Abstract
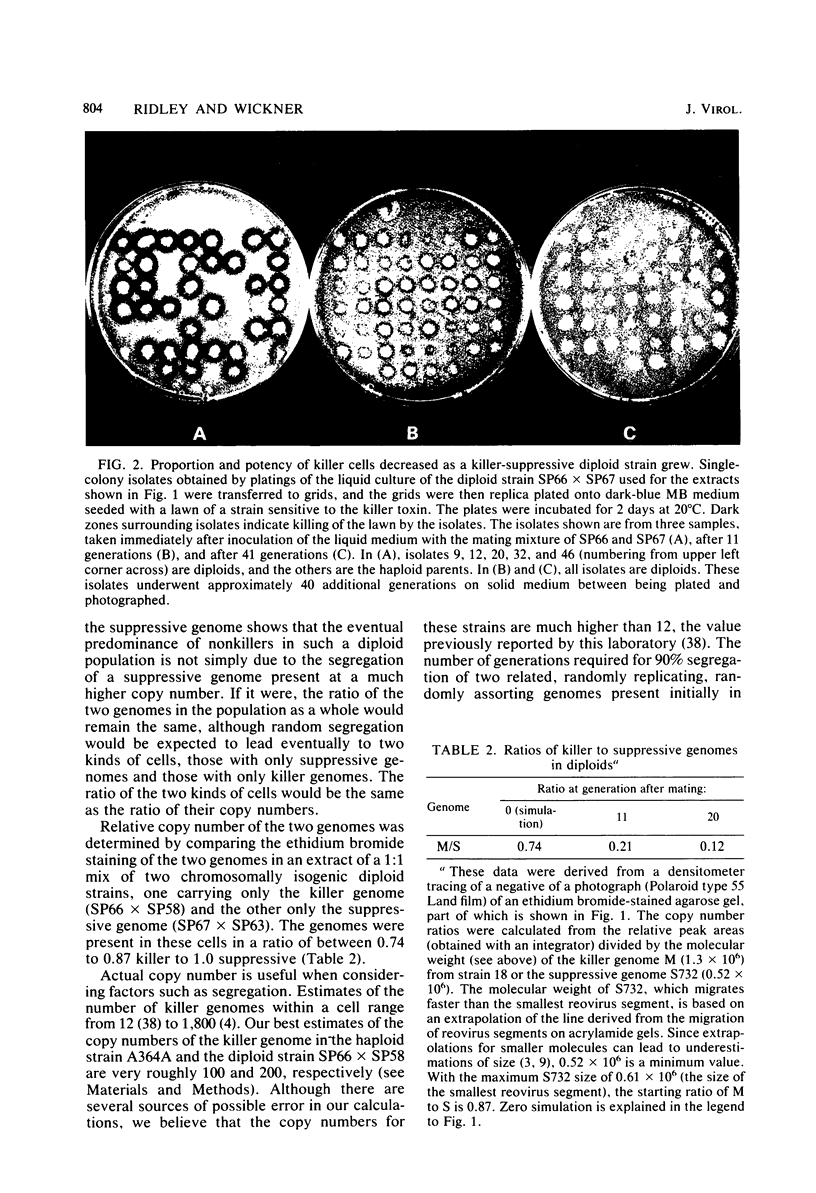
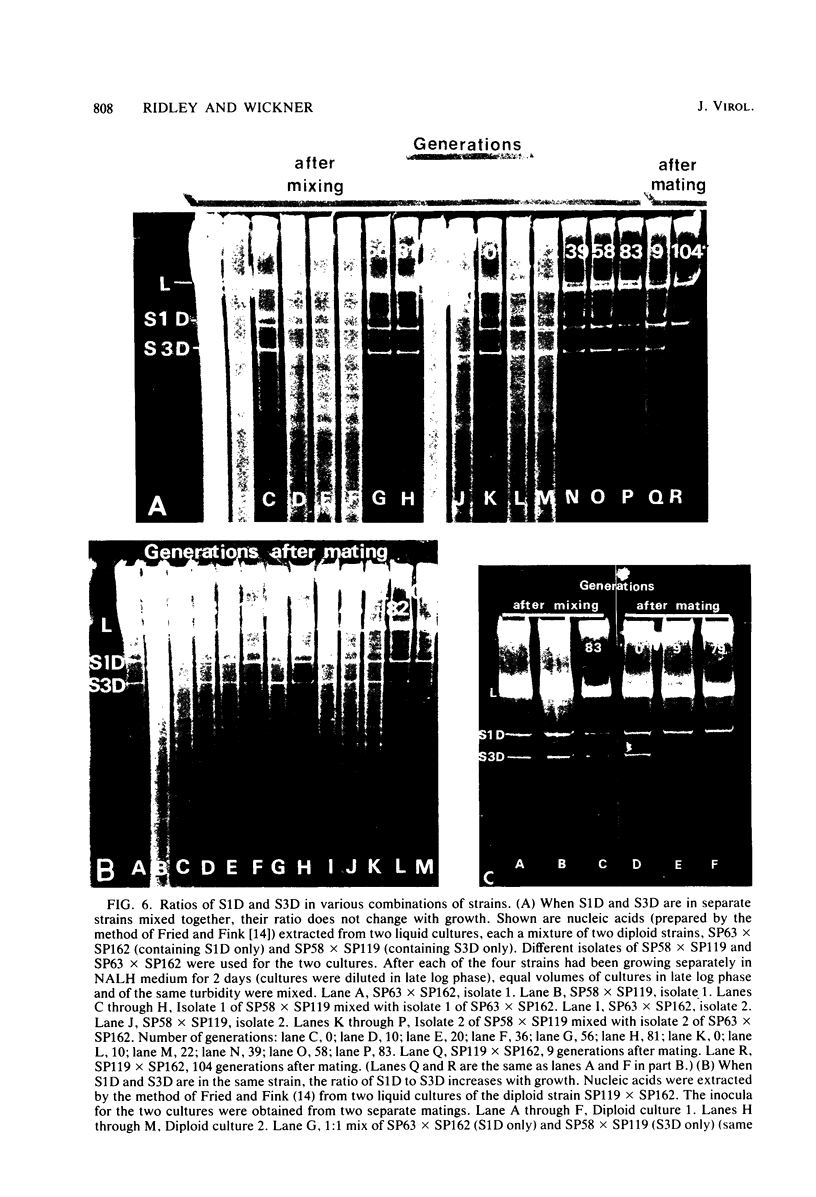
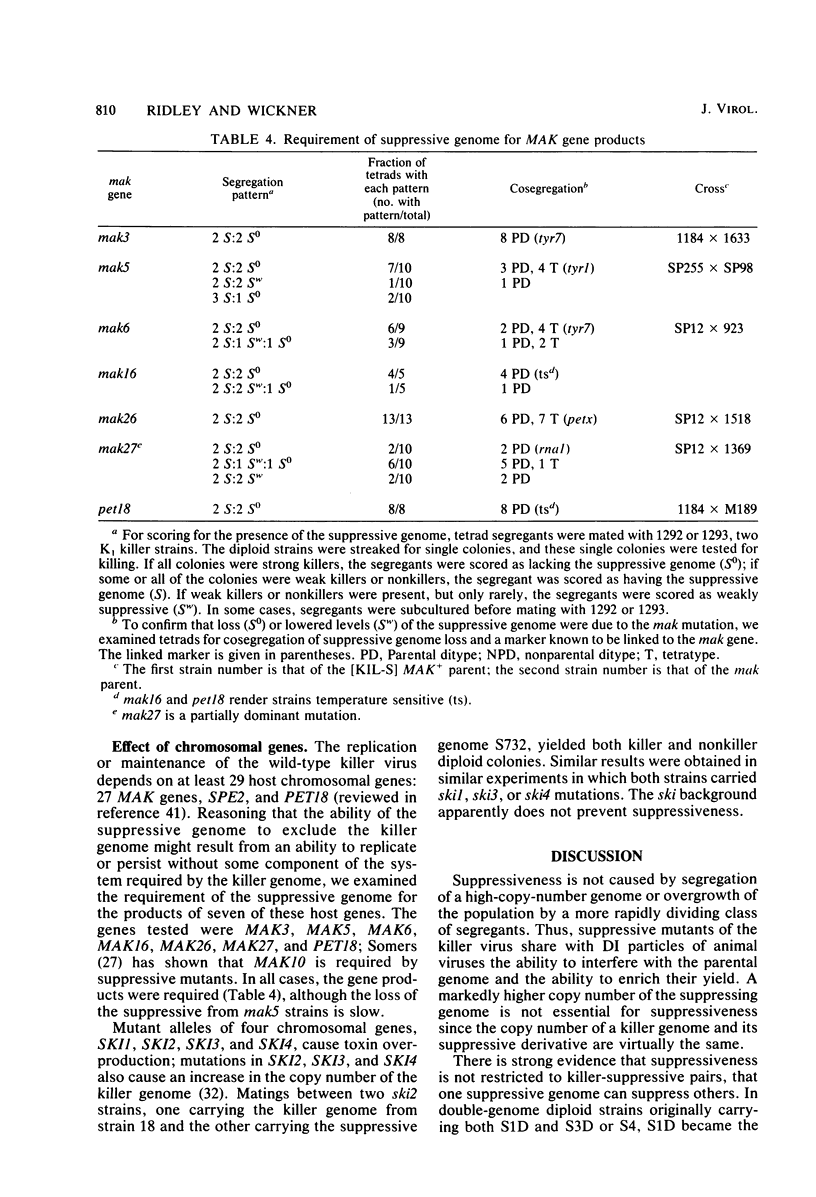
The K1 killer virus (or plasmid) of Saccharomyces cerevisiae is a noninfectious double-stranded RNA genome found intracellularly packaged in an icosahedral capsid. This genome codes for a protein toxin and for resistance to that toxin. Defective interfering virus mutants are deletion derivatives of the killer virus double-stranded RNA genome; such mutants are called suppressive. Unlike strains carrying the wild-type genome, strains with these deletion derivatives are neither toxin producers nor toxin resistant. If both the suppressive and the wildtype virus are introduced into the same cell, most progeny become toxin-sensitive nonkillers (J. M. Somers, Genetics 74:571-579, 1973). Diploids formed by the mating of a killer with a suppressive strain were grown in liquid culture, and RNA was extracted from samples taken up to 41 generations after the mating. The ratio of killer RNA to suppressive RNA decreased with increasing generations; by 41 generations the killer RNA was barely detectable. The copy numbers of the suppressive genome and its parental killer were virtually the same in isogenic strains, as were the growth rates of diploid strains containing either virus alone. Therefore, suppressiveness, not being due to segregation or overgrowth by faster growing segregants, is likely due to preferential replication or maintenance of the suppressive genome. Three suppressive viruses, all derivatives of the same killer virus (T. K. Sweeney et al., Genetics 84:27-42, 1976), did not coexist stably. The evidence strongly indicates that the largest genome of the three slowly suppressed both of the smaller genomes, showing that larger genomes can suppress smaller ones and that suppression can occur between two suppressives. Of 48 isolates of strains carrying the suppressive viruses, 5 had newly detectable RNA species, all larger than the original suppressive genomes. At least seven genes necessary for maintenance of the wild-type killer virus (MAK genes) were needed by a suppressive mutant. No effect of ski mutations (affecting regulation of killer virus double-stranded RNA replication) on suppressiveness was observed.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bevan E. A., Herring A. J., Mitchell D. J. Preliminary characterization of two species of dsRNA in yeast and their relationship to the "killer" character. Nature. 1973 Sep 14;245(5420):81–86. doi: 10.1038/245081b0. [DOI] [PubMed] [Google Scholar]
- Bostian K. A., Hopper J. E., Rogers D. T., Tipper D. J. Translational analysis of the killer-associated virus-like particle dsRNA genome of S. cerevisiae: M dsRNA encodes toxin. Cell. 1980 Feb;19(2):403–414. doi: 10.1016/0092-8674(80)90514-0. [DOI] [PubMed] [Google Scholar]
- Bozarth R. F., Harley E. H. The electrophoretic mobility of double-stranded RNA in polyacrylamide gels as a function of molecular weight. Biochim Biophys Acta. 1976 May 19;432(3):329–335. doi: 10.1016/0005-2787(76)90142-8. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. Preferential inclusion of extrachromosomal genetic elements in yeast meiotic spores. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5380–5384. doi: 10.1073/pnas.77.9.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenn J. A., Brennan V. E. Yeast viral double-stranded RNAs have heterogeneous 3' termini. Cell. 1980 Apr;19(4):923–933. doi: 10.1016/0092-8674(80)90084-7. [DOI] [PubMed] [Google Scholar]
- Bruenn J. A. Virus-like particles of yeast. Annu Rev Microbiol. 1980;34:49–68. doi: 10.1146/annurev.mi.34.100180.000405. [DOI] [PubMed] [Google Scholar]
- Bruenn J., Kane W. Relatedness of the double-stranded RNAs present in yeast virus-like particles. J Virol. 1978 Jun;26(3):762–772. doi: 10.1128/jvi.26.3.762-772.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck K. W., Ratti G. Molecular weight of double-stranded RNA: a re-examination of Aspergillus foetidus virus S RNA components. J Gen Virol. 1977 Oct;37(1):215–219. doi: 10.1099/0022-1317-37-1-215. [DOI] [PubMed] [Google Scholar]
- Bussey H. Physiology of killer factor in yeast. Adv Microb Physiol. 1981;22:93–122. doi: 10.1016/s0065-2911(08)60326-4. [DOI] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRAENKEL-CONRAT H., SINGER B., TSUGITA A. Purification of viral RNA by means of bentonite. Virology. 1961 May;14:54–58. doi: 10.1016/0042-6822(61)90131-3. [DOI] [PubMed] [Google Scholar]
- Franklin R. M. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1504–1511. doi: 10.1073/pnas.55.6.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Fink G. R. Electron microscopic heteroduplex analysis of "killer" double-stranded RNA species from yeast. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4224–4228. doi: 10.1073/pnas.75.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Krupnick D., Cryer D. R., Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970 Sep 14;52(2):323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- Harris M. S. Virus-like particles and double stranded RNA from killer and non-killer strains of Saccharomyces cerevisiae. Microbios. 1978;21(85-86):161–176. [PubMed] [Google Scholar]
- Herring A. J., Bevan E. A. Virus-like particles associated with the double-stranded RNA species found in killer and sensitive strains of the yeast Saccharomyces cerevisiae. J Gen Virol. 1974 Mar;22(3):387–394. doi: 10.1099/0022-1317-22-3-387. [DOI] [PubMed] [Google Scholar]
- Hopper J. E., Bostian K. A., Rowe L. B., Tipper D. J. Translation of the L-species dsRNA genome of the killer-associated virus-like particles of Saccharomyces cerevisiae. J Biol Chem. 1977 Dec 25;252(24):9010–9017. [PubMed] [Google Scholar]
- Ishii K., Hashimoto-Gotoh T., Matsubara K. Random replication and random assortment model for plasmid incompatibility in bacteria. Plasmid. 1978 Sep;1(4):435–445. doi: 10.1016/0147-619x(78)90002-1. [DOI] [PubMed] [Google Scholar]
- Kane W. P., Pietras D. F., Bruenn J. A. Evolution of defective-interfering double-stranded RNAs of the yeast killer virus. J Virol. 1979 Nov;32(2):692–696. doi: 10.1128/jvi.32.2.692-696.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Rao D. D., Huang A. S. Interference among defective interfering particles of vesicular stomatitis virus. J Virol. 1982 Jan;41(1):210–221. doi: 10.1128/jvi.41.1.210-221.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin A. J., Sipe J. D., Loh P. Separation of ten reovirus genome segments by polyacrylamide gel electrophoresis. J Virol. 1968 Oct;2(10):986–991. doi: 10.1128/jvi.2.10.986-991.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers J. M., Bevan E. A. The inheritance of the killer character in yeast. Genet Res. 1969 Feb;13(1):71–83. doi: 10.1017/s0016672300002743. [DOI] [PubMed] [Google Scholar]
- Somers J. M. Isolation of Suppressive Sensitive Mutants from Killer and Neutral Strains of SACCHAROMYCES CEREVISIAE. Genetics. 1973 Aug;74(4):571–579. doi: 10.1093/genetics/74.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. S., Wickner R. B. Co-curing of plasmids affecting killer double-stranded RNAs of Saccharomyces cerevisiae: [HOK], [NEX], and the abundance of L are related and further evidence that M1 requires L. J Bacteriol. 1982 May;150(2):545–551. doi: 10.1128/jb.150.2.545-551.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. S., Wickner R. B. Yeast L dsRNA consists of at least three distinct RNAs; evidence that the non-Mendelian genes [HOK], [NEX] and [EXL] are on one of these dsRNAs. Cell. 1982 Dec;31(2 Pt 1):429–441. doi: 10.1016/0092-8674(82)90136-2. [DOI] [PubMed] [Google Scholar]
- Sweeney T. K., Tate A., Fink G. R. A study of the transmission and structure of double stranded RNAs associated with the killer phenomenon in Saccharomyces cerevisiae. Genetics. 1976 Sep;84(1):27–42. doi: 10.1093/genetics/84.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-E A., Guerry P., Wickner R. B. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J Bacteriol. 1978 Dec;136(3):1002–1007. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh-E A., Wickner R. B. A mutant killer plasmid whose replication depends on a chromosomal "superkiller" mutation. Genetics. 1979 Apr;91(4):673–682. doi: 10.1093/genetics/91.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VON MAGNUS P. Incomplete forms of influenza virus. Adv Virus Res. 1954;2:59–79. doi: 10.1016/s0065-3527(08)60529-1. [DOI] [PubMed] [Google Scholar]
- Vodkin M., Katterman F., Fink G. R. Yeast killer mutants with altered double-stranded ribonucleic acid. J Bacteriol. 1974 Feb;117(2):681–686. doi: 10.1128/jb.117.2.681-686.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. "Killer character" of Saccharomyces cerevisiae: curing by growth at elevated temperature. J Bacteriol. 1974 Mar;117(3):1356–1357. doi: 10.1128/jb.117.3.1356-1357.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Chromosomal and nonchromosomal mutations affecting the "killer character" of Saccharomyces cerevisiae. Genetics. 1974 Mar;76(3):423–432. doi: 10.1093/genetics/76.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Deletion of mitochondrial DNA bypassing a chromosomal gene needed for maintenance of the killer plasmid of yeast. Genetics. 1977 Nov;87(3):441–452. doi: 10.1093/genetics/87.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Killer of Saccharomyces cerevisiae: a double-stranded ribonucleic acid plasmid. Bacteriol Rev. 1976 Sep;40(3):757–773. doi: 10.1128/br.40.3.757-773.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Leibowitz M. J. Chromosomal genes essential for replication of a double-stranded RNA plasmid of Saccharomyces cerevisiae: the killer character of yeast. J Mol Biol. 1976 Aug 15;105(3):427–443. doi: 10.1016/0022-2836(76)90102-9. [DOI] [PubMed] [Google Scholar]
- Wickner R. B., Leibowitz M. J. Two chromosomal genes required for killing expression in killer strains of Saccharomyces cerevisiae. Genetics. 1976 Mar 25;82(3):429–442. doi: 10.1093/genetics/82.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Toh-e A. [HOK], a new yeast non-Mendelian trait, enables a replication-defective killer plasmid to be maintained. Genetics. 1982 Feb;100(2):159–174. doi: 10.1093/genetics/100.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Twenty-six chromosomal genes needed to maintain the killer double-stranded RNA plasmid of Saccharomyces cerevisiae. Genetics. 1978 Mar;88(3):419–425. doi: 10.1093/genetics/88.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]








