J. The genetics of psoriasis 2001: the odyssey continues. Arch Dermatol 20011371447–1534. [DOI] [PubMed] [Google Scholar]
- 3.Elder J T, Nair R P, Weichenthal M, Stuart P, Voorhees J J, Christophers E, Jenisch S. The MHC genes in psoriasis. Curr Genomics 20056(1)39–43. [Google Scholar]
- 4.Elder J T, Nair R P, Guo S W, Henseler T, Christophers E, Voorhees J J. The genetics of psoriasis. Arch Dermatol 1994130(2)216–224. [PubMed] [Google Scholar]
- 5.Bowcock A M, Cookson W O. The genetics of psoriasis, psoriatic arthritis and atopic dermatitis. Hum Mol Genet 200413(Spec No 1)R43–R55. [DOI] [PubMed] [Google Scholar]
- 6.Tomfohrde J, Silverman A, Barnes R, Fernandez‐Vina M A, Young M, Lory D, Morris L, Wuepper K D, Stastny P, Menter A, Bowcock A M. Gene for familial psoriasis susceptibility mapped to the distal end of human chromosome 17q. Science 1994264(5162)1141–1145. [DOI] [PubMed] [Google Scholar]
- 7.Nair R P, Henseler T, Jenisch S, Stuart P, Bichakjian C K, Lenk W, Westphal E, Guo S W, Christophers E, Voorhees J J, Elder J T. Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome‐wide scan. Hum Mol Genet 19976(8)1349–1356. [DOI] [PubMed] [Google Scholar]
- 8.Enlund F, Samuelsson L, Enerback C, Inerot A, Wahlstrom J, Yhr M, Torinsson A, Martinsson T, Swanbeck G. Analysis of three suggested psoriasis susceptibility loci in a large Swedish set of families: confirmation of linkage to chromosome 6p (HLA region), and to 17q, but not to 4q. Hum Hered 199949(1)2–8. [DOI] [PubMed] [Google Scholar]
- 9.Zheng J, Jin S, Shi R. Confirmation of PSORS psoriasis susceptibility loci in a Chinese population. Arch Dermatol Res 2003295(1)14–18. [DOI] [PubMed] [Google Scholar]
- 10.Helms C, Cao L, Krueger J G, Wijsman E M, Chamian F, Gordon D, Heffernan M, Daw J A, Robarge J, Ott J, Kwok P Y, Menter A, Bowcock A M. A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. Nat Genet 200335(4)349–356. [DOI] [PubMed] [Google Scholar]
- 11.Cameron E R, Neil J C. The Runx genes: lineage‐specific oncogenes and tumor suppressors. Oncogene 200423(24)4308–4314. [DOI] [PubMed] [Google Scholar]
- 12.Prokunina L, Castillejo‐Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V, Brookes A J, Tentler D, Kristjansdottir H, Grondal G, Bolstad A I, Svenungsson E, Lundberg I, Sturfelt G, Jonssen A, Truedsson L, Lima G, Alcocer‐Varela J, Jonsson R, Gyllensten U B, Harley J B, Alarcon‐Segovia D, Steinsson K, Alarcon‐Riquelme M E. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet 200232(4)666–669. [DOI] [PubMed] [Google Scholar]
- 13.Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, Suzuki M, Nagasaki M, Ohtsuki M, Ono M, Furukawa H, Nagashima M, Yoshino S, Mabuchi A, Sekine A, Saito S, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet 200335(4)341–348. [DOI] [PubMed] [Google Scholar]
- 14.Prokunina L, Padyukov L, Bennet A, de Faire U, Wiman B, Prince J, Alfredsson L, Klareskog L, Alarcon‐Riquelme M. Association of the PD‐1.3A allele of the PDCD1 gene in patients with rheumatoid arthritis negative for rheumatoid factor and the shared epitope. Arthritis Rheum 200450(6)1770–1773. [DOI] [PubMed] [Google Scholar]
- 15.Capon F, Helms C, Veal C D, Tillman D, Burden A D, Barker J N, Bowcock A M, Trembath R C. Genetic analysis of PSORS2 markers in a UK dataset supports the association between RAPTOR SNPs and familial psoriasis. J Med Genet 200441(6)459–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hüffmeier U, Traupe H, Burkhardt H, Schurmeier‐Horst F, Lascorz J, Bohm B, Lohmann J, Stander M, Wendler J, Kelsch R, Baumann C, Kuster W, Wienker T F, Reis A. Lack of evidence for genetic association to RUNX1 binding site at PSORS2 in different German psoriasis cohorts. J Invest Dermatol 2005124(1)107–110. [DOI] [PubMed] [Google Scholar]
- 17.Elder J T, Voorhees J J. Psoriasis. In: Jameson JL, ed. Principles of molecular medicine. Vol 1. Totowa, NJ: Humana, 1998793–800.
- 18.Henseler T, Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol 198513(3)450–456. [DOI] [PubMed] [Google Scholar]
- 19.Nair R, Guo S, Jenisch S, Henseler T, Lange E M, Terhune M, Westphal E, Christophers E, Voorhees J J, Elder J T. Scanning chromosome 17 for psoriasis susceptibility: lack of evidence for a distal 17q locus. Hum Hered 199545219–230. [DOI] [PubMed] [Google Scholar]
- 20.Abecasis G R, Cherny S S, Cookson W O, Cardon L R. Merlin‐‐rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 200230(1)97–101. [DOI] [PubMed] [Google Scholar]
- 21.Kong A, Cox N J. Allele‐sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 199761(5)1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whittemore A S, Halpern J. A class of tests for linkage using affected pedigree members. Biometrics 199450(1)118–127. [PubMed] [Google Scholar]
- 23.Abecasis G R, Wigginton J E. Handling marker‐marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet 200577 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens M, Smith N J, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 200168(4)978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 200373(5)1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath S, Xu X, Lake S L, Silverman E K, Weiss S T, Laird N M. Family‐based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol 200426(1)61–69. [DOI] [PubMed] [Google Scholar]
- 27.Spielman R S, McGinnis R E, Ewens W J. Transmission test for linkage disequilibrium: the insulin gene region and insulin‐dependent diabetes mellitus (IDDM). Am J Hum Genet 199352(3)506–516. [PMC free article] [PubMed] [Google Scholar]
- 28.Martin E R, Monks S A, Warren L L, Kaplan N L. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 200067(1)146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin E R, Bass M P, Kaplan N L. Correcting for a potential bias in the pedigree disequilibrium test. Am J Hum Genet 200168(4)1065–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered 200050(4)211–223. [DOI] [PubMed] [Google Scholar]
- 31.Cluster 17 Collaboration Fine mapping of the psoriasis susceptibility gene PSORS1: a reassessment of risk associated with a putative risk haplotype lacking HLA‐Cw6. J Invest Dermatol 2005124(5)921–930. [DOI] [PubMed] [Google Scholar]
- 32.Laird N M, Horvath S, Xu X. Implementing a unified approach to family‐based tests of association. Genet Epidemiol 200019(Suppl 1)S36–S42. [DOI] [PubMed] [Google Scholar]
- 33.Knapp M. A note on power approximations for the transmission/disequilibrium test. Am J Hum Genet 199964(4)1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 199511(3)241–247. [DOI] [PubMed] [Google Scholar]
- 35.Thornton‐Wells T A, Moore J H, Haines J L. Genetics, statistics and human disease: analytical retooling for complexity. Trends Genet 200420(12)640–647. [DOI] [PubMed] [Google Scholar]
- 36.Knapp M. The transmission/disequilibrium test and parental‐genotype reconstruction: the reconstruction‐combined transmission/disequilibrium test. Am J Hum Genet 199964(3)861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGinnis R, Shifman S, Darvasi A. Power and efficiency of the TDT and case‐control design for association scans. Behav Genet 200232(2)135–144. [DOI] [PubMed] [Google Scholar]
- 38.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science 1996273(5281)1516–1517. [DOI] [PubMed] [Google Scholar]
- 39.Trembath R C, Clough R L, Rosbotham J L, Jones A B, Camp R D, Frodsham A, Browne J, Barber R, Terwilliger J, Lathrop G M, Barker J N. Identification of a major susceptibility locus on chromosome 6p and evidence for further disease loci revealed by a two stage genome‐wide search in psoriasis. Hum Mol Genet 19976(5)813–820. [DOI] [PubMed] [Google Scholar]
- 40.Veal C D, Clough R L, Barber R C, Mason S, Tillman D, Ferry B, Jones A B, Ameen M, Balendran N, Powis S H, Burden A D, Barker J N, Trembath R C. Identification of a novel psoriasis susceptibility locus at 1p and evidence of epistasis between PSORS1 and candidate loci. J Med Genet 200138(1)7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speckman R A, Wright Daw J A, Helms C, Duan S, Cao L, Taillon‐Miller P, Kwok P Y, Menter A, Bowcock A M. Novel immunoglobulin superfamily gene cluster, mapping to a region of human chromosome 17q25, linked to psoriasis susceptibility. Hum Genet 2003112(1)34–41. [DOI] [PubMed] [Google Scholar]
- 42.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001125(1–2)279–284. [DOI] [PubMed] [Google Scholar]
- 43.Sagoo G S, Tazi‐Ahnini R, Barker J W, Elder J T, Nair R P, Samuelsson L, Traupe H, Trembath R C, Robinson D A, Iles M M. Meta‐analysis of genome‐wide studies of psoriasis susceptibility reveals linkage to chromosomes 6p21 and 4q28‐q31 in Caucasian and Chinese Hans population. J Invest Dermatol 2004122(6)1401–1405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.