Abstract
The hoatzin is unique among known avian species because of the fermentative function of its enlarged crop. A small-bodied flying foregut fermenter is a paradox, and this bird provides an interesting model to examine how diet selection and the gut microbiota contribute to maximizing digestive efficiency. Therefore, we characterized the bacterial population in the crop of six adult hoatzins captured from the wild. A total of 1,235 16S rRNA gene sequences were grouped into 580 phylotypes (67% of the pooled species richness sampled, based on Good's coverage estimator, with CACE and Chao1 estimates of 1,709 and 1,795 species-level [99% identity] operational taxonomic units, respectively). Members of 9 of the ∼75 known phyla in Bacteria were identified in this gut habitat; the Firmicutes were dominant (67% of sequences, belonging to the classes Clostridia, Mollicutes, and Bacilli), followed by the Bacteroidetes (30%, mostly in the order Bacteroidales), Proteobacteria (1.8%), and Lentisphaerae, Verrucomicrobia, TM7, Spirochaetes, Actinobacteria, and Aminanaerobia (all <0.1%). The novelty in this ecosystem is great; 94% of the phylotypes were unclassified at the “species” level and thus likely include novel cellulolytic lineages.
The hoatzin (Opisthocomus hoazin) is one of the few folivorous birds and is the only known example of an animal with crop fermentation in the class Aves (16). The hoatzin is the only species in the family Opisthocomidae (47), which has traditionally been classified in the order Galliformes and is currently undefined and placed between the orders Cuculiformes and Musophagiformes (21, 47). Galliformes include a number of herbivorous grouse and partridges (49). These related herbivorous birds have distendible crops, well-developed gizzards, and large ceca where fermentation presumably occurs. In contrast, the hoatzin uses an extended foregut (Fig. 1) (enlarged crop and distal esophagus) for microbial fermentation of its leafy diet (17). A fermentation chamber that has a sufficient volume enables the retention of plant material until microbes can ferment it; the derived products, together with the microbial biomass, nourish the host (52). Despite its small body size, the hoatzin retains the digesta for as long as sheep do (15), and the concentration of fermentation products (volatile short-chain fatty acids) is equivalent to the concentrations found in the rumens of sheep and cows (16). As in the rumen, the fermenting microbes include bacteria, archaea, fungi, and ciliate protozoa (F. Godoy-Vitorino et al., presented at the 11th International Symposium on Microbial Ecology, Vienna, Austria, 20 to 25 August 2006).
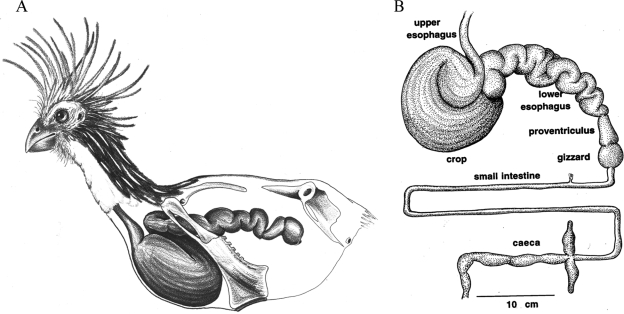
FIG. 1.
Schematic representation of the hoatzin digestive tract. (A) Location of the crop and expanded esophagus in the hoatzin body. The anterior sternum is much reduced to make room for the large crop. (Reprinted from Natural History [[17] with permission of the publisher.) (B) Extended complete digestive tract of the hoatzin. (Courtesy of Alejandro Grajal.)
Being a small-bodied flying foregut fermenter seems paradoxical. At a purely mechanical level, crop enlargement can require sternal modifications that restrict flight capabilities (16, 18). Moreover, the adult hoatzin is the smallest animal among known foregut fermenters, with an average weight of only 700 g. The minimum body size for a mammal with foregut fermentation is ∼3 kg (6); a small body size restricts the luminal volume needed for fermentation, and energy demand scales to metabolic weight (w0.75) (26). In addition, a bird must generate enough energy to maintain its body temperature, which can be as high as 43.5°C (3).
This paradox has been explained by considering that flying can facilitate extreme diet selectivity, which in turn could maximize fermentation rates and digestive efficiency (16). In this view, crop fermentation allows the hoatzin to be a unique browsing bird that exploits resources normally available only to specialized mammals and not to other avian species (16).
Foregut fermentation originated on several independent occasions during the evolution of herbivorous vertebrates (28, 29, 41). Species of the orders Artiodactyla, Edentata, and Primates have also evolved foregut fermentation (49). The Opisthocomiformes appeared in the Eocene, about 55 million years ago, according to fossil records from Argentina (5), at about the same time that primitive ungulates (Condylarths) migrated to South America (37). A browser, the hoatzin ancestor may have evolved during the Eocene at the same time as herbivorous mammalian browsers, such as extinct South American ungulates (37), arboreal primates (4), and sloths (48).
Almost two decades have passed since the first report describing foregut fermentation in the hoatzin appeared (16). Nonetheless, the microbial composition of the crop of this bird has remained elusive. In this study, we characterized the crop bacteria of the hoatzin using culture-independent molecular methods. The results reveal an unexpected degree of unclassified lineages that likely include novel cellulolytic bacteria.
MATERIALS AND METHODS
Sample collection.
Six adult hoatzins from different social groups were captured from the wild at Piñero Ranch in the savannas of Cojedes state in Venezuela (68°4′W, 8°82′N) with permits from the Venezuelan Ministry of Environment (#11-193) and from the UPR-IACUC (#601-2007). Crops with their contents from animals 1 to 6 (chen4, chen6, chen10, chen27, chen30, and chen32, respectively) were extracted in situ in the field, sealed anteriorly and posteriorly, and immediately frozen in liquid nitrogen. Samples were then stored in the lab at around −77°C for ≤1 month prior to DNA isolation.
Genomic DNA extraction.
DNA was extracted from crop contents using a modification of the method of Löffler et al. (32). The modification consisted of microbial disruption by bead beating (0.8 g of sterile 0.1-mm-diameter zirconium beads [Biospec Products, Bartlesville, OK] in 1 ml of TEN buffer [100 mM Tris, 10 mM EDTA; 2 M NaCl, pH 7.4] containing 200 mg of the contents from a single crop, with a bead beater [Biospec Products] set at 5,000 rpm for 1 min at room temperature). DNA samples were kept frozen until they were used.
PCR amplification of 16S rRNA genes.
PCR (two replicates per sample) were performed by using 50-μl mixtures containing 2 PuReTaq Ready-To-Go PCR beads (GE Healthcare Bio-Sciences Corp., Piscataway, NJ), ∼50 ng of DNA template, and 20 pmol of universal bacterial primers 8F and 1513R (38). PCR for two or three replicates per sample were performed using 30 cycles and an annealing temperature of 52°C, as described by Saitou and Nei (42).
Clone library construction and DNA sequencing.
PCR products were subsequently purified (PCR purification kit; Qiagen, Valencia, CA) and subcloned (pGEM-T Easy [Promega, Madison, WI], using Escherichia coli XL1-Blue [Stratagene, La Jolla, CA] or pCR4.0, a TOPO TA cloning kit, and TOP10 competent cells [Invitrogen, Carlsbad, CA]). Cloned amplicons were sequenced using vector-specific primers and an ABI 3730xl instrument (Applied Biosystems, Foster City, CA).
DNA sequence analyses and chimera detection.
Sequences were edited with Sequencher 4.6, aligned with the Greengenes database, and classified according to the Hugenholtz taxonomy (as of April 2007 [7]). Hoatzin crop 16S rRNA gene sequences and their closest neighbors in Greengenes were then imported into ARB (34). A BLAST script and the classification tool in Greengenes were used to determine similarity to GenBank sequences. Chimeras (n = 519) (checked with Bellerophon, version 3 [19]) and sequences corresponding to plant chloroplasts (n = 84) were excluded from further analysis.
Richness and coverage estimators.
To estimate richness, sequences were binned into “species-level” operational taxonomic units (OTUs). Since in species the level of similarity of 16S rRNA gene sequences can be as high as 98 to 99% (27), we chose a sequence identity level of 99% (with the hypervariable regions of the sequence masked using lanemaskPH [20] in DOTUR [43]). One representative sequence per OTU was randomly selected for calculating a phylogenetic tree using the neighbor-joining algorithm (42) in ARB (34). OTU abundance values were used to create a heat map for interindividual comparisons (TreeView) (http://rana.lbl.gov).
Collector's curves for observed and estimated (Chao1 and CACE) OTUs were computed using DOTUR (43) and EstimateS (R. K. Colwell, EstimateS: statistical estimation of species richness and shared species from samples, version 7, 2004 [http://viceroy.eeb.uconn.edu/estimates; www.purl.oclc.org/estimates]). Bacterial diversity was estimated with Shannon and Simpson indexes in DOTUR; the Shannon index takes into account the number and evenness of species (2), while the Simpson index estimates the probability that two randomly selected individuals belong to the same species (46). The Pielou evenness index (39) was also calculated for each animal's microbial community. Good's coverage index (12) was estimated using singleton sequences obtained from DOTUR.
Bacterial community comparisons.
Bacterial communities from the different hoatzin crops were compared using UniFrac (33). UniFrac analyses were based on an ARB neighbor-joining tree of 16S rRNA gene sequences, their frequencies, and their assigned environments (individual crops). Phylum-level comparisons between the hoatzin crop communities and the communities in other fermentative organs were also performed using previously described 16S rRNA gene data sets in which the sequences were more than 500 bp long and data sets containing more than 150 sequences from particular host species, including 425 sequences from the chicken intestine (53; P. T. Lan, M. Sakamoto, and Y. Benno, unpublished data from GenBank), 752 sequences from cows (25, 51; L. Cauquil, S. Combes, V. Montelis, J. Gordon, and T. Gidenne, unpublished data from GenBank; E. C. Shin, W. J. Lim, H. Kim, and H. D. Yun, unpublished data from GenBank), 176 sequences from yaks (1; S. Cai and X. Dong, unpublished data from GenBank), 180 sequences from deer (35, 50), 436 sequences from sheep (30, 40; S. Sawanon, S. Koike, and Y. Kobayashi, unpublished data from GenBank; T. Shinkai, N. Matsumoto, and Y. Kobayashi, unpublished data from GenBank), and 165 sequences from water buffalo (H. Mao, S. Ma, J. Chen, and W. Deng, unpublished data from GenBank).
Nucleotide sequence accession numbers.
Bacterial 16S rRNA gene sequences obtained in this study have been deposited in the GenBank database. The accession numbers for the 580 representative OTUs are EU344158 to EU344737, and the accession numbers for the remaining 655 cloned sequences are EU747884 to EU748538.
RESULTS
We used culture-independent, molecular methods to determine the composition of microbial communities harvested from the crops of six wild adult hoatzins. Each animal was from a different social group at Piñero Ranch in the savannas of Cojedes state in Venezuela (68°4′W, 8°82′N). For each crop sample, we constructed libraries of 16S rRNA genes amplified by PCR using bacterium-specific primers.
Sample representativeness and bacterial richness.
We obtained a total of 1,235 bacterial 16S rRNA gene sequences (166 to 332sequences/bird) (Table 1), which were binned into 580 OTUs (threshold cutoff for each OTU, ≥99% nucleotide sequence identity). Most sequences were full-length sequences (1,400 ± 50 bp); the exceptions were animal 4 sequences, whose average length was 800 bp. Microbial communities are complex, particularly those associated with the vertebrate gut; thus, 16S rRNA gene surveys seldom sample the complete diversity. Therefore, for each bird sample, we assessed richness (actual diversity) using the Chao1 and CACE estimators and coverage (how well a sample represents the population) using Good's coverage estimator. We chose 99% sequence identity (with hypervariable regions removed) as a cutoff for differentiation of “species” (10). The results (Table 1) indicate that, on average, 67% of the pooled species richness (55% for each individual) was sampled.
TABLE 1.
Diversity indices for bacterial communities in individual hoatzin crops
| Animal | No. of sequences | No. of OTUs | % of OTUs unique to one bird | Shannon diversity indexa | Simpson diversity indexb | Pielou's evenness indexc | Good's coverage index (%)d |
|---|---|---|---|---|---|---|---|
| 1 | 332 | 183 | 60 | 4.72 | 0.013 | 0.91 | 65 |
| 2 | 189 | 120 | 60 | 4.49 | 0.013 | 0.94 | 52 |
| 3 | 191 | 125 | 57 | 4.36 | 0.023 | 0.90 | 52 |
| 4 | 179 | 125 | 67 | 4.51 | 0.013 | 0.93 | 44 |
| 5 | 166 | 76 | 71 | 3.59 | 0.061 | 0.83 | 66 |
| 6 | 178 | 116 | 65 | 4.34 | 0.016 | 0.91 | 51 |
| All | 1,235 | 580 | 65 | 5.79 | 0.006 | 0.91 | 67 |
The Shannon diversity index takes into account the number and evenness of species. A higher Shannon-Weaver diversity index is associated with greater diversity (2, 45).
The Simpson diversity index estimates the probability that two randomly selected individuals belong to the same species (46).
Pielou's evenness index is a measure of how evenly distributed abundance is among the species that are in a community (39) and ranges from 0 to 1 (eveness to uneveness).
Good's coverage index is the sum of the probabilities of the observed classes calculated as follows: [1 − (n/N)] × 100, where n is the number of singleton sequences and N is the total number of sequences (12).
The UniFrac metric was employed to compare foregut bacterial communities from the six individual birds. The results indicate that the overall phylogenetic compositions of the communities from six hoatzins differed significantly (UniFrac significance for pairwise comparisons, P < 0.05). The communities all had similar high levels of species richness and evenness, with the exception of hoatzin 5 (Table 1). There was very little overlap in species-level OTU representation between birds (Fig. 2A), and the individual representation of bacterial phyla was also heterogeneous (Fig. 2B). The majority (80.3%) of OTUs were unique to one bird; 13.4% of the OTUs were shared by two birds, 4.2% were shared by three birds, 1.4% were shared by four birds, and 0.5% were shared by five birds. No OTU was shared by all six birds. When we used a 97% cutoff value, we still found that the majority of the OTUs were unique to one animal (70.2%), and only one OTU was shared by all six individuals (see Fig. S3 in the supplemental material).
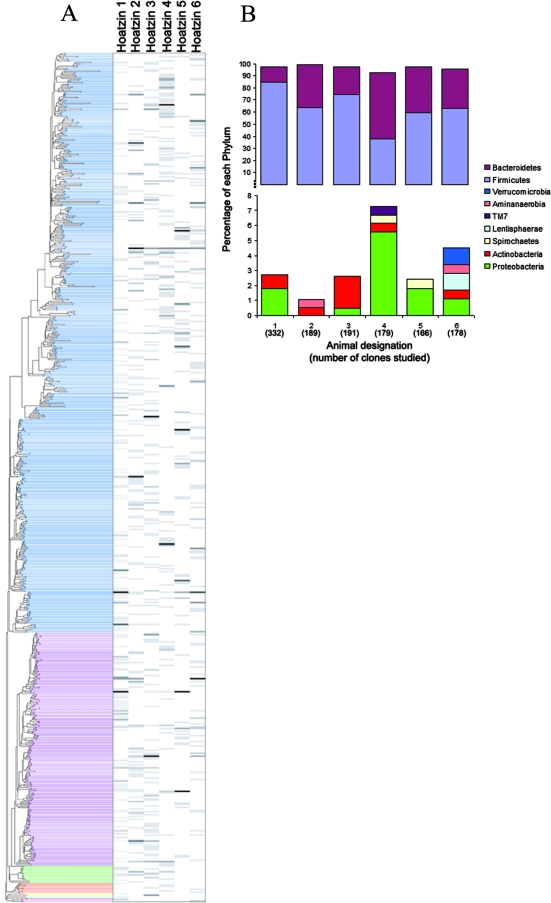
FIG. 2.
Phylogenetic analysis of the hoatzin crop bacteria. (A) Tree of the 580 identified bacterial OTUs, showing relative clone abundance for six individual animals. In the neighbor-joining tree, the lines on the left indicate OTUs and the columns on the right show data for animals. The phyla are color coded as described in Fig. S1 in the supplemental material and (from top to bottom) are Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Spirochaetes, Lentisphaerae, Verrucomicrobia, TM7, and Aminanaerobia. The relative clone abundance for each OTU in each animal is indicated by a shade of gray (white, 0%; black, 100%). (B) Distribution of the 1,235 16S rRNA gene sequences in nine bacterial phyla for individual hoatzins. Data for the two phyla represented most often, Bacteroidetes and Firmicutes, are shown in the top panel, and data for the other seven phyla are shown in the bottom panel. Proteobacteria were the predominant bacteria in animal 4 but were absent in animal 2, whereas Verrucomicrobia and Lentisphaerae were detected only in animal 6.
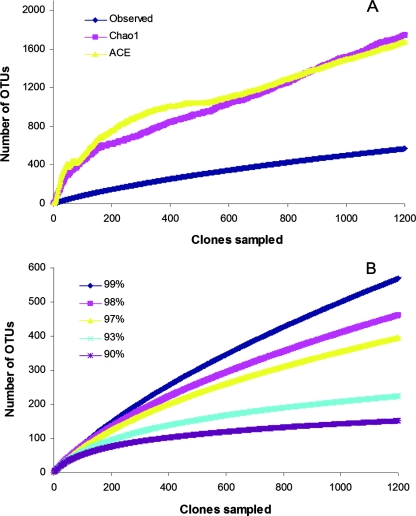
Collector's curves for Chao1 and CACE richness estimators (Fig. 3A) and rarefaction curves for observed richness (Fig. 3B) revealed that the diversity inherent in the communities was far from circumscribed by this study; the estimated richness was 1,709 using the CACE estimator and 1,795 OTUs using the Chao1 estimator.
FIG. 3.
Collector's and rarefaction curves of OTU richness for the 1,235 16S rRNA gene sequences from the pooled hoatzin crops. (A) Collector's curves of observed and estimated species-level OTU richness using a 99% identity cutoff value. The richness was estimated to be 1,795 OTUs using the Chao1 estimator and 1,709 OTUs using the CACE estimator. (B) Rarefaction at identity cutoff values of 90 to 99%. There was a notable increase in the number of OTUs with only a small increase in the cutoff value (580 OTUs at a cutoff value of 99% and 470, 402, 226, and 152 OTUs at cutoff values of 98, 97, 93, and 90%, respectively), reflecting greater richness at lower phylogenetic branching depth.
Bacterial community composition.
The relatedness of the hoatzin crop sequences to known small-subunit rRNA gene sequences was determined by BLAST comparisons with the >100,000 sequences currently in the Greengenes database in order to find the closest match for a representative sequence from each hoatzin OTU (see Table S1 in the supplemental material). Overall, the bacterial sequences represented only 9 of the ∼75 known phyla in the domain Bacteria (see Fig. S1 in the supplemental material), similar to the results for the rumens of cow and sheep (12 and 11 phyla, respectively) (see Fig. S2 in the supplemental material). Bacteria belonged principally to the Firmicutes (67% of sequences) and the Bacteroidetes (30%) (see Fig. S1 in the supplemental material). Other phyla represented included the Proteobacteria (1.8%) and the Lentisphaerae, Verrucomicrobia, TM7, Spirochaetes, Actinobacteria, and Aminanaerobia (Synergystes plus Dethiosulfovibrio) (all <0.1%).
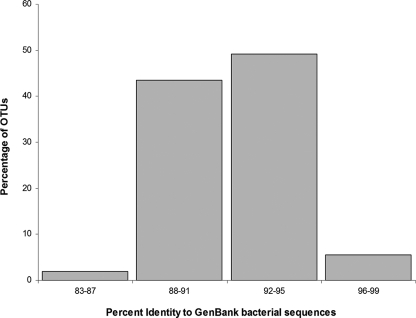
Remarkably, most OTUs exhibited less than 95% identity with any previously described 16S rRNA gene sequence (Fig. 4); of the 580 OTUs, 545 (94%) were unclassified at the species level, while 327 (56%), 147 (25%), 70 (12%), and 46 (8%) were unclassified at the genus, family, order, and class levels, respectively (Table 2). Within the Firmicutes phylum, the class Clostridia was the most abundant (329/396 OTUs; 83%); it was represented by the order Clostridiales almost exclusively, including the family Lachnospiraceae (248 OTUs, 75%), candidate group RC6 (46 OTUs, 16%), the family Peptostreptococcaceae (31 OTUs, 9.5%), and the family Clostridiaceae (4 OTUs, 1%). The class Mollicutes was represented by 27 OTUs (7%), while there were only three Bacilli OTUs (<1%). The Bacteroidetes phylum contained 159 OTUs, all of which belonged to order Bacteroidales, including the family Prevotellaceae (82 OTUs, 51%), candidate group rf14 (36 OTUs, 23%), and a few other candidate groups in the Hugenholtz taxonomy (http://greengenes.lbl.gov/cgi-bin/nph-classify.cgi). The Proteobacteria contained 12 OTUs, 11 of which were Gammaproteobacteria or Betaproteobacteria (the remaining OTU belonged to the Epsilonproteobacteria). The OTUs whose best match was a rumen bacterium (49% of the OTUs) had 16S rRNA gene sequence identities ranging from 88 to 95% with bacteria such as Butyrivibrio fibrisolvens, Eubacterium cellulosolvens, or Bacteroides xylanolyticus (see Table S1 in the supplemental material).
FIG. 4.
Distribution of species-level OTU frequencies in relation to known sequences. Most of the hoatzin crop bacterial OTUs have ≤95% identity to previously reported sequences.
TABLE 2.
Numbers of unclassified crop bacterial OTUs for each phylum
| Phylum | No. of unclassified crop bacterial OTUs at different levels
|
||||
|---|---|---|---|---|---|
| Class | Order | Family | Genus | Species | |
| Firmicutes | 46 | 68 | 70 | 229 | 371 |
| Bacteroidetes | 73 | 83 | 156 | ||
| Proteobacteria | 10 | 10 | |||
| Actinobacteria | 1 | 3 | 3 | 3 | |
| Aminanaerobia | 2 | ||||
| Verrucomicrobia | |||||
| Lentisphaerae | 1 | 1 | |||
| Spirochaetes | 1 | 1 | 1 | 1 | |
| TM7 | 1 | ||||
| Total | 46 (8)a | 70 (12) | 147 (25) | 327 (56) | 545 (94) |
The numbers in parentheses are percentages.
DISCUSSION
The hoatzin consumes a diet high in fiber (9). Previous work has shown that, like microbes in the rumen, hoatzin crop microbes break down plant polysaccharides from the leafy diet (14, 24) and detoxify plant chemicals (11, 24). The characterization of crop bacteria in this study revealed a dominance of Bacteroidetes and Firmicutes, a feature which appears to be typical for the vertebrate gut (31). In fact, the dominance of these phyla has also been described in the rumens of Artiodactyla mammals (1, 51) and in the ceca of chickens (53) and turkeys (44). The only order within Bacteroidetes found in the crop, Bacteroidales, is a known gut-adapted lineage (31). We found no crop 16S rRNA gene sequences that shared ≥95% identity with the main rumen cellulolytic genera Ruminococcus, Bacteroides, Prevotella, and Butyrivibrio (22, 23). Furthermore, no members of the Fibrobacteria phylum, which includes cellulolytic bacteria common to rumens, were identified. The conspicuous absence of known cellulolytic bacteria and the discovery of many novel unclassified sequences related to rumen bacteria raise the possibility that many of the unclassified lineages may be cellulolytic. The hoatzin's body temperature is ∼39°C (13), while Artyodactyla have body temperatures below this value (3). The gut temperature is likely to increase with fermentative activity; however, the crop temperature is 39°C ± 1°C (n = 4 animals) (our unpublished observation).
Despite the partial coverage obtained in this survey, the 580 OTUs (or even the 402 OTUs that could be classified using a less stringent threshold cutoff, ≥97% 16S rRNA gene sequence identity) represent greater richness than that described in the more deeply sampled human colon (10). The majority of the OTUs were unique in each individual, perhaps due in part to the partial sample coverage. The high level of bacterial diversity is likely related to the high dietary diversity of the hoatzin, which feeds on leaves of members of the Fabaceae, Sterculiaceae, Rutaceae, and Vitaceae (9).
While cellulose digestion is a very important function of the rumen (52), cellulose is not a major digested polysaccharide in the crop (8). Hemicellulose degradation appears to be more important than cellulolysis in the hoatzin crop (24), as it is in other selective browsers, such as the howler monkey (36), which chooses young leaves and buds with thin cell walls and high cellular contents (9). However, the metabolic activities of the hoatzin crop microbiota remain elusive; the surprising diversity and novelty suggest that comparative metagenomic sequencing of whole-community microbial DNA prepared from the hoatzin and other foregut fermenters would be a worthwhile undertaking.
The results may provide important new insights into (i) how gut microbial communities are affected by diet and host phylogeny, (ii) the degree to which glycobiomes (genes involved in the acquisition and metabolism of carbohydrates) vary among communities that consist of seemingly divergent collections of organismal lineages, and (iii) how fermentation by the crop microbiota may provide sufficient energy from both cellular contents and cell wall polysaccharides to satisfy the hoatzin's energetic requirements.
Supplementary Material
Acknowledgments
This work was supported by NSF grants IOS 0716911, NSF:DDIG 0709840 (to M.G.D.B.), and HRD0206200 (to UPR-CREST); by grants from the W. M. Keck Foundation and Ellison Medical Foundation (to J.I.G.) and an INBRE grant, which supported the High Performance Computing Facility and H. Ortiz-Zuazaga; and by grant P20 RR-016470 from NCRR/NIH.
We acknowledge Daniel Ayala for providing the BLAST script and Falk Warnecke, Todd DeSantis, Philip Hugenholtz, and Leslie Dethlefsen for helpful suggestions concerning the use of sequence analysis software. We also thank Alejandro Grajal for providing his drawing of the hoatzin digestive tract. The logistic field support provided by Hato Piñero personnel and José and Antonio González from Hato Mataclara and the collection permits provided by the Venezuelan Ministry of Environment are deeply appreciated.
Footnotes
Published ahead of print on 8 August 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.An, D., X. Dong, and Z. Dong. 2005. Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos taurus) estimated by 16S rDNA homology analyses. Anaerobe 11:207-215. [DOI] [PubMed] [Google Scholar]
- 2.Chao, A., and T.-J. Shen. 2003. Nonparametric estimation of Shannon's index of diversity when there are unseen species in sample. Environ. Ecol. Stat. 10:429-443. [Google Scholar]
- 3.Clarke, A., and P. Rothery. 2008. Scaling of body temperature in mammals and birds. Funct. Ecol. 22:58-67. [Google Scholar]
- 4.Collinson, M. E., and J. J. Hooker. 1991. Fossil evidence of interactions between plants and plant-eating mammals. Philos. Trans. R. Soc. Lond. B 333:197-208. [DOI] [PubMed] [Google Scholar]
- 5.Cracraft, J. 1971. A new family of hoatzin-like birds (order Opisthocomiformes) from the Eocene of South America. Ibis 113:229-233. [Google Scholar]
- 6.Demment, M. W., and P. J. Van Soest. 1985. A nutritional explanation for body-size patterns of ruminant and nonruminant herbivores. Am. Nat. 125:641-672. [Google Scholar]
- 7.DeSantis, T. Z., Jr., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domínguez-Bello, M. G., M. Lovera, P. Suárez, and F. Michelangeli. 1993. Microbial digestive symbionts of the crop of the hoatzin (Opisthocomus hoazin): the only foregut fermenter avian. Physiol. Zool. 66:374-383. [Google Scholar]
- 9.Domínguez-Bello, M. G., F. Michelangeli, M. C. Ruiz, A. Garcia, and E. Rodriguez. 1994. Ecology of the folivorous hoatzin (Opisthocomus hoazin) on the Venezuelan plains. Auk 111:643-651. [Google Scholar]
- 10.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Amado, M. A., F. Michelangeli, P. Gueneau, and M. E. Perez. 2007. Bacterial detoxification of saponins in the crop of the avian foregut fermenter Opisthocomus hoazin. J. Anim. Feed Sci. 16:82-85. [Google Scholar]
- 12.Good, I. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 13.Grajal, A. 1991. Digestive efficiency of the hoatzin (Opisthocomus hoatzin), a folivorous bird with foregut fermentation. University of Florida, Gainesville.
- 14.Grajal, A. 1995. Structure and function of the digestive tract of the hoatzin (Opisthocomus hoazin): a folivorous bird with foregut fermentation. Auk 112:20-28. [Google Scholar]
- 15.Grajal, A., and O. Parra. 1995. Passage rates of digesta markers in the gut of the hoatzin, a folivorous bird with foregut fermentation. Condor 97:675-683. [Google Scholar]
- 16.Grajal, A., S. Strahl, R. Parra, M. G. Dominguez, and A. Neher. 1989. Foregut fermentation in the hoatzin, a neotropical leave-eating bird. Science 245:1236-1238. [DOI] [PubMed] [Google Scholar]
- 17.Grajal, A., and S. Strahl. 1991. A bird with the guts to eat leaves. Nat. His. 100:48-55. [Google Scholar]
- 18.Hedges, S. B., M. D. Simmons, M. A. van Dijk, G. J. Caspers, W. W. de Jong, and C. G. Sibley. 1995. Phylogenetic relationships of the hoatzin, an enigmatic South American bird. Proc. Natl. Acad. Sci. USA 92:11662-11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 20.Hugenholtz, P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3:REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes, J. M., and A. J. Baker. 1999. Phylogenetic relationships of the enigmatic hoatzin (Opisthocomus hoazin) resolved using mitochondrial and nuclear gene sequences. Mol. Biol. Evol. 16:1300-1307. [DOI] [PubMed] [Google Scholar]
- 22.Hungate, R. E. 1950. The anaerobic mesophilic cellulolytic bacteria. Bacteriol. Rev. 14:1-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hungate, R. E. 1966. The rumen and its microbes. Academic Press, New York, NY.
- 24.Jones, R. J., M. A. G. Amado, and M. G. Dominguez-Bello. 2000. Comparison of the digestive ability of crop fluid from the folivorous hoatzin (Opisthocomus hoazin) and cow rumen fluid with seven tropical forages. Anim. Feed Sci. Technol. 87:287-296. [Google Scholar]
- 25.Karnati, S. K., J. T. Sylvester, S. M. Noftsger, Z. Yu, N. R. St.-Pierre, and J. L. Firkins. 2007. Assessment of ruminal bacterial populations and protozoal generation time in cows fed different methionine sources. J. Dairy Sci. 90:798-809. [DOI] [PubMed] [Google Scholar]
- 26.Kleiber, M. 1961. The fire of life. Wiley, New York, NY.
- 27.Konstantinidis, K. T., and J. M. Tiedje. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 102:2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornegay, J. R. 1996. Molecular genetics and evolution of stomach and nonstomach lysozymes in the hoatzin. J. Mol. Evol. 42:676-684. [DOI] [PubMed] [Google Scholar]
- 29.Kornegay, J. R., J. W. Schilling, and A. C. Wilson. 1994. Molecular adaptation of a leaf-eating bird: stomach lysozyme of the hoatzin. Mol. Biol. Evol. 11:921-928. [DOI] [PubMed] [Google Scholar]
- 30.Larue, R., Z. Yu, V. A. Parisi, A. R. Egan, and M. Morrison. 2005. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ. Microbiol. 7:530-543. [DOI] [PubMed] [Google Scholar]
- 31.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Löffler, F. E., K. M. Ritalahti, and J. M. Tiedje. 1997. Dechlorination of chloroethenes is inhibited by 2-bromoethanesulfonate in the absence of methanogens. Appl. Environ. Microbiol. 63:4982-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackie, R. I., R. I. Aminov, W. Hu, A. V. Klieve, D. Ouwerkerk, M. A. Sundset, and Y. Kamagata. 2003. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl. Environ. Microbiol. 69:6808-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milton, K., and R. H. McBee. 1983. Structural carbohydrate digestion in a new world primate, Alouatta palliata Gray. Comp. Biochem. Physiol. 74:29-31. [DOI] [PubMed] [Google Scholar]
- 37.Muizon, C., and R. L. Cifelli. 2000. The “condylarths” (archaic Ungulata, Mammalia) from the early Palaeocene of Tiupampa (Bolivia): implications on the origin of the South American ungulates. Geodiversitas 22:47-150. [Google Scholar]
- 38.Pei, Z., E. Bini, L. Yang, M. Zhou, F. Francois, and M. Blaser. 2004. Bacterial biota in the human distal esophagus. Proc. Natl. Acad. Sci. USA 101:4250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pielou, E. C. 1966. The measurement of diversity in different types of biological collections. J. Theor. Biol. 13:131-144. [Google Scholar]
- 40.Rattray, R. M., and A. M. Craig. 2007. Molecular characterization of sheep ruminal enrichments that detoxify pyrrolizidine alkaloids by denaturing gradient gel electrophoresis and cloning. Microb. Ecol. 54:264-275. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz, M., M. Dominguez-Bello, and F. Michelangeli. 1994. Gastric lysozyme in the hoatzin (Opisthocomus hoazin), an avian folivore. Experientia 50:499-501. [Google Scholar]
- 42.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 43.Schloss, P., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scupham, J. A., T. G. Patton, E. Bent, and D. O. Bayles. 2008. Comparison of the cecal microbiota of domestic and wild turkeys. Microb. Ecol. 56:322-331. [DOI] [PubMed] [Google Scholar]
- 45.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication. University of Illinois Press, Urbana, IL.
- 46.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 47.Sorenson, M. D., E. Oneal, J. Garcia-Moreno, and D. P. Mindell. 2003. More taxa, more characters: the hoatzin problem is still unresolved. Mol. Biol. Evol. 20:1484-1498. [DOI] [PubMed] [Google Scholar]
- 48.Springer, M. S., W. J. Murphy, E. Eizirik, and S. J. O'Brien. 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. USA 100:1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens, C. E., and I. D. Hume. 1998. Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol. Rev. 78:393-427. [DOI] [PubMed] [Google Scholar]
- 50.Sundset, M., K. Praesteng, I. Cann, S. Mathiesen, and M. R. I. 2007. Novel rumen bacterial diversity in two geographically separated sub-species of reindeer. Microb. Ecol. 54:424-438. [DOI] [PubMed] [Google Scholar]
- 51.Tajima, K., R. I. Aminov, T. Nagamine, H. Matsui, M. Nakamura, and Y. Benno. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Soest, P. J. 1994. Nutritional ecology of the ruminant, 2nd ed. Comstock, Ithaca, NY.
- 53.Zhu, X. Y., T. Zhong, Y. Pandya, and R. D. Joerger. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.