Abstract
Background
Recent work suggests that multiple genes and several environmental risk factors influence risk for non‐syndromic oral clefts, one of the most common birth defects in humans. Advances in high‐throughput genotyping technology now make it possible to test multiple markers in many candidate genes simultaneously.
Methods
We present findings from family based association tests of single nucleotide polymorphism (SNP) markers in 64 candidate genes genotyped using the BeadArray approach in 58 case‐parent trios from Maryland (USA) to illustrate how multiple markers in multiple genes can be analysed. To assess whether these genes were expressed in human craniofacial structures relevant to palate and lip development, we also analysed data from the Craniofacial and Oral Gene Expression Network (COGENE) consortium, and searched public databases for expression profiles of these genes.
Results
Thirteen candidate genes showed significant evidence of linkage in the presence of disequilibrium, and ten of these were found to be expressed in relevant embryonic tissues: SP100, MLPH, HDAC4, LEF1, C6orf105, CD44, ALX4, ZNF202, CRHR1, and MAPT. Three other genes showing statistical evidence (ADH1C, SCN3B, and IMP5) were not expressed in the embryonic tissues examined here.
Conclusions
This approach demonstrates how statistical evidence on large numbers of SNP markers typed in case‐parent trios can be combined with expression data to identify candidate genes for complex disorders. Many of the genes reported here have not been previously studied as candidates for oral clefts and warrant further investigation.
Keywords: candidate gene, gene expression, oral cleft, single nucleotide polymorphism, transmission disequilibrium test
As a group, oral clefts are one of the most common birth defects and represent a major public health problem. Although >300 malformation syndromes can include an oral cleft, non‐syndromic forms account for ∼70% of cases with cleft lip with or without cleft palate (CL/P) and ∼50% of cases with cleft palate only (CP).1 Current research suggests multiple levels of aetiological heterogeneity underlie non‐syndromic oral clefts: different genes (locus heterogeneity) and different mutations in one gene (allelic heterogeneity) may influence risk for oral clefts, plus causal genes may interact with one another and/or with environmental risk factors (for example, maternal smoking).2
A number of candidate genes and chromosomal regions for oral clefts have been identified using linkage approaches.3 However, linkage studies may miss evidence of genes with modest effects on risk, while tests for linkage disequilibrium (LD) should provide greater statistical power.1 Among candidate genes for oral clefts recognised in multiple studies, only the homeobox, msh‐like 1 (MSX1) gene has been shown to be causal: 2% of 917 non‐syndromic CL/P cases can be attributed to different mutations in this one gene.4 Recently, Zucchero et al5 reported 11.6% of 387 Filipino cases with non‐syndromic CL/P can be attributed to different variants in the interferon regulatory factor 6 (IRF6) gene, the causal gene in van der Woude syndrome. An Italian study confirmed the importance of IRF6.6
Family based association tests, including the transmission disequilibrium test (TDT), evaluate the independence of transmission of markers and phenotypes across families.7 Haplotypes of several single nucleotide polymorphism (SNPs) together also can provide more information than single marker analysis.8 With high‐throughput genotyping technologies, the scope of candidate gene studies can be considerably broadened.9 Here, we describe analysis of 64 candidate genes genotyped with an average of 4.3 SNPs per gene on a sample of 58 case‐parent trios from Maryland (USA). Additionally, expression data for these genes from the Craniofacial and Oral Gene Expression Network (COGENE) consortium were examined.
Methods
Subjects
Infants born with isolated, non‐syndromic CL/P or CP and their parents have been ascertained through treatment centres in Maryland and Washington, DC under a protocol approved by the Johns Hopkins University IRB. After written consent was obtained from parents, ethnicity and other data were obtained through structured interviews.2 Both subjects and their parents provided blood samples. Fifty eight non‐syndromic CL/P or CP case‐parent trios (35 complete and 23 with one parent missing) were genotyped for the current study.
Candidate genes and SNP selection
A set of 274 SNP markers in or near 64 different candidate genes from six chromosomal regions was genotyped (see appendix). Four chromosomal regions (2q37, 4q21–25, 6p23–25, and 11p11–13) were chosen based on positive evidence of linkage to a CL/P locus.3,10,11 Association between a specific mutation (W185X) in the poliovirus receptor‐related 1 (PVRL1) gene and ectodermal dysplasia syndrome with CL/P (CLPED1) prompted inclusion of the 11q23–25 region.12 Chromosome 17q21–25 corresponds to the mouse clf1 locus13 which has some previous evidence for linkage in humans.3 Candidate genes in these six chromosomal regions were selected based on available genome sequence data and/or current knowledge from animal or human studies. SNP markers of interest were obtained from literature review and the SNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) using a NorthStar searchlet program (available from http://geneticsoftwareinnovations.com/). SNPs with high “design scores” as provided by Illumina (San Diego, CA), heterozygosity above 0.1, and HapMap validation (see www.hapmap.org/index.html.en) were chosen for inclusion in the final marker panel.
DNA preparation and SNP genotyping
Genomic DNA samples were prepared from peripheral blood by protein precipitation as described previously.14 Concentration of DNA was determined using the PicoGreen dsDNA Quantitation Kit (Molecular Probes, Eugene, OR). A 3 μg aliquot of DNA was genotyped for SNP markers using Golden Gate chemistry on Sentrix Array Matrices (Illumina) at the Johns Hopkins SNP Center. Two duplicates and four CEPH control DNA samples were included on each plate.
Statistical analysis
At each SNP, minor allele frequency, heterozygosity (HET), and a test for Hardy‐Weinberg equilibrium (HWE) were computed among parents.15 Pair‐wise LD was computed as both D′ and r2 for all SNPs within a gene, extending up to 20 kb beyond both ends.16 Preliminary analyses of LD patterns and haplotype blocks were performed using Haploview (http://www.broad.mit.edu/mpg/haploview/index.php/).17
Each SNP was tested individually using the family based association test (FBAT) program (http://www.biostat.harvard.edu/˜fbat/default.html). Sliding windows of haplotypes consisting of two, three, four, and five SNPs were tested for each individual gene using the haplotype command (HBAT).18 Empirical p values for observed versus expected transmission were obtained using permutation tests, and these were further corrected for multiple comparisons using two methods, the Bonferroni correction and the principal components (spectral decomposition) method.19 Subgroups of these case‐parent trios were re‐analysed separately as a check for aetiological heterogeneity.
Candidate gene expression analysis
To assess whether these 64 genes were expressed in eight human craniofacial structures relevant to normal palate and lip development, we analysed data from the COGENE consortium (http://hg.wustl.edu/COGENE/), and searched other public databases for expression profiles.20 Four embryonic structures (4th week pharyngeal arch 1, 5th week pharyngeal arch 1, 6th week maxilla, and 8.5th week palatine shelves) are of the palate lineage and active genes may contribute to the pathogenesis of CL/P or CP alone. Four other embryonic structures (4th week frontonasal prominence, 5th week frontonasal prominence, 6th week median nasal prominence, and 8.5th week upper lip) are relevant to development of the upper lip, and genes expressed here may contribute to formation of cleft lip (CL). Data from both Affymetrix GeneChip analysis (Affymetrix, Santa Clara, CA) and Serial Analysis of Gene Expression (SAGE; http://www.sagenet.org/) were used to assess gene expression patterns.21 For Affymetrix microarray analysis, information from both perfect‐match and mis‐match probes was included to perform background correction and normalisation. A proprietary algorithm was used to classify whether expression was present (detection of transcripts representing at least 1:100 000–1:300 000 of the total transcripts in a sample) or absent.22 For SAGE analysis, a gene was considered expressed if its corresponding tags were observed at least twice, since a single count could merely represent sequencing errors.
Results
Probands in this study consisted of 8, 32, and 18 unrelated infants with non‐syndromic CL, CLP, and CP, respectively. Among these, 51 families (88%) were European American (four African American, one Filipino American, and two inter‐racial families were included). Examining duplicated samples revealed a very high reproducibility for SNP genotypes (99.98%).
Among 274 SNPs genotyped in 58 trios, 42 (15%) had minor allele frequencies and 24 (9%) had heterozygosity levels too low (<10%) to be informative. Six SNPs (2%) were dropped because they showed significant deviation from HWE (p<0.01).15 Many intragenic or flanking markers near a candidate gene showed high LD, and some genes showed virtually complete LD (r2>0.95) for all or most SNPs (for example, ADH1C). Markers become redundant when there is such complete LD.16 Table 1 presents measures of pairwise LD between all markers in each gene showing statistically significant evidence of linkage and LD.
Table 1 Characteristics of SNP markers genotyped for 13 candidate genes showing significant evidence of linkage in the presence of LD in 58 case‐parent trios.
| Gene (region) | SNP | Physical location* | Distance (bp) | HET† | HWE‡ (p) | r2\D′§ | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M5 | M6 | M7 | M8 | M9 | M10 | M11 | M12 | M13 | ||||||
| SP100 | rs890674 | 231105656 | – | 0.28 | 0.67 | – | 0.45 | 0.07 | 0.18 | 0.07 | ||||||||
| (2q37.1) | rs1678203 | 231154555 | 48 899 | 0.50 | 0.89 | 0.03 | – | 0.66 | 0.78 | 0.38 | ||||||||
| rs1678158 | 231189295 | 34 740 | 0.32 | 0.87 | 0.00 | 0.22 | – | 1.00 | 1.00 | |||||||||
| rs33079 | 231200123 | 10 828 | 0.22 | 0.57 | 0.00 | 0.04 | 0.03 | – | 0.88 | |||||||||
| rs890669 | 231210736 | 10 613 | 0.47 | 0.63 | 0.00 | 0.08 | 0.26 | 0.10 | – | |||||||||
| MLPH | rs880931 | 238178741 | – | 0.26 | 0.97 | – | 0.95 | 0.79 | ||||||||||
| (2q37.3) | rs1463795 | 238194392 | 15 651 | 0.37 | 0.57 | 0.46 | – | 0.08 | ||||||||||
| rs8260 | 238244324 | 49 932 | 0.26 | 0.03 | 0.03 | 0.00 | – | |||||||||||
| HDAC4 | rs1466094 | 239694849 | – | 0.47 | 0.65 | – | 0.86 | 0.08 | 0.05 | 0.19 | 0.08 | 0.26 | 0.12 | 0.06 | 0.06 | 0.20 | 0.01 | 0.18 |
| (2q37.2) | rs1055333 | 239706228 | 11 379 | 0.37 | 0.89 | 0.52 | – | 0.05 | 0.78 | 1.00 | 0.12 | 0.09 | 0.03 | 0.15 | 0.15 | 0.07 | 0.07 | 0.17 |
| rs2121980 | 239791807 | 85 579 | 0.48 | 0.85 | 0.00 | 0.00 | – | 0.79 | 0.57 | 0.29 | 0.15 | 0.10 | 0.43 | 0.43 | 0.10 | 0.17 | 0.68 | |
| rs2100171 | 239805757 | 13 950 | 0.23 | 1.00 | 0.00 | 0.03 | 0.19 | – | 0.79 | 0.42 | 0.38 | 1.00 | 1.00 | 1.00 | 0.53 | 0.23 | 0.26 | |
| rs686606 | 239837349 | 31 592 | 0.16 | 0.35 | 0.01 | 0.04 | 0.06 | 0.40 | – | 1.00 | 0.69 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.62 | |
| rs1962113 | 239844371 | 7022 | 0.33 | 0.02 | 0.01 | 0.01 | 0.07 | 0.01 | 0.05 | – | 0.03 | 0.64 | 0.54 | 0.54 | 0.31 | 0.21 | 0.16 | |
| rs291329 | 239880676 | 36 305 | 0.49 | 1.00 | 0.06 | 0.00 | 0.02 | 0.04 | 0.09 | 0.00 | – | 0.91 | 0.89 | 0.89 | 0.26 | 0.02 | 0.30 | |
| rs1015458 | 239892906 | 12 230 | 0.23 | 1.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.01 | 0.03 | 0.19 | – | 1.00 | 1.00 | 0.10 | 0.22 | 0.22 | |
| rs1403607 | 239912287 | 19 381 | 0.20 | 1.00 | 0.00 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.14 | 0.80 | – | 1.00 | 0.09 | 0.24 | 0.24 | |
| rs1403608 | 239918126 | 5839 | 0.20 | 1.00 | 0.00 | 0.01 | 0.01 | 0.02 | 0.01 | 0.01 | 0.14 | 0.80 | 1.00 | – | 0.09 | 0.24 | 0.24 | |
| rs2018414 | 239995925 | 77 799 | 0.39 | 0.41 | 0.01 | 0.00 | 0.01 | 0.02 | 0.05 | 0.09 | 0.02 | 0.00 | 0.09 | 0.00 | – | 0.83 | 0.83 | |
| rs2176046 | 240031867 | 35 942 | 0.17 | 1.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.01 | 0.01 | 0.00 | 0.04 | 0.24 | 0.06 | 0.18 | – | 1.00 | |
| rs925736 | 240039566 | 7699 | 0.44 | 1.00 | 0.01 | 0.00 | 0.10 | 0.00 | 0.02 | 0.00 | 0.02 | 0.00 | 0.24 | 0.00 | 0.00 | 0.05 | – | |
| ADH1C | rs1614972 | 100615333 | – | 0.39 | 0.30 | – | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||
| (4q22) | rs1662058 | 100619937 | 4604 | 0.53 | 0.56 | 0.32 | – | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||||||
| rs904096 | 100620762 | 825 | 0.53 | 0.54 | 0.31 | 1.00 | – | 1.00 | 1.00 | 1.00 | 1.00 | |||||||
| rs1789911 | 100620956 | 194 | 0.53 | 0.54 | 0.31 | 1.00 | 1.00 | – | 1.00 | 1.00 | 1.00 | |||||||
| rs1693482 | 100621143 | 187 | 0.53 | 0.54 | 0.31 | 1.00 | 1.00 | 1.00 | – | 1.00 | 1.00 | |||||||
| rs1662051 | 100624417 | 3274 | 0.53 | 0.54 | 0.31 | 1.00 | 1.00 | 1.00 | 1.00 | – | 1.00 | |||||||
| rs980972 | 100636425 | 12 008 | 0.52 | 0.61 | 0.32 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | – | |||||||
| LEF1 | rs1291490 | 109351616 | – | 0.20 | 1.00 | – | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |||||||
| (4q23–25) | rs898518 | 109374428 | 22 812 | 0.49 | 0.34 | 0.10 | – | 1.00 | 1.00 | 0.95 | 1.00 | |||||||
| rs744369 | 109383839 | 9411 | 0.50 | 0.53 | 0.10 | 0.98 | – | 1.00 | 0.98 | 1.00 | ||||||||
| rs1460405 | 109397188 | 13 349 | 0.13 | 1.00 | 0.01 | 0.07 | 0.08 | – | 1.00 | 1.00 | ||||||||
| rs956237 | 109404564 | 7376 | 0.49 | 0.51 | 0.12 | 0.79 | 0.81 | 0.07 | – | 1.00 | ||||||||
| rs922168 | 109415008 | 10 444 | 0.35 | 0.08 | 0.05 | 0.50 | 0.51 | 0.02 | 0.43 | – | ||||||||
| C6orf105 | rs210910 | 11826113 | – | 0.31 | 0.08 | – | 1.00 | 1.00 | ||||||||||
| (6p24.1) | rs210903 | 11832528 | 6415 | 0.36 | 0.35 | 0.70 | – | 0.74 | ||||||||||
| rs210895 | 11839849 | 7321 | 0.40 | 0.03 | 0.70 | 0.55 | – | |||||||||||
| CD44 | rs353612 | 35136227 | – | 0.29 | 0.45 | – | 0.92 | 0.16 | 0.15 | |||||||||
| (11pter‐p13) | rs353636 | 35141306 | 5079 | 0.34 | 0.45 | 0.70 | – | 0.37 | 0.32 | |||||||||
| rs713330 | 35180521 | 39 215 | 0.40 | 1.00 | 0.00 | 0.00 | – | 0.07 | ||||||||||
| rs13347 | 35209848 | 68 542 | 0.27 | 1.00 | 0.00 | 0.02 | 0.00 | – | ||||||||||
| ALX4 | rs729287 | 44236666 | – | 0.50 | 1.00 | – | 0.08 | 0.50 | ||||||||||
| (11p11.2) | rs879238 | 44258914 | 22 248 | 0.48 | 0.70 | 0.00 | – | 0.24 | ||||||||||
| rs1869480 | 44276233 | 17 319 | 0.15 | 1.00 | 0.06 | 0.22 | 0.00 | |||||||||||
| SCN3B | rs2027767 | 123005969 | – | 0.33 | 0.23 | – | 0.18 | 0.10 | 1.00 | 0.17 | 0.17 | 0.17 | 0.06 | |||||
| (11q24.1) | rs1274205 | 123013111 | 7142 | 0.21 | 1.00 | 0.02 | – | 1.00 | 0.88 | 0.01 | 0.01 | 0.01 | 0.32 | |||||
| rs1783901 | 123018716 | 5605 | 0.30 | 0.43 | 0.01 | 0.04 | – | 1 | 0.65 | 0.65 | 0.65 | 0.74 | ||||||
| rs1148107 | 123020997 | 2281 | 0.08 | 1.00 | 0.01 | 0.29 | 0.01 | – | 1.00 | 1.00 | 1.00 | 0.85 | ||||||
| rs1720340 | 123024127 | 3130 | 0.21 | 1.00 | 0.02 | 0.00 | 0.24 | 0.01 | – | 1.00 | 1.00 | 1.00 | ||||||
| rs1783902 | 123025214 | 1087 | 0.21 | 1.00 | 0.02 | 0.00 | 0.24 | 0.01 | 1.00 | – | 1.00 | 1.00 | ||||||
| rs1720328 | 123026420 | 1206 | 0.21 | 1.00 | 0.02 | 0.00 | 0.24 | 0.01 | 1.00 | 1.00 | – | 1.00 | ||||||
| rs1148085 | 123032279 | 5859 | 0.39 | 0.87 | 0.00 | 0.05 | 0.05 | 0.12 | 0.05 | 0.05 | 0.05 | – | ||||||
| ZNF202 | rs558021 | 123090819 | – | 0.44 | 0.26 | – | 1.00 | 1.00 | 1.00 | |||||||||
| (11q23.3) | rs481168 | 123099450 | 8631 | 0.04 | 1.00 | 0.03 | – | 1.00 | 1.00 | |||||||||
| rs10904 | 123101265 | 1815 | 0.43 | 0.39 | 0.29 | 0.01 | – | 1.00 | ||||||||||
| rs679597 | 123103014 | 1749 | 0.34 | 0.40 | 0.73 | 0.06 | 0.21 | – | ||||||||||
| CRHR1 | rs171441 | 41249125 | – | 0.15 | 0.39 | – | 1.00 | 0.95 | 0.89 | 0.77 | 0.93 | |||||||
| (17q12–22) | rs242937 | 41254149 | 5024 | 0.56 | 0.24 | 0.15 | – | 1.00 | 1.00 | 0.78 | 0.86 | |||||||
| rs242936 | 41254990 | 841 | 0.20 | 0.93 | 0.81 | 0.17 | – | 1.00 | 0.80 | 0.94 | ||||||||
| rs242950 | 41266434 | 11 444 | 0.20 | 0.83 | 0.75 | 0.16 | 0.95 | – | 0.78 | 1.00 | ||||||||
| rs878887 | 41268363 | 1929 | 0.37 | 0.91 | 0.02 | 0.17 | 0.03 | 0.03 | – | 0.87 | ||||||||
| IMP5 | rs242943 | 41280192 | 11 829 | 0.32 | 0.60 | 0.42 | 0.21 | 0.48 | 0.52 | 0.06 | – | |||||||
| (17q21.31) | rs962885 | 41291420 | 11 228 | 0.42 | 0.49 | – | 1.00 | 1.00 | 0.89 | 1.00 | 0.88 | 1.00 | 1.00 | 1.00 | ||||
| MAPT | rs916793 | 41310477 | 19 057 | 0.37 | 0.91 | 0.17 | – | 1.00 | 1.00 | 1.00 | 0.90 | 1.00 | 1.00 | 1.00 | ||||
| (17q21.1) | rs1864325 | 41333623 | 23 146 | 0.37 | 0.91 | 0.17 | 1.00 | – | 1.00 | 1.00 | 0.90 | 1.00 | 1.00 | 1.00 | ||||
| rs1467967 | 41342006 | 8383 | 0.46 | 0.63 | 0.72 | 0.16 | 0.16 | – | 1.00 | 0.93 | 1.00 | 1.00 | 1.00 | |||||
| rs1467970 | 41354402 | 12 396 | 0.37 | 0.97 | 0.18 | 1.00 | 1.00 | 0.17 | – | 0.90 | 1.00 | 1.00 | 1.00 | |||||
| rs242556 | 41358078 | 3676 | 0.34 | 0.32 | 0.42 | 0.08 | 0.08 | 0.51 | 0.08 | – | 0.90 | 0.90 | 0.90 | |||||
| rs754512 | 41411483 | 53 405 | 0.37 | 0.91 | 0.17 | 1.00 | 1.00 | 0.16 | 1.00 | 0.08 | – | 1.00 | 1.00 | |||||
| rs1052553 | 41429726 | 18 243 | 0.37 | 0.91 | 0.17 | 1.00 | 1.00 | 0.16 | 1.00 | 0.08 | 1.00 | – | 1.00 | |||||
| rs9468 | 41457408 | 27 682 | 0.37 | 0.91 | 0.17 | 1.00 | 1.00 | 0.16 | 1.00 | 0.08 | 1.00 | 1.00 | – | |||||
*Physical location determined based on Build 35; †observed heterozygosity; ‡p value for test of Hardy‐Weinberg equilibrium; §two measures of pair‐wise LD: D′ is above the diagonal, r2 is below the diagonal, M1–M13 (from the first SNP marker).
Tests of association for candidate genes
The number of SNPs genotyped per gene and results from both single marker and haplotype TDT performed for all 58 trios are summarised in the appendix. Test statistics were not computed when <10 informative families were available for an individual marker. Among the 64 candidate genes examined here, 13 yielded nominally significant p values for a single marker and/or haplotype, and these results are presented in table 2. Seven genes (MLPH, HDAC4, ADH1C, C6orf105, ALX4, SCN3, and IMP5) yielded significance at the 5% level, even after correcting for multiple comparisons. Even when two separate methods to adjust empiric p values for multiple SNP markers in determining statistical significance were used, five (HDAC4, ADH1C, ALX4, SCN3B, and IMP5) of the seven genes showed statistically significant evidence of linkage in the presence of disequilibrium. Findings on selected genes are discussed below.
Table 2 Gene expression for 13 candidate genes showing significant evidence of linkage in the presence of LD in 58 cleft case‐parent trios.
| Gene (region) | SNP | Allele* | T/N† | p value‡ | Expression§ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Haplotype | Palate lineage | Lip lineage | ||||||||||
| 1 | 2 | 1 | 2 | 3 | 4 | 5 | Affy | SAGE | Affy | SAGE | |||
| SP100 | rs890674 | G | A | 14/4 | 0.049 | 0.033 | W5PA1 | A | A | A | |||
| (2q37.1) | rs1678203 | G | A | 19/11 | W85PS | ||||||||
| rs1678158 | G | A | 13/8 | ||||||||||
| rs33079 | G | A | 7/4 | ||||||||||
| rs890669 | A | G | 14/12 | ||||||||||
| MLPH | rs880931 | G | A | 11/4 | 0.039 | 0.019 | NA | W5PA1 | NA | A | |||
| (2q37.3) | rs1463795 | C | G | 16/6 | 0.049 | ||||||||
| rs8260 | A | G | 10/5 | ||||||||||
| HDAC4 | rs1466094 | G | A | 14/11 | 0.006 | W4PA1 | W4PA1 | W5FNP | W4FNP | ||||
| (2q37.2) | rs1055333 | G | C | 15/7 | 0.036 | 0.004 | 0.013 | W5PA1 | W5PA1 | W5FNP | |||
| rs2121980 | G | A | 20/10 | 0.049 | 0.012 | 0.001 | 0.009 | 0.014 | W6MAX | ||||
| rs2100171 | A | C | 7/5 | ||||||||||
| rs686606 | G | A | 7/3 | 0.038 | |||||||||
| rs1962113 | A | C | 16/4 | 0.028 | |||||||||
| rs291329 | G | A | 18/10 | ||||||||||
| rs1015458 | A | G | 10/4 | ||||||||||
| rs1403607 | G | A | 9/3 | ||||||||||
| rs1403608 | G | A | 9/3 | ||||||||||
| rs2018414 | G | A | 15/8 | ||||||||||
| rs2176046 | G | A | 6/5 | ||||||||||
| rs925736 | A | G | 14/13 | ||||||||||
| ADH1C | rs1614972 | G | A | 19/9 | 0.002 | 0.002 | 0.002 | 0.004 | A | A | A | A | |
| (4q22) | rs1662058 | A | G | 24/6 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | ||||
| rs904096 | A | C | 25/8 | 0.005 | 0.007 | 0.010 | 0.010 | 0.015 | |||||
| rs1789911 | A | G | 24/8 | 0.008 | 0.011 | 0.009 | 0.016 | ||||||
| rs1693482 | G | A | 24/8 | 0.008 | 0.006 | 0.010 | |||||||
| rs1662051 | C | A | 25/8 | 0.005 | 0.007 | ||||||||
| rs980972 | C | A | 24/8 | 0.008 | |||||||||
| LEF1 | rs1291490 | G | A | 8/6 | A | W5PA1 | W4FNP | W5FNP | |||||
| (4q23–25) | rs898518 | A | C | 20/8 | 0.040 | 0.030 | 0.046 | W5FNP | |||||
| rs744369 | G | A | 22/8 | 0.028 | 0.036 | ||||||||
| rs1460405 | G | A | 5/2 | 0.038 | |||||||||
| rs956237 | A | G | 22/8 | 0.033 | 0.033 | ||||||||
| rs922168 | G | A | 13/7 | ||||||||||
| C6orf105 | rs210910 | G | A | 17/7 | 0.050 | 0.035 | 0.028 | W5PA1 | A | A | A | ||
| (6p24.1) | rs210903 | G | A | 20/7 | 0.020 | 0.048 | |||||||
| rs210895 | A | G | 18/12 | ||||||||||
| CD44 | rs353612 | G | A | 13/5 | 0.020 | 0.023 | W4PA1 | W4PA1 | W5FNP | W4FNP | |||
| (11pter‐p13) | rs353636 | C | A | 15/7 | 0.041 | 0.038 | W5PA1 | W5PA1 | W85UL | W5FNP | |||
| rs713330 | A | G | 19/9 | W6MAX | |||||||||
| rs13347 | G | A | 18/9 | W85PS | |||||||||
| ALX4 | rs729287 | G | A | 19/15 | 0.016 | 0.030 | 0.023 | NA | A | NA | A | ||
| (11p11.2) | rs879238 | G | A | 16/14 | |||||||||
| rs1869480 | C | G | 6/2 | ||||||||||
| SCN3B | rs2027767 | G | A | 14/11 | 0.031 | 0.005 | 0.007 | 0.008 | NA | A | A | A | |
| (11q24.1) | rs1274205 | C | G | 12/1 | 0.005 | 0.001 | 0.004 | 0.001 | 0.001 | ||||
| rs1783901 | G | A | 13/5 | 0.046 | 0.009 | 0.004 | 0.004 | 0.004 | |||||
| rs1148107 | A | C | 8/1 | 0.039 | 0.046 | 0.049 | |||||||
| rs1720340 | A | G | 11/6 | ||||||||||
| rs1783902 | C | A | 11/6 | ||||||||||
| rs1720328 | C | G | 11/6 | ||||||||||
| rs1148085 | A | G | 15/12 | ||||||||||
| ZNF202 | rs558021 | T | C | 16/8 | W4PA1 | A | W4FNP | A | |||||
| (11q23.3) | rs481168 | A | C | 2/1 | W5PA1 | W5FNP | |||||||
| rs10904 | A | G | 16/10 | W6MAX | |||||||||
| rs679597 | A | G | 18/7 | 0.048 | W85PS | ||||||||
| CRHR1 | rs171441 | T | C | 10/3 | W4PA1 | NA | W4FNP | NA | |||||
| (17q12–22) | rs242937 | T | C | 17/14 | W5PA1 | W5FNP | |||||||
| rs242936 | T | C | 11/4 | 0.035 | 0.046 | 0.037 | W6MAX | W6MNP | |||||
| rs242950 | A | G | 10/4 | 0.045 | W85PS | W85UL | |||||||
| rs878887 | T | C | 16/7 | 0.046 | 0.024 | 0.025 | 0.024 | ||||||
| IMP5 | rs242943 | A | G | 16/7 | 0.028 | 0.031 | 0.021 | NA | A | NA | A | ||
| (17q21.31) | rs962885 | C | T | 15/11 | 0.021 | 0.019 | 0.022 | 0.032 | |||||
| MAPT | rs916793 | T | C | 16/8 | 0.041 | 0.025 | W5PA1 | A | W85UL | A | |||
| (17q21.1) | rs1864325 | T | C | 16/8 | 0.048 | 0.025 | 0.025 | W85PS | |||||
| rs1467967 | A | G | 19/13 | 0.026 | 0.025 | 0.026 | |||||||
| rs1467970 | T | G | 16/8 | ||||||||||
| rs242556 | T | A | 12/11 | ||||||||||
| rs754512 | T | A | 16/8 | ||||||||||
| rs1052553 | A | G | 16/8 | ||||||||||
| rs9468 | T | C | 16/8 | ||||||||||
*Overtransmitted alleles are in bold type and the major allele is listed first; †transmission/non‐transmission counts from heterozygous parents; ‡significant p values from single SNP TDT and global p values from haplotype TDT with two, three, four, and five SNPs; §the embryonic structures analysed were: W4PA1, 4th week pharyngeal arch 1; W5PA1, 5th week pharyngeal arch 1; W6MAX, 6th week maxilla; W85PS, 8.5th week palatine shelves; W4FNP, 4th week frontonasal prominence; W5FNP, 5th week frontonasal prominence; W6MNP, 6th week median nasal prominence; W85UL, 8.5th week upper lip. A, absent; NA, not available.
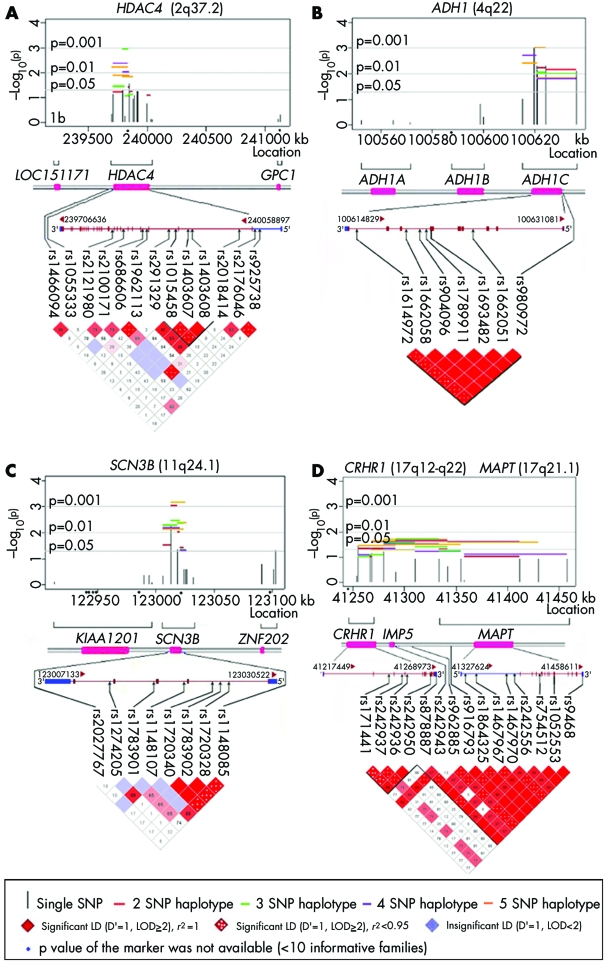
In chromosome 2q37, the most significant evidence of linkage and LD was observed for histone deacetylase 4 (HDAC4) (lowest p = 0.001). Since rs1962113 deviated slightly from HWE (p = 0.02), the evidence for linkage and LD from haplotypes in fig 1A (horizontal lines) may be largely due to preferential transmission of allele G at SNP rs2121980 (see table 2).
Figure 1 Results of SNP and haplotype analysis for selected candidate genes showing significant evidence of linkage and LD in 58 case‐parent trios, along with patterns of pairwise LD between markers. (A) HDAC4; (B) ADH1C; (C) SCN3B; (D) gene cluster consisting of CRHR1, IMP5, and MAPT. In each plot, the –log10 (empiric p value) for the overall χ2 test for an individual SNP (vertical line) and for sliding windows of haplotypes of two to five SNPs (horizontal lines) is presented. Nominal significance levels are denoted by grey lines (5%, 1%, and 0.1%).
Among seven genes in 4q21–25, alcohol dehydrogenase 1C gamma polypeptide (ADH1C) showed the strongest evidence of linkage and LD for both single markers (lowest p = 0.001) and haplotypes (lowest p = 0.0007) (see fig 1B). Six of eight SNPs in ADH1C formed a block of complete LD, including a non‐synonymous variant (rs1693482) which leads to substitution of arginine (R) to glutamine (Q) in exon 3.
As shown in fig 1C, two adjacent SNPs (rs1274205 and rs1783901) in the sodium channel, voltage‐gated, type III, beta (SCN3B) gene appeared to be responsible for much of the statistical evidence from multiple haplotypes (lowest p = 0.0007) in this gene. Nominally significant evidence of linkage and LD was observed from multiple haplotypes composed of markers in and across three adjacent genes in 17q21–22: corticotropin releasing hormone receptor 1 (CRHR1), intramembrane protease 5 (IMP5), and microtubule‐associated protein τ (MAPT) (lowest p = 0.019) (fig 1D).
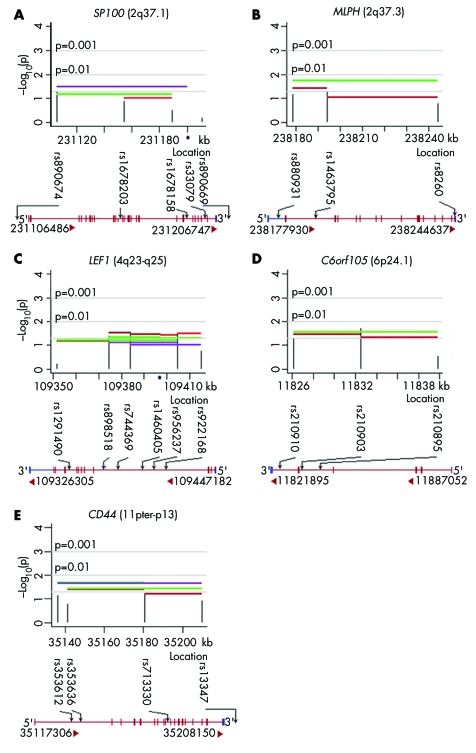
Figure 2 summarises analyses of five genes yielding nominally significant evidence for linkage and LD from individual SNPs and haplotypes, which also showed positive expression results (see table 2). These genes are nuclear antigen Sp100 (SP100), melanophilin (MLPH), lymphoid enhancer‐binding factor 1 (LEF1), chromosome 6 open reading frame 105 (C6orf105), and CD44 antigen (CD44). Two additional genes yielded statistical evidence: aristaless‐like homeobox 4 (ALX4) and zinc finger protein 202 (ZNF202). None of the four SNPs genotyped in ZNF202 were in LD with SNPs in the nearby SCN3B gene.
Figure 2 Plots of –log10 (p value) for five genes yielding significant evidence of linkage in the presence of LD from both individual SNP and haplotypes in 58 case‐parent trios and showing expression in the lip and palate lineages.
The case‐parent trio design minimises problems of confounding due to population stratification but cannot eliminate other sources of aetiological heterogeneity. Therefore, 40 CL/P trios were re‐analysed separately as a check for heterogeneity (see appendix). Of the 13 genes showing statistical evidence for linkage and LD, six genes (SP100, MLPH, HDAC4, C6orf105, CD44, and SCN3B) showed no change in the strength of evidence, while seven yielded slightly weaker evidence (ADH1C, LEF1, ALX4, ZNF202, CRHR1, IMP5, and MAPT). Only ALX4, ZNF202, and the CRHR‐IMP5‐MAPT cluster would have been overlooked if the analysis was limited to this smaller subgroup. Three genes showed nominal statistical significance in the CL/P subgroup but not in the total group (SP110, FLJ12584, and WNT11).
Candidate gene expression analysis
Gene expression information could be obtained for 37 of these 64 genes (58%) due to the presence of probe sets on the Affymetrix U95Av2 chips used for microarray analysis. For SAGE analysis, gene expression information could be obtained for 53 genes (83%) since identifiable tag sequences were assigned to these genes. Gene expression information from 36 genes (56%) was available from both microarray and SAGE analyses.
In our microarray analysis, gene expression data were available for four structures in the palate lineage and four structures in the lip lineage (described above). In SAGE analysis, gene expression data were only available for two structures (4th week pharyngeal arch 1 and 5th week pharyngeal arch 1) in the palate lineage, and two structures (4th week frontonasal prominence and 5th week frontonasal prominence) in the lip lineage. When both palate and lip lineages were considered, 29 genes were expressed in at least one stage by microarray, and 24 genes were expressed in at least one stage by SAGE analysis. A total of 39 genes were expressed in either lineage by either expression analysis (see appendix).
Nine of the 13 candidate genes showing significant evidence of linkage and LD (SP100, MLPH, C6orf105, ZNF202, CRHR1, HDAC4, LEF1, CD44, and MAPT) were expressed in either the developing palate or lip as documented by one of these two expression analyses and three were found to be expressed by both microarray and SAGE (table 2). LEF1 expression was relatively low compared to HDAC4 and CD44, which showed moderate to high expression (with more than 50 out of 50 000 tags in each of the four SAGE libraries, and present in four to six of eight structures by microarray analyses). SP100, C6orf105, and MAPT showed lower expression than ZNF202 by microarray analysis, possibly explaining why these genes were not detected by SAGE. We did not detect expression by either method for four genes showing statistical evidence of linkage and LD (ADH1C, SCN3B, ALX4, and IMP5), but the last three of these could not have been detected by microarray because their probe sets were not on the chip. Searching public databases revealed that the mouse homolog of ALX4 is expressed in the frontonasal prominence at E9.5,23 the stage corresponding to human 4th to 5th week of embryonic development. Thus, ALX4 could be expressed during human 4th and/or 5th week frontonasal prominence development. Six genes (DGKD, COL6A3, LRRFIP1, WNT11, NSF, and USP36) among nine showing marginal statistical significance (0.05<p<0.1) were also expressed in these key embryonic structures. Expression data for all 64 candidate genes at the different developmental stages are listed in the appendix.
Discussion
Many new candidate genes were tested in this analysis, although several recognised candidate genes (for example, IRF6, TGFA, MSX1, etc) were not included. Subgroup analyses of 40 CL/P trios showed little evidence of aetiological heterogeneity, although some genes yielded weaker statistical evidence (as would be expected due to smaller sample sizes). Clearly, the combination of statistical and expression evidence for any candidate gene in this study raises the possibility that it could play some aetiological role for oral clefts, but lack of statistical evidence from this modest sample of case‐parent trios does not preclude aetiological importance.
We have only partial knowledge of the biological function of these genes showing both statistical and expression evidence. HDAC4 represses transcription, and Hdac4‐null mice displayed premature ossification of developing bones.24 The ADH1C gene product is involved in both ethanol and retinol oxidation. Maternal alcohol consumption has been shown to increase risk of non‐syndromic oral clefts, while retinol may be a teratogen for CP.25 Recently, it was suggested the Ile350Val variant at ADH1C may protect against oral clefts, but there was no significant evidence for an effect of fetal genotype or interaction with maternal alcohol consumption.26 The gamma 2 protein (corresponding to the 272Gln‐350Val haplotype) results in slower ethanol oxidisation than the gamma 1 protein (272Arg‐350Ile haplotype).27 Our study showed significant over‐transmission of the A allele at rs1693482 (which results in a Gln at amino acid 272) to affected children compared to the frequencies of A allele among mothers and fathers (0.44 v 0.41 and 0.38, respectively; p = 0.008).
The SCN3B gene codes for an auxiliary component regulating ion‐conducting alpha subunits.28PVRL1 is located 3.9 Mb away from the 3′ end of SCN3B, and has been reported to be associated with oral clefts.12 However, the statistical evidence of association in our study (lowest p = 0.0007) is the first suggestion that SCN3B may be involved in oral clefts. The CRHR1 gene product binds to corticotropin‐releasing hormone,29 while MAPT transcripts are differentially expressed in the nervous system.30 Although these genes are homologs of genes in the mouse clf1 region,13 and both are expressed during craniofacial development, neither has been suggested to control risk for oral clefts.
SP100, a nuclear autoimmune antigen, acts as an antagonist for ETS1 mediated cell proliferation and differentiation in humans.31 The gene for MLPH, a critical component of the melanosome transport machinery, is located 1.16 Mb away from D2S338, a marker which showed evidence for linkage in 26 CL/P multiplex families.10,32 Two other genes (ASB18 and IQCA) near D2S338 showed no evidence for linkage and LD, and were not expressed in either palate or lip lineages. The transcription factor LEF1 is regulated by TGFβ3 which appears to play a major role in transformation of the medial edge epithelial seam into mesenchyme.33 Furthermore, LEF1 binds in response to stimulation through the WNT signalling pathway. Juriloff et al recently presented evidence that insertion of a transposable element near the 3′ end of Wnt9b may cause the high incidence of CLP in A/WySn mice.13 In our study, both WNT3 and WNT11 genes yielded only marginal statistical evidence and only WNT 11 was expressed in 6th week median nasal prominence. The WNT9B gene showed no evidence of association in these 58 trios and expression of this gene could not be tested.
Chromosomal region 6p23–25 has been identified from previous genome scans, and genes such as the bone morphogenetic protein 6 (BMP6) and endothelin1 (EDN1), which are involved in neural crest development, have been suggested as candidate genes.3 While markers in these two genes failed to show any evidence of linkage and LD here, the combination of statistical evidence and expression of C6orf105 suggested this putative gene may play some aetiological role. CD44 is an integral cell membrane glycoprotein with a postulated role in matrix adhesion and proliferation of mesenchymal cells.34 The Alx4 gene product is a potent transcriptional activator expressed at sites of epithelial‐mesenchymal interaction. Alx3/Alx4 double mutants show nasal clefts in mice, even though homozygotes for a null allele at Alx3 are indistinguishable from wild type mice.23
The presence of a gene transcript in the appropriate embryonic tissue validates selecting a gene as a candidate for oral clefts, but its absence does not necessarily mean the gene cannot influence risk. Since approximately 50 000 tags were sequenced for each SAGE library, only moderately to highly expressed genes could be detected, and the AffyMetrix Genechip can only analyse genes with included probe sets. For example, ALX4 may not have been detected because it is expressed below the detection limit of our SAGE libraries, and no probes were included on the chip, even though published mouse data suggest ALX4 could be expressed in the developing lip.
Another possibility is that a candidate gene might act in organs other than the developing face. For example, the ADH1C and SCN3B genes are expressed in response to exposure to ethanol (primarily in the liver) and cellular stress, respectively. No published expression data on embryonic craniofacial development were available for SCN3B and IMP5 (or their mouse homologs). Theses genes may be indirectly involved in the pathogenesis, or they may be a surrogate for some nearby causal gene in the statistical analysis described here.
Fixing an arbitrary significance level to account for multiple testing may be too conservative, since multiple genes may have small effects on risk for complex and heterogeneous disorders such as oral clefts. Furthermore, SNPs over small physical distances are often correlated.35 Given our small sample size, measuring significance becomes a concern and false negative results are more plausible than false positives. Therefore, marginal results from TDT analyses presented here require replication in additional studies. It is worth noting that five genes (HDAC4, ADH1C, ALX4, SCN3B, and IMP5) yielded significance even after Bonferroni correction, which is generally considered overly conservative.19
Most of the genes showing evidence of linkage and LD in this study have not been extensively studied. Ultimately, our results must be replicated in other studies, but this study does illustrate how large amounts of SNP data obtained from high‐throughput genotyping methods can be tested for linkage and LD. In addition, our results from family based association tests were generally validated through expression data. The novel candidate genes identified here provide a starting point for future studies.
Acknowledgements
We thank all participants who donated samples for the Maryland oral cleft study (1992–2003), in addition to all the staff at Johns Hopkins Hospital and McKusick‐Nathans Institute of Genetic Medicine who were involved in this research. In particular, we appreciate the technical assistance of Audrey Grant, Shu Zhang, and Catherine Tran.
Electronic‐database information
The following URLs were mentioned in this article: SNP database: http://www.ncbi.nlm.nih.gov/projects/SNP/; the NorthStar searchlet program: http:// geneticsoftwareinnovations.com/; Haploview: http://www.broad.mit.edu/mpg/haploview/index.php/; FBAT program: http://www.biostat.harvard.edu/˜fbat/default.html; COGENE consortium: http://hg.wustl.edu/COGENE/; and Serial Analysis of Gene Expression (SAGE): http://www.sagenet.org/
Abbreviations
CL - cleft lip
CL/P - cleft lip with or without cleft palate
COGENE - Craniofacial and Oral Gene Expression Network
CP - cleft palate only
HET - observed heterozygosity
HWE - Hardy‐Weinberg equilibrium
LD - linkage disequilibrium
SAGE - Serial Analysis of Gene Expression
SNP - single nucleotide polymorphism
TDT - transmission disequilibrium test
Appendix
TableA1 Summary of TDT for SNPs and haplotypes in 64 candidate genes for non‐syndromic oral cleft in 58 trios, and gene expression in eight structures relevant to craniofacial development.
| Gene | SNP (n) | TDT (p value) | Expression | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 58 Trios* | 40 Trios with CL/P† | Palate lineage | Lip lineage | ||||||
| SNP | Haplotype | SNP | Haplotype | Affy‡ | SAGE§ | Affy‡ | SAGE§ | ||
| Chromosome 2q37 (26 genes/115 SNPs) | |||||||||
| SP110 | 3 | <0.05 | <0.05 | A | A | A | A | ||
| SP100 | 5 | <0.05 | <0.05 | <0.05 | 4 | A | A | A | |
| SPATA3 | 3 | NA | 2 | NA | A | ||||
| FLJ12584 | 4 | <0.05 | NA | 1, 2 | NA | 5, 6 | |||
| PDE6D | 3 | A | A | 6 | 5, 6 | ||||
| EIF4EL3 | 3 | NA | 1, 2 | NA | 6 | ||||
| TNRC15 | 3 | 2 | 1, 2 | 6, 7 | 6 | ||||
| NGEF | 4 | NA | A | NA | A | ||||
| DGKD | 6 | <0.1 | 1–3 | 1, 2 | 6, 7 | 6 | |||
| USP40 | 4 | <0.1 | <0.1 | NA | A | NA | A | ||
| TRPM8 | 6 | NA | A | NA | A | ||||
| CENTG2 | 9 | 1, 2 | 2 | 6 | A | ||||
| ASB18 | 4 | NA | NA | NA | NA | ||||
| IQCA | 4 | NA | A | NA | A | ||||
| COL6A3 | 3 | <0.1 | <0.1 | 1–3 | 1, 2 | 6–8 | 5, 6 | ||
| MLPH | 3 | <0.05 | <0.05 | <0.05 | NA | 2 | NA | A | |
| LRRFIP1 | 4 | <0.1 | <0.1 | 2 | A | 6 | A | ||
| FLJ40411 | 4 | NA | NA | NA | NA | ||||
| NCE2 | 3 | NA | 2 | NA | 5, 6 | ||||
| ASB1 | 4 | A | 1 | A | 5 | ||||
| LOC151171 | 3 | NA | NA | NA | NA | ||||
| HDAC4 | 13 | <0.05 | <0.01 | <0.05 | <0.05 | 1–3 | 1, 2 | 6 | 5, 6 |
| GPC1 | 3 | 1–3 | A | 5–7 | 6 | ||||
| KIF1A | 3 | A | A | A | A | ||||
| PASK | 3 | <0.1 | 1–4 | A | 6 | A | |||
| FARP2 | 8 | A | A | A | A | ||||
| Chromosome 4q21–q25 (7 genes/29 SNPs) | |||||||||
| MGC46496 | 3 | NA | NA | NA | NA | ||||
| ADH1A | 3 | 1, 2 | A | A | A | ||||
| ADH1B | 3 | A | A | 6 | A | ||||
| ADH1C | 8 | <0.01 | <0.001 | <0.05 | <0.05 | A | A | A | A |
| SLC39A8 | 3 | 1–3 | A | 6 | A | ||||
| UBE2D3 | 3 | 1–3 | A | 5–7 | 4 | ||||
| LEF1 | 6 | <0.05 | <0.05 | <0.1 | <0.1 | A | 2 | 5, 6 | 4 |
| Chromosome 6p23–p25 (6 genes/20 SNPs) | |||||||||
| SEC5L1 | 3 | NA | A | NA | A | ||||
| GMDS | 3 | A | 1 | 8 | A | ||||
| BMP6 | 3 | A | A | A | A | ||||
| C6orf105 | 3 | <0.05 | <0.05 | <0.05 | <0.1 | 2 | A | A | A |
| EDN1 | 4 | NA | NA | NA | NA | ||||
| JARID2 | 4 | 1–3 | 1, 2 | 5–7 | 3, 4 | ||||
| Chromosome 11p11–p13 (6 genes/22 SNPs) | |||||||||
| C11orf8 | 3 | 2, 3 | A | 6, 7 | 3, 4 | ||||
| ABTB2 | 3 | 3 | 2 | 6 | 3 | ||||
| CD44 | 4 | <0.1 | <0.05 | <0.05 | <0.05 | 1–4 | 1, 2 | 6, 8 | 3, 4 |
| ALX4 | 4 | <0.05 | <0.05 | NA | A | NA | A | ||
| LOC220074 | 5 | NA | 1 | NA | A | ||||
| WNT11 | 3 | <0.1 | <0.05 | <0.1 | A | A | 7 | A | |
| Chromosome 11q23–q25 (5 genes/29 SNPs) | |||||||||
| KIAA1959 | 5 | <0.1 | <0.1 | NA | NA | NA | NA | ||
| KIAA1201 | 9 | A | A | A | A | ||||
| SCN3B | 8 | <0.01 | <0.001 | <0.01 | NA | A | A | A | |
| ZNF202 | 4 | <0.05 | 1–4 | A | 5, 6 | A | |||
| LOC338661 | 3 | NA | NA | NA | NA | ||||
| Chromosome 17q21–q25 (14 genes/59 SNPs) | |||||||||
| CRHR1 | 5 | <0.05 | <0.05 | 1–4 | NA | 5–8 | NA | ||
| IMP5 | 2 | <0.1 | <0.05 | NA | A | NA | A | ||
| MAPT | 9 | <0.05 | 2, 4 | A | 8 | A | |||
| LOC284058 | 5 | NA | 1, 2 | NA | 4 | ||||
| NSF | 3 | <0.1 | <0.1 | 1–3 | A | 5, 6 | A | ||
| WNT3 | 5 | <0.1 | NA | A | NA | A | |||
| WNT9B | 3 | NA | NA | NA | NA | ||||
| ITGB3 | 5 | 1 | A | A | A | ||||
| DLX4 | 3 | 2 | A | A | A | ||||
| KCNJ16 | 5 | NA | NA | NA | NA | ||||
| KCNJ2 | 4 | 2 | A | A | A | ||||
| LOC400618 | 3 | NA | NA | NA | NA | ||||
| USP36 | 4 | <0.1 | NA | 1, 2 | NA | 3, 4 | |||
| TIMP2 | 3 | A | 1, 2 | A | 3, 4 | ||||
*58 trios including four African American, one Filipino American, and two inter‐racial families; †40 trios with CL/P; ‡for Affymetrix GeneChip analysis (Affy), the structures analyzed are: 1) 4th week pharyngeal arch 1, 2) 5th week pharyngeal arch 1, 3) 6th week maxilla, 4) 8.5th week palatine shelves, 5) 4th week frontonasal prominence, 6) 5th week frontonasal prominence, 7) 6th week median nasal prominence, and 8) 8.5th week upper lip; A, absent call; NA, not available because no probe sets were on the chip; §for Serial Analysis of Gene Expression (SAGE), the structures analyzed are: 1) 4th week pharyngeal arch 1, 2) 5th week pharyngeal arch 1, 3) 4th week frontonasal prominence, 4) 5th week frontonasal prominence; A, absent; NA, not available because appropriate tag sequence information was omitted.
Footnotes
This research was supported by P60‐DE13078 and R01‐DE014581, NO1‐DE92630 from the National Institute of Dental and Craniofacial Research.
Competing interests: none declared
The following URLs were mentioned in this article: SNP database: http://www.ncbi.nlm.nih.gov/projects/SNP/; the NorthStar searchlet program: http:// geneticsoftwareinnovations.com/; Haploview: http://www.broad.mit.edu/mpg/haploview/index.php/; FBAT program: http://www.biostat.harvard.edu/˜fbat/default.html; COGENE consortium: http://hg.wustl.edu/COGENE/; and Serial Analysis of Gene Expression (SAGE): http://www.sagenet.org/
References
- 1.Lidral A C, Murray J C. Genetic approaches to identify disease genes for birth defects with cleft lip/palate as a model. Birth Defects Res A Clin Mol Teratol 200470893–901. [DOI] [PubMed] [Google Scholar]
- 2.Beaty T H, Maestri N E, Hetmanski J B, Wyszynski D F, Vanderkolk C A, Simpson J C, McIntosh I, Smith E A, Zeiger J S, Raymond G V, Panny S R, Tifft C J, Lewanda A F, Cristion C A, Wulfsberg E A. Testing for interaction between maternal smoking and TGFA genotype among oral cleft cases born in Maryland 1992–1996. Cleft Palate Craniofac J 199734447–454. [DOI] [PubMed] [Google Scholar]
- 3.Marazita M L, Murray J C, Lidral A C, Arcos‐Burgos M, Cooper M E, Goldstein T, Maher B S, Daack‐Hirsch S, Schultz R, Mansilla M A, Field L L, Liu Y E, Prescott N, Malcolm S, Winter R, Ray A, Moreno L, Valencia C, Neiswanger K, Wyszynski D F, Bailey‐Wilson J E, Albacha‐Hejazi H, Beaty T H, McIntosh I, Hetmanski J B, Tuncbilek G, Edwards M, Harkin L, Scott R, Roddick L G. Meta‐analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. Am J Hum Genet 200475161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jezewski P A, Vieira A R, Nishimura C, Ludwig B, Johnson M, O'Brien S E, Daack‐Hirsch S, Schultz R E, Weber A, Nepomucena B, Romitti P A, Christensen K, Orioli I M, Castilla E E, Machida J, Natsume N, Murray J C. Complete sequencing shows a role for MSX1 in non‐syndromic cleft lip and palate. J Med Genet 200340399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zucchero T M, Cooper M E, Maher B S, Daack‐Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira A R, Orioli I M, Castilla E E, Moreno L, Arcos‐Burgos M, Lidral A C, Field L L, Liu Y E, Ray A, Goldstein T H, Schultz R E, Shi M, Johnson M K, Kondo S, Schutte B C, Marazita M L, Murray J C. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med 2004351769–780. [DOI] [PubMed] [Google Scholar]
- 6.Scapoli L, Palmieri A, Martinelli M, Pezzetti F, Carinci P, Tognon M, Carinci F. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am J Hum Genet 200576180–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spielman R S, McGinnis R E, Ewens W J. Transmission test for linkage disequilibrium: the insulin gene region and insulin‐dependent diabetes mellitus (IDDM). Am J Hum Genet 199352506–516. [PMC free article] [PubMed] [Google Scholar]
- 8.Lu X, Niu T, Liu J S. Haplotype information and linkage disequilibrium mapping for single nucleotide polymorphisms. Genome Res 2003132112–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliphant A, Barker D L, Stuelpnagel J R, Chee M S. BeadArray technology: enabling accurate, cost‐effective approach to high‐throughput genotyping. Biotechniques 2002(Suppl)56–8, 60-1. [PubMed]
- 10.Zeiger J S, Hetmanski J B, Beaty T H, VanderKolk C A, Wyszynski D F, Bailey‐Wilson J E, de Luna R O, Perandones C, Tolarova M M, Mosby T, Bennun R, Segovia M, Calda P, Pugh E W, Doheny K, McIntosh I. Evidence for linkage of nonsyndromic cleft lip with or without cleft palate to a region on chromosome 2. Eur J Hum Genet 200311835–839. [DOI] [PubMed] [Google Scholar]
- 11.Schultz R E, Cooper M E, Daack‐Hirsch S, Shi M, Nepomucena B, Graf K A, O'Brien E K, O'Brien S E, Marazita M L, Murray J C. Targeted scan of fifteen regions for nonsyndromic cleft lip and palate in Filipina families. Am J Med Genet A 2004125A17–22. [DOI] [PubMed] [Google Scholar]
- 12.Sozen M A, Suzuki K, Tolarova M M, Bustos T, Fernandez Iglesias J E, Spritz R A. Mutation of PVRL1 is associated with sporadic, non‐syndromic cleft lip/palate in northern Venezuela. Nat Genet 200129141–142. [DOI] [PubMed] [Google Scholar]
- 13.Juriloff D M, Harris M J, Dewell S L, Brown C J, Mager D L, Gagnier L, Mah D G. Investigations of the genomic region that contains the clf1 mutation, a causal gene in multifactorial cleft lip and palate in mice. Birth Defects Res A Clin Mol Teratol 200573103–113. [DOI] [PubMed] [Google Scholar]
- 14.Bellus G A, Hefferon T W, Ortiz de Luna R I, Hecht J T, Horton W A, Machado M, Kaitila I, McIntosh I, Francomano C A. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet 199556368–373. [PMC free article] [PubMed] [Google Scholar]
- 15.Schaid D J, Jacobsen S J. Biased tests of association: comparisons of allele frequencies when departing from Hardy‐Weinberg proportions. Am J Epidemiol 1999149706–711. [DOI] [PubMed] [Google Scholar]
- 16.Ardlie K G, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet 20023299–309. [DOI] [PubMed] [Google Scholar]
- 17.Barrett J C, Fry B, Maller J, Daly M J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 200521263–265. [DOI] [PubMed] [Google Scholar]
- 18.Horvath S, Xu X, Laird N M. The family based association test method: strategies for studying general genotype‐‐phenotype associations. Eur J Hum Genet 20019301–306. [DOI] [PubMed] [Google Scholar]
- 19.Nyholt D R. A simple correction for multiple testing for single‐nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 200474765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai J, Ash D, Kotch L E, Jabs E W, Attie‐Bitach T, Auge J, Mattei G, Etchevers H, Vekemans M, Korshunova Y, Tidwell R, Messina D N, Winston J B, Lovett M. Gene expression in pharyngeal arch 1 during human embryonic development. Hum Mol Genet 200514903–912. [DOI] [PubMed] [Google Scholar]
- 21.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Serial analysis of gene expression. Science 1995270484–487. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Wong W H. Model‐based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A 20019831–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beverdam A, Brouwer A, Reijnen M, Korving J, Meijlink F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development 20011283975–3986. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Dowling M, Yang X J, Kao G D. Caspase‐mediated specific cleavage of human histone deacetylase 4. J Biol Chem 200427934537–34546. [DOI] [PubMed] [Google Scholar]
- 25.Sulik K K, Cook C S, Webster W S. Teratogens and craniofacial malformations: relationships to cell death. Development 1988103(Suppl)213–231. [DOI] [PubMed] [Google Scholar]
- 26.Chevrier C, Perret C, Bahuau M, Nelva A, Herman C, Francannet C, Robert‐Gnansia E, Cordier S. Interaction between the ADH1C polymorphism and maternal alcohol intake in the risk of nonsyndromic oral clefts: an evaluation of the contribution of child and maternal genotypes. Birth Defects Res A Clin Mol Teratol 200573114–122. [DOI] [PubMed] [Google Scholar]
- 27.Hines L M, Stampfer M J, Ma J, Gaziano J M, Ridker P M, Hankinson S E, Sacks F, Rimm E B, Hunter D J. Genetic variation in alcohol dehydrogenase and the beneficial effect of moderate alcohol consumption on myocardial infarction. N Engl J Med 2001344549–555. [DOI] [PubMed] [Google Scholar]
- 28.Adachi K, Toyota M, Sasaki Y, Yamashita T, Ishida S, Ohe‐Toyota M, Maruyama R, Hinoda Y, Saito T, Imai K, Kudo R, Tokino T. Identification of SCN3B as a novel p53‐inducible proapoptotic gene. Oncogene 2004237791–7798. [DOI] [PubMed] [Google Scholar]
- 29.Contoreggi C, Rice K C, Chrousos G. Nonpeptide corticotropin‐releasing hormone receptor type 1 antagonists and their applications in psychosomatic disorders. Neuroendocrinology 200480111–123. [DOI] [PubMed] [Google Scholar]
- 30.Heutink P. Untangling tau‐related dementia. Hum Mol Genet 20009979–986. [DOI] [PubMed] [Google Scholar]
- 31.Yordy J S, Moussa O, Pei H, Chaussabel D, Li R, Watson D K. SP100 inhibits ETS1 activity in primary endothelial cells. Oncogene 200524916–931. [DOI] [PubMed] [Google Scholar]
- 32.Menasche G, Ho C H, Sanal O, Feldmann J, Tezcan I, Ersoy F, Houdusse A, Fischer A, de Saint Basile G. Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5A F‐exon deletion (GS1). J Clin Invest 2003112450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nawshad A, LaGamba D, Hay , eds. Transforming growth factor beta (TGFbeta) signalling in palatal growth, apoptosis and epithelial mesenchymal transformation (EMT). Arch Oral Biol 200449675–689. [DOI] [PubMed] [Google Scholar]
- 34.Sherman L, Wainwright D, Ponta H, Herrlich P. A splice variant of CD44 expressed in the apical ectodermal ridge presents fibroblast growth factors to limb mesenchyme and is required for limb outgrowth. Genes Dev 1998121058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao D C, Gu C. False positives and false negatives in genome scans. Adv Genet 200142487–498. [DOI] [PubMed] [Google Scholar]