Abstract
The enterohepatic Helicobacter species Helicobacter hepaticus colonizes the murine intestinal and hepatobiliary tract and is associated with chronic intestinal inflammation, gall stone formation, hepatitis, and hepatocellular carcinoma. Thus far, the role of H. hepaticus motility and flagella in intestinal colonization is unknown. In other, closely related bacteria, late flagellar genes are mainly regulated by the sigma factor FliA (σ28). We investigated the function of the H. hepaticus FliA in gene regulation, flagellar biosynthesis, motility, and murine colonization. Competitive microarray analysis of the wild type versus an isogenic fliA mutant revealed that 11 genes were significantly more highly expressed in wild-type bacteria and 2 genes were significantly more highly expressed in the fliA mutant. Most of these were flagellar genes, but four novel FliA-regulated genes of unknown function were identified. H. hepaticus possesses two identical copies of the gene encoding the FliA-dependent major flagellin subunit FlaA (open reading frames HH1364 and HH1653). We characterized the phenotypes of mutants in which fliA or one or both copies of the flaA gene were knocked out. flaA_1 flaA_2 double mutants and fliA mutants did not synthesize detectable amounts of FlaA and possessed severely truncated flagella. Also, both mutants were nonmotile and unable to colonize mice. Mutants with either flaA gene knocked out produced flagella morphologically similar to those of wild-type bacteria and expressed FlaA and FlaB. flaA_1 mutants which had flagella but displayed reduced motility did not colonize mice, indicating that motility is required for intestinal colonization by H. hepaticus and that the presence of flagella alone is not sufficient.
The Helicobacter species have been expanding during the last few years as an important family of rather host-specific, obligate host-associated ɛ-proteobacteria in the gastrointestinal tracts of mammals and birds. Phylogenetic analysis of 55 different helicobacters has revealed three different branches of the genus that encompass gastric, enterohepatic, and unsheathed species (51). The gastric species mostly inhabit the stomach, and the enterohepatic species colonize the enterohepatic tract and the biliary tree (9, 46). Helicobacter hepaticus, which persistently colonizes the murine cecum and colon and other parts of the intestinal tracts of mice and other small rodents, is the best-studied species of the enterohepatic helicobacters (46). All Helicobacter species possess flagella as prominent motility organelles and are vigorously motile (42, 51). Flagellar motility of the gastric Helicobacter species, in particular of H. pylori, has been intensively investigated (36). Flagella of the gastric Helicobacter species H. pylori, H. mustelae, and H. felis are important or even essential for colonization of and persistence in their natural niche, the stomach mucus (2, 7, 19, 24, 36). The formation of the flagellar apparatus in H. pylori is regulated in a particular manner, different from other bacteria such as Enterobacteriaceae or Vibrio spp. (34), and the structure of helicobacter flagella is characterized by complex flagellar filaments which consist of two highly divergent flagellin subunits, FlaA and FlaB (19, 20).
Thus far, flagella of enterohepatic Helicobacter species have not been investigated although flagellar morphological differences have been used as one hallmark of distinction between Helicobacter species. H. hepaticus is the only enterohepatic Helicobacter species whose complete genome sequence is known (46). Therefore, it is available as a resource for comparative analysis of flagellar function, structure, and regulation between gastric and enterohepatic Helicobacter species (8, 46). Genetic evidence from H. hepaticus suggests that the flagellar genes and components and flagellar regulatory genes are closely related to those of gastric species such as H. pylori (45). Furthermore, flagellar genes of H. hepaticus, as in H. pylori, are scattered all over the bacterial chromosome, with few small clusters of functionally linked genes visible. H. hepaticus is able, during persistent colonization, to cause chronic intestinal inflammation or cancer in susceptible murine hosts (9, 10, 51). It is therefore widely used as a model to characterize the immune status and the underlying potential of an infectious etiology associated with chronic inflammatory bowel disease, which, in humans, may become manifest as Crohn's disease or ulcerative colitis (3, 26). Each H. hepaticus bacterium carries two flagella which are localized in a characteristic bipolar manner and possess a flagellar sheath. It is unknown for any enterohepatic Helicobacter species whether motility and/or flagella play a role in infection and persistent colonization. In H. hepaticus, genes encoding the σ28 (FliA) and σ54 (RpoN) subunits of the RNA polymerase enzyme, which in H. pylori are central regulators that govern the ordered transcription of flagellar genes, were identified by homology searches (46). In various bacteria, including H. pylori and Escherichia coli, the FliA sigma factor is predominantly responsible for the transcription of late flagellar structural components such as the filament subunits, the flagellins. Moreover, the flaA and flaB genes, which probably encode the major and minor flagellin subunits that form the multimeric flagellar filament, are present in the H. pylori and H. hepaticus genomes. One particular feature common to all of the H. hepaticus genomes analyzed is the occurrence of two identical copies of the flaA gene, including its promoter region (flaA_1 and flaA_2), which are present in two distinct and separate locations within the genome (46). The complete sequence identity of the two promoters and flaA copies indicates a very recent gene duplication in this bacterial species.
The aim of our present study was to investigate the role of the sigma factor FliA of H. hepaticus in gene regulation, motility, and flagellum formation. Furthermore, by generating fliA and flaA single mutants and flaA_1 flaA_2 double mutants of H. hepaticus, we assessed how defects in motility and flagellar biosynthesis affect the colonization and persistence of H. hepaticus in the murine intestine. Notably, FliA proved to be the central regulator of late flagellar genes in H. hepaticus. In addition, fliA and flaA double and single mutants were not able to colonize mice, even in short-term experiments, although flaA single mutants possessed flagella morphologically similar to those of the wild type. Therefore, we suggest that functional and complete flagella and motility are of prime importance for both the initial and persistent colonization of the mouse intestinal tract by H. hepaticus. Moreover, it is suggested that flagella and motility are as important in the host-dependent life cycle of enterohepatic helicobacters as they are in gastric helicobacters.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. hepaticus ATCC 51449 was grown under specific microaerobic conditions by evacuating air-tight incubation jars four times to −0.2 × 105 Pa and equilibrating with a gas mixture of 80% N2, 10% CO2, and 10% H2. Bacteria were routinely cultured on blood agar plates (Columbia agar base II; Oxoid, Wesel, Germany) supplemented with 10% horse blood and antibiotics (10 mg/liter vancomycin, 2,500 U/liter polymyxin B, 5 mg/liter trimethoprim, 4 mg/liter amphotericin B). For liquid cultures, bacteria were grown in brain heart infusion (BHI) broth supplemented with 10% horse serum and antibiotics as indicated above. For growth of mutants, the plates were additionally supplemented with 20 mg/liter chloramphenicol (CM) or 10 mg/liter gentamicin (GM).
Motility assays.
Bacteria grown on blood agar plates for 24 h were stabbed into swarm agar plates (0.28 g/liter brucella broth, 0.3% Bacto agar, 10% horse serum, 10 mg/liter vancomycin, 2,500 U/liter polymyxin B, 5 mg/liter trimethoprim, 4 mg/liter amphotericin B). The zone of motility within the agar around the inoculated spot (motility halo) was examined and photographed after a 5-day incubation period. Bacteria were also observed in wet mounts in BHI broth for their swimming abilities.
Electron microscopy.
For the preparation of specimens for electron microscopy, bacteria were grown on blood agar plates for 24 h at low density. A copper electron microscopy grid coated with Formvar and carbon was placed onto the agar plate for 1 min. Afterwards, bacteria which became adherent to the grid were lifted from the plate and fixed with 1% phosphotungstic acid (pH = 7.0) for 30 s. The specimens were viewed and photographed in a Zeiss EM10 electron microscope.
Preparation of sheared flagellins and bacterial crude lysates.
For the preparation of whole bacterial lysates, the bacteria were grown to the indicated optical density at 600 nm (OD600) in liquid cultures or on blood agar plates for 24 h. Afterwards, bacteria grown in liquid cultures were centrifuged at 10,000 × g for 1 min at 4°C and resuspended in lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA, Complete protease inhibitor cocktail [Roche]). Bacteria grown on plates were directly resuspended in lysis buffer. For lysis, bacteria were sonicated for 2 to 3 min at 4°C at a power setting of 5 (Branson Sonifier). For the preparation of flagellins, bacteria grown on plates for 24 h were resuspended in 1 ml of 0.9% NaCl and pelleted at 3,000 × g for 10 min at 4°C. The bacterial pellet was then resuspended in 600 μl of 0.9% NaCl and the bacteria were gently sheared by pushing the suspension 20 times through a 26-gauge syringe needle. The intact bacteria were separated from the sheared-off material by centrifugation (9,000 × g at 4°C) for 20 min, and the supernatant was transferred to a fresh tube. To pellet flagellins, the supernatant was afterwards centrifuged at 40,000 × g at 4°C for 1 h in an ultracentrifuge. The pellet after ultracentrifugation, which is predominantly composed of flagellar material as shown by electron microscopy, was resuspended in a suitable volume of phosphate-buffered saline.
Protein methods and Western blotting.
Protein concentrations were commonly determined by a bicinchoninic acid test (Pierce, Rockford, IL) performed according to the manufacturer's instructions. Equal amounts of proteins were separated in 11.5% Tris-glycine sodium dodecyl sulfate (SDS)-polyacrylamide gels and then transferred onto BA83 nitrocellulose membranes with a 0.2-μm pore size (Schleicher & Schuell, Germany). The membranes were blocked with 5% skim milk in Tris-buffered saline supplemented with 0.05% Tween 20. The membranes were then incubated overnight at 4°C or at room temperature for 2 h with a rabbit polyclonal antiserum directed against H. pylori FlaA or FlaB (21) or intact H. pylori flagellins (20) and afterwards with a peroxidase-conjugated goat-anti-rabbit immunoglobulin G antibody (1:20,000 dilution; Dianova) at room temperature for 1 h. Detection was performed by the chemiluminescence substrate Enhanced SuperSignal West (Pierce).
Molecular biological methods.
DNA purification, cloning, and PCR were performed by standard methods.
RNA preparations.
Bacteria were grown in liquid cultures and harvested by centrifuging the cultures at 20,000 × g for 1 min at 4°C. The pellets were flash-frozen in liquid nitrogen and afterwards stored at −80°C. For the preparation of RNA, up to 3 × 109 bacteria were resuspended in 850 μl RLT buffer from the RNeasy kit (Qiagen) supplemented with 10 μl/ml β-mercaptoethanol and mixed with glass beads in 2 ml FastRNA tubes with lysing matrix B (MP Biomedicals). The bacteria were then lysed in a FastPrep FP220 shaker (MP Biomedicals) for 45 s at a speed of 6.5 m/s. Further purification was performed according to the protocol of the RNeasy kit (Qiagen). Residual DNA was digested with RNase-free DNase I (Roche), and the RNA was again purified according to the RNeasy kit RNA cleanup protocol. The RNA was tested for DNA contamination with primers C97_Heli16S_fw and C05_Heli16S_rv (GCTATGACGGGTATCC and ACTTCACCCCAGTCGCTG, respectively), amplifying a fragment of the H. hepaticus 16S rRNA gene. The quality and quantity of the RNA were checked by agarose gel electrophoresis and OD260 and OD280 measurement with a GeneQuant photometric instrument (Bio-Rad).
Microarray experiments.
Ten to 18 μg of total RNA of H. hepaticus ATCC 51449 and the isogenic fliA mutant was reverse transcribed and labeled with Cy3 or Cy5 fluorescent dye with random primers, SuperScript II reverse transcriptase (Invitrogen), and Cy3-dCTP- or Cy5-dCTP-labeled nucleotides (Amersham). Afterwards, the RNA was hydrolyzed by alkaline lysis. The labeled cDNA was purified with a PCR cleaning kit (Qiagen), dried, and solubilized in hybridization buffer (50% formamide, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.5% SDS, 50 mM NaHPO3 [pH = 8.0], 5× Denhardt's solution). The H. hepaticus whole-genome microarray (MWG Biotech GmbH, now available from Ocimum Inc.) consists of specific 50-mer oligonucleotides (one for each open reading frame) which allow the detection of 1,863 out of 1,875 open reading frames of the H. hepaticus genome (46). The microarrays were blocked with 4× SSC-0.1% SDS-1% bovine serum albumin, washed five times with double-distilled H2O, and dried by centrifugation. The labeled samples were then applied to the microarray in a humid hybridization chamber (Corning, NY) and incubated at 42°C for 16 to 20 h. Afterwards, the microarrays were washed successively in 2× SSC-0.1% SDS and 1× SSC-0.1% SDS and two times in 0.5× SSC. The hybridized slides were scanned in a confocal fluorescence scanner (Affymetrix 428). Fluorescence intensities were determined by the program ImaGene 5.0 with the following settings: segmentation method, fixed circle; signal low, 0.65; signal high, 0.95; background low, 0.05; background high, 0.95; background buffer, 3.0, background width, 3.0; empty spot threshold, 3.0. Data were normalized by the MAVI program (version Pro 2.5.1; MWG Biotech) (34). In order to minimize bias due to labeling and dye differences, for all experiments, the samples from each biological replicate were hybridized onto two microarrays with a switch of the fluorescent dye between the two samples (dye flip). The complete microarray data series (six arrays) is accessible at NCBI Gene Expression Omnibus in MIAME-compatible format (GEO series accession number GSE12123).
cDNA synthesis and semiquantitative PCR.
Total RNA was transcribed into cDNA by adding 3 ng/μl random primers (Invitrogen) to 0.5 μg or 1 μg RNA in a total volume of 11 μl, denaturing at 65°C for 10 min, and annealing of primers for 10 min at 20°C. Afterwards, 4 μl of Superscript buffer (Invitrogen), 2 μl of 0.1 M dithiothreitol, 1 μl deoxynucleoside triphosphates (10 mM each), 1 μl RNAseOut (Invitrogen), and 1 μl SuperScriptII (Invitrogen) were added to the reaction mixture, which was incubated at 42°C for 2 h. The reaction was terminated by heating the samples to 75°C for 15 min. To each reaction mixture, 30 μl H2O was added to increase the total volume to 50 μl. For semiquantitative reverse transcription-PCR (RT-PCR), 1.5 μl of cDNA per reaction mixture was amplified with gene-specific oligonucleotides (oligonucleotide sequences and conditions for PCRs are available on request).
Construction of mutants of H. hepaticus ATCC 51449. (i) fliA mutant.
The fliA gene of H. hepaticus ATCC 51449 was amplified by PCR with primers Hh_FliA-1 and Hh-FliA-2 (TTAGGATCCGGATTGACATTTTCCATTACC and TATGGATCCTCTATATCAGCAGTATCGC, respectively; amplified fragment of 1,096 bp containing BamHI restriction sites [underlined]) and cloned into pUC18 cut with BamHI. The resulting plasmid, pSUS2000, was reverse amplified with primers Hh_FliA-3 and Hh_FliA-4 (AATAGATCTCAAGCATTGCTCCATTCACTCG and ATAAGATCTGTCCTTTCTCGTGCTAATCG, respectively [BglII sites underlined]). Thereby, a 21-bp deletion was introduced. A gene encoding a CM acetyltransferase of Campylobacter coli mediating resistance to CM was amplified from pBHcp8 (14) with primers pCAT_1 and pCAT_2 (AACAGCTATGACCATGATTACG and AGAGGATCCGATATCGCATGCCTGCAGAG, respectively [BamHI site underlined]) and cloned into the BglII site of the inverse amplified PCR product. The resulting plasmid, pSUS2002, was used for transformation into H. hepaticus ATCC 51449.
(ii) flaA_1 mutant.
A random mutagenesis approach, initiated to generate a mutant bank of H. hepaticus genes in E. coli before the complete genome sequence became available, was used to generate a CM acetyltransferase (cm) cassette insertion mutation in flaA_1 (HH1364). To accomplish this, mass excision of a genomic H. hepaticus lambda ZAP II library (53) was performed according to the manufacturer's recommendations with the ExAssist helper phage (Stratagene). In vivo excision and recircularization yielded an H. hepaticus genomic library in pBluescript SK(−). Plasmid DNA was isolated from this library, digested with HpaI, and ligated to a HincII-digested cm cassette from pBHpC8 (14). Ligated plasmid was used to transform E. coli DH5α by high-voltage electroporation, followed by selection for CM resistance on LB agar containing 20 μg/ml CM. Plasmid was isolated from individual clones, sequenced, and used to transform E. coli ER1793. Plasmid from a clone with flaA homology was used to generate isogenic H. hepaticus flaA_1 mutants after plasmid transformation of strain ATCC 51449, which were selected on CM-containing plates. The cm cassette insertion in flaA_1 occurred at the very beginning of the flaA_1 coding region, downstream of the first 30 coding nucleotides. Thus, expression of FlaA_1 is virtually abolished in the mutant.
(iii) flaA_2 mutant.
A flaA_2 gene (HH1653) mutant was constructed by PCR amplifying exclusively the flaA_1 gene and a region upstream of the flaA_1 gene and including the cm cassette inserted in the 5′ coding region of flaA_1, from genomic DNA of the flaA_1 mutant (see above), with primers Hh_FlaA_3 and Hh_FlaA_repeat1_reverse (TAGCCGATTTGTATTACAGCC and GTGATAAGTCTAGCAAGTGC, respectively). This longer PCR product was used as the template in a subsequent nested PCR to amplify flaA_1, containing the cm cassette but excluding any upstream sequences, with primers Hh_FlaA_3 and Hh_FlaA_4 (TTACTTCAATTAGTTAAGTAACC), resulting in a PCR product with 100% homology to the flaA_1 and flaA_2 genes, with the exception of the cm cassette. This final PCR product was used for transformation into H. hepaticus ATCC 51449. The resultant clones were checked by PCR for whether they had incorporated the cm cassette into the flaA_1 gene or, as intended, into flaA_2.
(iv) flaA_1 flaA_2 double mutant.
Mutagenesis was performed by a quick PCR and ligation strategy without cloning. For this purpose, a fragment of 481 bp including the 5′ part of flaA_2 and a part upstream of flaA_2 with no homology to flaA_1 and with a KpnI restriction site inserted (primers Hh_FlaA2_7 and Hh_FlaA2_8 (CACTTGCAAGGTTACAACTGC and CGCGGTACCGATTGTCCATCTTGAGCTGC, respectively [KpnI restriction site underlined]) and a fragment of 564 bp including the 3′ part of flaA_2 and a part downstream of flaA_2 with no homology to flaA_1 and with a BamHI restriction site inserted (primers Hh_FlaA2_9 and Hh_FlaA2_10 (CGCGGATCCTATAATGCTGTTATTGCATCTGG and CAATAAATCCTGCCACAAATGC, respectively [BamHI restriction site underlined]) were amplified by PCR from genomic DNA of H. hepaticus ATCC 51449. A GM resistance (gm) cassette was amplified from plasmid pUC18-Gm (5) with primers Gm_1 and Gm_2 (TATGGTACCCGGGTGACTAACTA and AGAGGATCCCCGTGTCATTA, respectively [KpnI and BamHI restriction sites underlined]). The flaA_2-specific PCR products were digested with KpnI and BamHI, respectively, and the gm cassette was digested with KpnI and BamHI. The three digested PCR products were afterwards ligated with each other, and the ligation product was used as the template for a PCR with primers Hh_FlaA2_7 and Hh_FlaA2_10, which anneal to the most 5′ and 3′ parts of the two PCR products obtained before from the genomic DNA of ATCC 51449. The product of the desired length, containing the two sequences homologous to the genome of ATCC 51449 surrounding the gm cassette (1,887 bp), was gel purified. The gm cassette was oriented in the same direction as flaA_2 to minimize polar effects and because the gm cassette does not possess its own promoter sequence. In this way, the gm cassette was supposed to be cotranscribed as one mRNA from the promoter of the disrupted flaA_2 gene after integration into the genome of H. hepaticus ATCC 51449. The purified PCR product was then used for transformation into the flaA_1 mutant of H. hepaticus ATCC 51449 and resulted in the selection of several CM and GM double-resistant clones.
Transformation of H. hepaticus.
H. hepaticus was transformed by electroporation as previously described (13). Briefly, bacteria grown for 24 h on plates were harvested into 10% glycerol in distilled H2O and washed twice. An 80-μl volume of bacteria was mixed with plasmid or PCR product and subjected to electroporation in a 0.2-cm electroporation cuvette (Bio-Rad) at a voltage of 2.5 kV, a resistance of 200 Ω, and a capacitance of 25 μF. Afterwards, 100 μl of BHI broth was added and the bacteria were incubated for 24 h on nonselective plates. Then, the bacteria were transferred onto selective plates containing CM or GM and cultivated until resistant clones grew. Mutant clones were then characterized by PCR, protein expression, and electron microscopy.
Murine infection experiments.
C57BL/6 mice were obtained from Charles River (Wilmington, MA) from certified Helicobacter-free breeding colonies and were again tested in our laboratory to be free of any Helicobacter species, by PCR from feces with the following primer pairs: Helicobacter genus-specific C97_Heli16S_fw and C05_Heli16S_rv (11) and H. hepaticus-specific B38 and B39 (GCATTTGAAACTGTTACTCTG and CTGTTTTCAAGCTCCCC, respectively) (41). All animal experiments were conducted in accordance with the German Animal Welfare Law and with European Communities Council Directive 86/609/EEC for the protection of animals used for experimental purposes. All experiments were approved by the local institutional animal care and research advisory committee and authorized by the local district authority. Mice were fed a sterile pellet diet and water ad libitum and were housed in microisolator cages. For colonization experiments, animals were infected on 3 successive days with 2 × 108 bacteria or were mock infected (by application of BHI medium only). Colonization levels were monitored by regular detection of H. hepaticus by PCR with H. hepaticus-specific primers as indicated above on genomic DNA prepared from feces. Genomic DNA from feces was prepared after homogenization of the feces in S.T.A.R. buffer (Roche) supplemented with 10% chloroform, by using the Qiagen QiaAmp DNA Stool Kit Mini as indicated by the manufacturer. At the end of the experiments, animals were euthanized and serial dilutions of tissue homogenates prepared from the tip of the cecum were plated on blood agar plates supplemented with antibiotics (see above) to obtain colony counts of H. hepaticus. Furthermore, genomic DNA was prepared from the cecum tip and the proximal colon (Qiagen QiaAmp Tissue Kit) and examined for H. hepaticus-specific DNA content by PCR with the primer pairs mentioned above. The limit of detection by PCR, assessed with appropriate control DNA dilutions used for spiking, was about 10 genomic copies of H. hepaticus. Detection by PCR was less sensitive than detection by colony counting after plating.
For all experiments, six animals per experimental group were employed, with three male and three female mice in each group (see Table 2). In a first set of experiments, animals were infected with wild-type H. hepaticus or the fliA mutant for 4 and 8 weeks. This experiment was repeated once with an infection duration of 4 weeks. In a second set of experiments, animals were infected with wild-type H. hepaticus, the flaA_1 flaA_2 double mutant, and the flaA_1 gene mutant for 4 weeks.
TABLE 2.
Experimental infection of C57BL/6 mice by wild-type H. hepaticus and flagellar mutant bacteriaa
| Expt | Duration (wk) |
H. hepaticus ATCC 51449
|
fliA mutant
|
P value |
flaA_1 mutant (clone 1)
|
P value |
flaA_1 flaA_2 mutant
|
P value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of animals | No. colonized/total | No. of animals | No. colonized/total | No. of animals | No. colonized/total | No. of animals | No. colonized/total | |||||
| 1a | 4 | 6 | 6/6 | 6 | 0/6 | 0.002 | ||||||
| 1b | 8 | 5 | 5/5 | 6 | 0/6 | 0.002 | ||||||
| 2 | 4 | 6 | 6/6 | 3 | 0/3 | 0.012 | ||||||
| Overall | 17 | 17/17 | 15 | 0/15 | <0.001 | |||||||
| 3 | 4 | 6 | 6/6 | 6 | 0/6 | 0.002 | ||||||
| 4 | 4 | 6 | 6/6 | 6 | 0/6 | 0.002 | ||||||
For each experiment, the duration of the experiment, the number of animals per group, and the number of colonized animals/total number of animals are indicated. The P values were calculated by Fisher's exact test between animals inoculated with wild-type bacteria and those inoculated with the respective flagellar mutants.
Statistical analyses.
Statistically significantly regulated genes in the microarray experiments were determined with the program Significance Analysis of Microarrays (SAM; Stanford University, Palo Alto, CA) (49) and the following settings: delta = 0.364, leading to a false discovery rate of 0 (q value of ≤0.05; for settings and algorithms, see the SAM manual at http://www-stat.stanford.edu/∼tibs/SAM/). Data on the colonization levels of H. hepaticus in mice were examined for statistical significance with Fisher's exact test.
RESULTS
Construction of H. hepaticus fliA, flaA_1, and flaA_2 mutants and flaA_1 flaA_2 double mutants.
For the analysis of flagellar regulation and the role of motility in H. hepaticus in colonization, first a knockout mutation in the gene encoding the σ factor FliA or σ28 (HH1146) was constructed by disruption with a CM resistance cassette in the direct orientation (see Materials and Methods).
Since FliA was assumed to strongly affect transcriptional regulation at the same time as flagellar biosynthesis, we also constructed several different mutants with only structural defects in late flagellin genes, which are not involved in transcriptional regulation. Interestingly, H. hepaticus possesses two copies of the gene encoding the main subunit of flagellin, FlaA (HH1364 and HH1653 [flaA_1 and flaA_2, respectively]), with identical sequences including the promoter region. To further examine the possible role of this gene duplication, mutants lacking the expression of either one (flaA_1 or flaA_2 single mutant) or both (flaA_1 flaA_2 double mutant) of the flaA genes were generated by allelic exchange mutagenesis (for details, see Materials and Methods). The flaA_1 disruption mutant contained a CM resistance marker inserted after the first 30 coding nucleotides of flaA_1. A flaA_2 knockout mutant was constructed by amplifying the flaA_1 gene including the inserted CM resistance cassette from the already existing flaA_1 mutant. This PCR product was used for transformation into H. hepaticus ATCC 51449, and several flaA_2 insertion mutants were identified among the resulting clones by PCR.
A second selective marker had not been established for H. hepaticus before. For the construction of a mutant in which both copies of flaA were knocked out, a promoterless gm cassette (5) was successfully established as a novel resistance marker for H. hepaticus (for details, see Materials and Methods). Mutagenesis of flaA_2 for constructing the double mutant was performed by a quick PCR and ligation strategy (see Materials and Methods). The final PCR product was used for transformation of the previously generated flaA_1 mutant of H. hepaticus ATCC 51449, and three resulting colonies, which were both GM and CM resistant, were tested and verified to carry the correct integration of the gm cassette in the flaA_2 gene.
Flagellar and motility phenotype of H. hepaticus fliA and flagellin mutants.
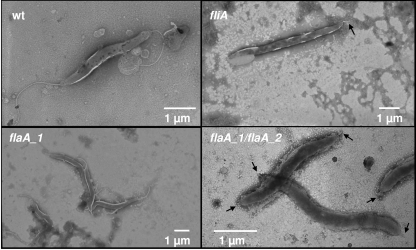
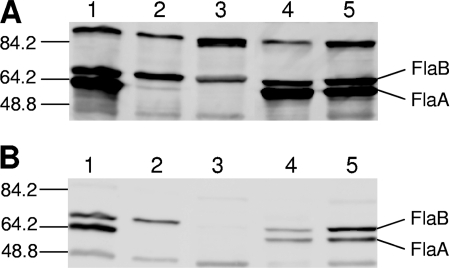
The mutants deficient in the flagellar σ28 factor FliA and the flaA single and double mutants were first characterized phenotypically by light and electron microscopy. Wild-type H. hepaticus bacteria possess one flagellum at each pole (9). In contrast, the fliA mutants did not display complete flagellar structures (Fig. 1). Some bacteria possessed no flagella at all, while most bacteria (79 of 100 counted bacteria) exhibited a short, truncated flagellum at one or both poles. Otherwise, the fliA mutant bacteria did not show any obvious morphological differences compared to wild-type bacteria. The motility of the fliA mutant was impaired, as demonstrated by motility agar stab assays (Fig. 2), and no translational motility was observed in wet mounts (not shown), in contrast to wild-type bacteria, which displayed strong motility in both assays. In order to determine the expression of flagellins in the different mutants, Western blot assays with various antisera raised against recombinant or native H. pylori flagellins were performed. All of these sera also recognized H. hepaticus flagellins with high sensitivity. According to our Western blot analysis (Fig. 3), the fliA mutant did not synthesize detectable amounts of FlaA protein, while similar amounts of the minor flagellar filament subunit, FlaB, were present in the fliA mutant compared to wild-type bacteria.
FIG. 1.
Electron microscopic pictures of H. hepaticus ATCC 51449 and flagellar mutants. Electron microscopic pictures of wild-type (wt) H. hepaticus ATCC 51449, the fliA and flaA_1 (clone 1) single mutants, and the flaA_1 flaA_2 double mutant were prepared. A representative picture of each strain is shown. Black arrows indicate truncated flagella.
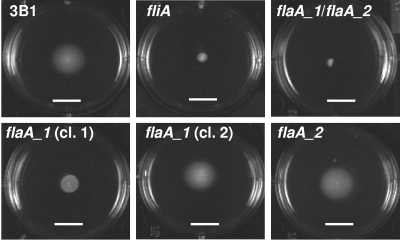
FIG. 2.
Plate motility assays of wild type and flagellar mutant H. hepaticus ATCC 51449. Wild-type H. hepaticus; the fliA, flaA_1, and flaA_2 single mutants; and the flaA_1 flaA_2 double mutant were inoculated into motility agar and incubated for 5 days. A representative picture of each strain from at least three independent assays is shown. For the flaA_1 mutant, where different motility was observed with different clones, representative experiments with two different clones (cl. 1 and cl. 2) are depicted. The white bars equal 1 cm.
FIG. 3.
Expression of FlaA and FlaB by H. hepaticus ATCC 51449 and flagellar mutants. Ten micrograms of lysate of wild-type or mutant H. hepaticus was separated by SDS-polyacrylamide gel electrophoresis and Western immunoblotted with an antibody (AK179; 1:2,000 dilution) raised against native purified H. pylori flagella (A) or against recombinantly expressed H. pylori FlaB (1:1,000 dilution) (B). Lanes: 1, wild-type H. hepaticus; 2, fliA mutant; 3, flaA_1 flaA_2 double mutant; 4, flaA_1 mutant; 5, flaA_2 mutant. The values on the left are molecular masses in kilodaltons.
Similar to the fliA mutant, the flaA_1 flaA_2 double mutant had only very short truncated flagella (85 bacteria out of 100 possessed short flagella) at one or, predominantly, both poles but otherwise appeared to be morphologically similar to wild-type bacteria (Fig. 1). Like the fliA mutant, the flaA double mutant did not show any motility halo when incubated in motility agar (Fig. 2) and swimming was not observed in wet mounts. In Western blot assays, as expected, no FlaA was detected (Fig. 3). An antibody raised against recombinant H. pylori FlaB detected very low levels of FlaB in lysates of the H. hepaticus flaA_1 flaA_2 double mutant (Fig. 3B), while an antibody directed against native Helicobacter flagella detected some, although less, FlaB in lysates of the flaA double mutant compared to lysates of wild-type bacteria (Fig. 3A). In addition, a slight gel migration change was observed for FlaB in the double mutant. These findings suggested that FlaB was differently posttranslationally modified in the flaA double mutant compared to wild-type bacteria. In contrast, there was no obvious difference in the detection of FlaB in wild-type bacteria with both antisera (Fig. 3A). In fliA mutants, differential immunodetection of FlaB with the two different antisera was observed in some experiments (not shown), although less than in the flaA_1 flaA_2 double mutant.
Both flaA_1 and flaA_2 single mutants were morphologically similar to wild-type bacteria, possessing long flagella at both poles (Fig. 1). In motility agar plates, four different clones of the flaA_2 mutant showed slightly reduced motility compared to wild-type bacteria (Fig. 2). On the other hand, different clones of the flaA_1 single mutant displayed motility to different extents in motility plates. Of seven clones tested, one (clone 2) was almost as motile as the wild type (swimming ability in wet mounts and in agar motility tests [Fig. 2]) while the other six were strongly impaired in motility in agar (Fig. 2) and did not swim in wet mounts. Sequence determination of flaA_1 including the noncoding upstream sequences from all different flaA_1 clones did not reveal any nucleotide differences between those clones or from the wild-type strain that could explain the different motility phenotype. Different clones of flaA_2 mutants or wild-type bacteria, in contrast, all displayed similar motility within each group. In all flaA single mutant clones, the amounts of FlaA and FlaB detected by Western immunoblotting with antibodies directed against recombinant or native flagellins appeared to be similar (Fig. 3A and B). Also, all flaA single mutants had similar amounts of FlaA and FlaB compared to wild-type bacteria.
Definition of the FliA regulon of H. hepaticus by microarray analysis.
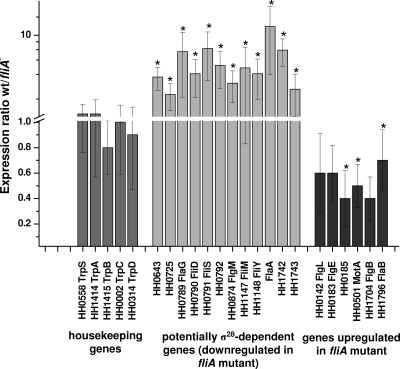
The transcriptional regulon, which is under the control of the σ28 transcription factor (FliA) in H. hepaticus, was determined with microarrays. An oligonucleotide-based whole-genome microarray by MWG Biotech was used that can detect 1,863 of the proposed 1,875 open reading frames of H. hepaticus ATCC 51449 (46). Wild-type H. hepaticus ATCC 51449 and the isogenic fliA mutant were grown in liquid culture to an OD600 of 0.7 to 0.8 (mid-logarithmic growth phase). Cy3- or Cy5-labeled cDNA synthesized from these two bacterial preparations was competitively hybridized onto the microarrays. Three independent biological experiments were performed (six microarrays, including dye flip experiments). In the subsequent data analysis, overall 11 transcripts were found to be significantly more highly expressed in wild-type bacteria than in the fliA mutant by the microarray statistics program SAM (see Materials and Methods). These genes were predicted to belong to seven operons due to their genomic arrangements (Table 1 and Fig. 4) (46). Among the transcripts which were more abundant in wild-type bacteria were the late flagellar genes flaA, flgM, fliD, and fliS. Also downregulated in the fliA mutants in comparison to wild-type bacteria were transcripts of fliM and fliY, which probably are organized in an operon together with fliA. Additionally, the transcripts HH0789 (flaG) and HH0792 (a presumed fliT homolog), which are in the same proposed operon as fliD and fliS, and genes HH0643, HH0725 (putative methyl-accepting chemotaxis protein), and HH1742, which is of unknown function (see Discussion), showed significantly higher expression in wild-type bacteria than in the fliA mutant (Fig. 4). Transcript amounts of HH1008, which encodes an NH3-dependent NAD+ synthetase (nadE), and the open reading frame HH1743, which is of unknown function, also were, on average, more than twofold reduced in the fliA mutant than in wild-type bacteria, although due to a high standard deviation, their differential expressions were not significant at the SAM settings used (see Materials and Methods). Regulation of selected downregulated genes in the fliA mutant (HH0643, HH0789, flgM, fliM, fliY, and flaA) was confirmed by semiquantitative RT-PCR assays performed with the same total RNA samples (Table 1). Differential regulation of two transcripts, HH0725 and HH1008 (nadE), could not be validated by semiquantitative RT-PCR, whereas for all of the other genes the pattern of regulation indicated by the microarray data could be confirmed (Table 1). Two genes were significantly more highly expressed in the fliA mutant than in wild-type bacteria (HH0185 and flgB; Fig. 4), which were verified by semiquantitative RT-PCR (Table 1). Transcript amounts of the two flagellar genes flgI and flaB, which appeared to be slightly more highly expressed in the fliA mutant than in wild-type bacteria according to the microarrays (although the difference was not significant by SAM analysis), were additionally examined and confirmed to be upregulated in the fliA mutant by semiquantitative RT-PCR.
TABLE 1.
Microarray analysis of transcripts differentially regulated between wild-type H. hepaticus and the fliA mutanta
| Group and open reading frame(s) | Gene(s) | Mean wild-type/fliA expression ratiob | SD | Significantly regulated (SAM)c | Semiquantitative RT-PCRd |
|---|---|---|---|---|---|
| Proposed σ28-dependent genes | |||||
| HH0643 | 12.8 | 9.47 | Yes | Confirmed | |
| HH0724 | NPe | No | NDf | ||
| HH0725 | 1.9 | 0.69 | Yes | Not confirmed | |
| HH0789 | flaG | 6.3 | 4.54 | Yes | Confirmed |
| HH0790 | fliD | 3.4 | 1.66 | Yes | ND |
| HH0791 | fliS | 6.9 | 4.08 | Yes | ND |
| HH0792 | 4.3 | 2.07 | Yes | ND | |
| HH0874 | flgM | 2.6 | 1.09 | Yes | Confirmed |
| HH1008 | nadE | 2.4 | 1.09 | No | Not confirmed |
| HH1009 | lpxK | 1.3 | 0.43 | No | ND |
| HH1010 | bcp | 1.2 | 0.26 | No | ND |
| HH1144 | 0.8 | 0.40 | No | Not regulated | |
| HH1145 | 0.7 | 0.32 | No | Not regulated | |
| HH1146 | fliA | 1.8 | 0.85 | No | ND |
| HH1147 | fliM | 4.0 | 3.17 | Yes | Confirmed |
| HH1148 | fliY | 3.4 | 1.74 | Yes | Confirmed |
| HH1364, HH1653 | flaA_1, flaA_2 | 12.8 | 9.47 | Yes | Confirmed |
| HH1742 | 6.6 | 2.43 | Yes | Confirmed | |
| HH1743 | 2.2 | 1.17 | No | ND | |
| Proposed σ54-dependent genes | |||||
| HH0140 | 0.9 | 0.35 | No | ND | |
| HH0141 | 0.7 | 0.25 | No | ND | |
| HH0142 | flgI | 0.6 | 0.31 | No | Confirmed |
| HH0183 | flgE | 0.6 | 0.22 | No | ND |
| HH0184 | flgD | 1.0 | 0.28 | No | ND |
| HH0185 | 0.4 | 0.22 | Yes | Confirmed | |
| HH0186 | 0.7 | 0.60 | No | ND | |
| HH0501 | motA | 0.5 | 0.17 | Yes | Not confirmed |
| HH0502 | motB | 0.8 | 0.25 | No | ND |
| HH0503 | 1.1 | 0.21 | No | ND | |
| HH0504 | 0.7 | 0.33 | No | ND | |
| HH1407 | flgB | 0.4 | 0.17 | Yes | Confirmed |
| HH1408 | flgC | 0.8 | 0.39 | No | ND |
| HH1409 | fliE | 1.1 | 0.29 | No | ND |
| HH1410 | 1.0 | 0.20 | No | ND | |
| HH1411 | 1.0 | 0.44 | No | ND | |
| HH1796 | flaB | 0.7 | 0.24 | No | Confirmed |
Gene groups belonging to proposed operons are grouped together. All genes belonging to each of the proposed operons had the same predicted transcriptional orientation. The upper part of the table lists the genes with higher transcript amounts in the wild type, and the lower part shows the genes with higher transcript amounts in the fliA mutant.
Ratios higher than 2 indicate increased transcript levels in the wild type, and ratios lower than 0.5 indicate reduced transcript levels in wild-type bacteria.
Genes significantly regulated according to a SAM analysis.
For genes whose regulation was validated by semiquantitative RT-PCRs, it is indicated whether the microarray results were or were not confirmed.
NP, not present on array.
ND, not determined.
FIG. 4.
Results of the microarray analyses of wild-type and fliA mutant H. hepaticus. H. hepaticus whole-genome microarrays were competitively hybridized with cDNAs of both wild-type (wt) and fliA mutant H. hepaticus. The ratio of the mean spot intensity of the wild type versus the fliA mutant is depicted for selected genes. The mean ratio and standard deviation were calculated from six independent experiments (three biological experiments with two arrays each). Shown are the two groups of genes which are differentially regulated between the wild-type and fliA mutant bacteria and, as a control, five tryptophan metabolism housekeeping genes. Asterisks indicate genes that are significantly differently regulated according to statistical analysis with the SAM software package (see Materials and Methods).
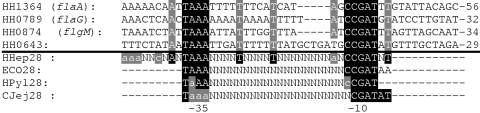
σ28 promoter consensus sequences, which are recognized by FliA, are well established for the closely related H. pylori bacteria or for other bacteria such as E. coli. Therefore, we analyzed the promoter sequences of putative operons with genes downregulated in the fliA mutant for the existence of σ28 promoter-like sequences. Upstream of HH1364 (flaA_1), HH1653 (flaA_2), HH0789 (flaG), HH0874 (flgM), and HH0643, sequences were detected which corresponded to the σ28 promoter consensus sequences of H. pylori or E. coli (Fig. 5). In H. pylori, the spacer between the −35 and −10 boxes has predominantly a length of 13 bp (21, 52). In contrast, the length of the σ28 promoter spacers in H. hepaticus appeared to be less conserved since it varied between 15 and 19 bp. No σ28 promoter-like sequences were detected upstream of the genes HH1008 and HH1742. Upstream of the gene HH0724, which is not present on the microarray and is probably in one operon with HH0725, a σ28 promoter-like sequence was also identified, although it was less conserved than for the other FliA-dependent genes.
FIG. 5.
Alignment of proposed σ28 promoter sequences of H. hepaticus. The highly conserved −10 and −35 boxes are white on a black background. Other highly conserved nucleotides are highlighted with gray. Consensus σ28 promoter sequences of H. hepaticus (Hhep28) and E. coli (ECO28) are shown, and C. jejuni (CJej28) and H. pylori (HPyl28) sequences are given for comparison. Less conserved residues in the consensus sequences are indicated by lowercase letters. The upstream distance to the transcriptional start of the respective gene is shown at the right side of the alignment.
FliA is required for H. hepaticus colonization in experimental infections of C57BL/6 mice.
As described above, a fliA knockout mutant yielded nonmotile bacteria that possess either no or strongly truncated flagella. It is not known which role flagella or motility may play in intestinal colonization by enterohepatic Helicobacter species. Therefore, we asked whether fliA mutant bacteria are still capable of colonizing C57BL/6 mice. In a first colonization study, 12 mice in each group were sham infected or infected with wild-type H. hepaticus ATCC 51449 or with the isogenic fliA mutant. The mice were euthanized after 4 and 8 weeks (Table 2), respectively. Colonization was examined by bacterial culture from homogenates of the cecum tip and by PCR on genomic DNA prepared from homogenates of cecal tissue and the proximal colon. Mice colonized with wild-type H. hepaticus yielded high bacterial counts with, on average, 1.4 × 109 bacteria per g cecum after 4 weeks and 6.8 × 108 bacteria per g cecum after 8 weeks (CFU count differences are not statistically significant). In contrast, none of the mice which were inoculated with the fliA mutant tested positive for H. hepaticus in culture and PCR at 4 and 8 weeks after the initial inoculation (Table 2). To follow the course of infection at early time points in live animals, fecal pellets were collected at 1 and 2 weeks postinfection and examined for the presence of H. hepaticus by PCR. While all of the mice infected with wild-type H. hepaticus tested positive in the fecal PCR at all times, none of the mice inoculated with the fliA mutant were positive for the presence of H. hepaticus at any time point. All of the sham-infected mice were negative for the presence of H. hepaticus at all time points, except for one mouse which was probably cross-contaminated because it was colonized with wild-type H. hepaticus at 4 weeks.
In a second set of in vivo experiments, mice were sham infected or infected with wild-type H. hepaticus and the fliA mutant and their infection status was analyzed at 4 weeks postinfection (Table 2). Three mice in the fliA mutant group died of unrelated causes before and during the experiment, so that only three mice could be evaluated. All of the mice infected with wild-type bacteria tested positive for H. hepaticus in culture and for H. hepaticus DNA in PCR. In contrast, none of the mice inoculated with the fliA mutant became successfully infected with H. hepaticus according to PCR assay of DNA from fecal pellets at 1 week postinoculation, and negative results when using culture and PCR assay of DNA from cecal and colonic tissues at 4 weeks postinoculation.
Role of motility in the colonization ability of H. hepaticus.
As demonstrated in the experimental mouse infections, H. hepaticus fliA mutants were not able to colonize C57B/L6 mice. This raised the question of whether this effect was indeed due to the lack of flagella and impaired motility observed in this mutant or, in addition, may be due to or enhanced by differential regulation of other σ28-dependent genes. To answer this question, C57B/L6 mice were inoculated with wild-type H. hepaticus and the flaA_1 flaA_2 double mutant, which does not express any FlaA and therefore exhibits a severe flagellar structure defect (Fig. 1) and loss of motility (Fig. 2). For each group, six mice were inoculated and monitored for infection until 4 weeks postinoculation. While all of the animals inoculated with wild-type bacteria were successfully colonized, none of the mice infected with the flaA_1 flaA_2 mutant tested positive for H. hepaticus in both PCR and culture from cecal tissue homogenates. To assess the effect of the lack of a single copy of flaA on colonization, we also inoculated a group of mice with a flaA_1 mutant clone (see above) which possessed flagella with a morphology similar to that of the wild type but showed diminished motility (clone 1; Fig. 2). Like the flaA double mutant and the fliA mutant, flaA_1 clone 1 bacteria did not colonize mice (Table 2). Fecal monitoring for H. hepaticus by PCR, starting at 1 week postinfection and at any further subsequent week, and CFU counting in cecal tissue homogenate at 4 weeks after inoculating the mice detected no infection with either the flaA double mutant or the flaA_1 clone 1 mutant (Table 2).
DISCUSSION
Aims of the present study were to gain insight into transcriptional control by the H. hepaticus sigma factor FliA and to investigate the role of motility and of FliA-dependent genes in murine intestinal colonization. We first analyzed the transcriptional regulation of fliA-dependent genes. Overall, 11 genes exhibited significantly higher transcript levels in wild-type bacteria than in the FliA mutant. Most of these genes (flaA, flgM, flaG, fliD, fliS, and HH0792/fliT) were well-known flagellar genes which had also been reported before to be dependent on the FliA sigma factor in H. pylori (21) and Campylobacter jejuni (4). Upstream of most of the above genes, including the two copies of flaA, highly conserved σ28-like promoter sequences were detected (Fig. 5). flaA transcription was exclusively dependent on σ28 in other bacteria (21). However, in the H. hepaticus σ28 mutant, small amounts of flaA transcript were observed by semiquantitative RT-PCR (not shown). This may indicate residual transcriptional activation of flaA independently of σ28, by a different weakly transcribed unidentified promoter. Also upstream of flgM, a σ28 promoter sequence was detected, which explained the flgM downregulation seen in the H. hepaticus fliA mutant. In other bacteria such as Enterobacteriaceae, Pseudomonas, or H. pylori, transcriptional activation of flgM is dependent on more than one transcription factor (6, 21, 34). At 46 bp upstream of the small open reading frame HH0873 of unknown function preceding H. hepaticus flgM, an additional σ54-like promoter sequence (TTGGTA-N6-TTGCAT) was indeed identified. A proposed operon encoding FlaG and the flagellar accessory genes and chaperones FliD (flagellar cap), FliS, and FliT (34) was downregulated in the fliA mutant. These genes were shown to be partially dependent on both FliA and RpoN in C. jejuni and H. pylori (4, 21, 34). For H. hepaticus, no predicted promoter sequences other than that for σ28 could be identified upstream of this operon.
The flagellar genes flgI, flgE, flgB, and flaB were expressed at higher transcript levels in the fliA mutant than in wild-type bacteria. Upstream of all of these genes, highly conserved σ54 promoter sequences were identified. The upregulation of these genes in the fliA mutant was probably indirect, dependent on factors other than FliA. It is assumed that a feedback repression loop of active FliA on σ54-dependent genes is inactivated in fliA mutants, since similar effects on σ54-dependent genes were previously reported in H. pylori and C. jejuni (4, 34, 43).
Besides genes encoding proteins directly involved in the biosynthesis or function of flagella, some novel genes were dependent on H. hepaticus σ28 (Table 1). HH0725, which was downregulated in the fliA mutant, possesses a methyl-accepting chemotaxis protein domain and is therefore very likely related to motility functions. No complete σ28 promoter consensus was found upstream of this gene. The product of another proposed FliA-dependent gene, HH0643, may include a methyltransferase domain according to HHpred. In addition, the predicted protein HH0643 contains a domain of unknown function related to glycosyltransferases (DUF1975/COG2849 family). Homologs of this protein are present in various Enterobacteriaceae. A novel proposed σ28-dependent operon is composed of the genes HH1742 and HH1743. The encoded proteins are predicted to be secreted or membrane proteins because they contain high-scoring signal peptides for secretion (SignalP). Homologs of HH1742 of unknown function have been identified in the δ-proteobacterium MLMS-1 and in Wolinella succinogenes, while homologs to HH1743 are present in H. pylori and Helicobacter acinonychis. HH1742 contains a large domain similar to a bacterial outer membrane porin (PorB; HHpred, probability of 99.6%). HH1743 possesses a domain belonging to the family of YfaZ precursors (Pfam P value, 0.059) of unknown function. YfaZ was recently characterized in E. coli as an outer membrane protein (29).
Novel nonflagellar genes regulated by σ28 could, for instance, represent novel virulence-associated genes, possibly secreted by the flagellar type III secretion system. In C. jejuni, novel σ28-dependent virulence factors were identified via a regulatory screen, some of which were secreted in a flagellum-dependent manner (4, 17, 38). Therefore, we asked whether the novel σ28-regulated genes of H. hepaticus might be involved in virulence. HH0725- and HH0643-like genes are widespread in other bacteria, and HH0725 contains a methyl-accepting chemotaxis protein domain. HH1742 and HH1743 are probably secreted or membrane proteins. We performed comparative DNA sequence analysis of HH0643, HH0725, and HH1742 PCR products from all 13 different H. hepaticus isolates from our strain collection (not shown) and found only low interstrain sequence diversity, while the genes were present in all strains. Taken together, these analyses indicate that the proteins are more likely associated with signaling, protein modification, or transport and motility functions rather than being candidates for pathogen-specific novel virulence factors.
FliA-dependent regulons have been investigated by microarray analysis in two other members of the closely related Campylobacterales, H. pylori (21) and C. jejuni (4, 18, 23). Well-characterized FliA-dependent flagellum-related genes in H. pylori and C. jejuni largely overlap genes detected by the present analysis of H. hepaticus. The complement of FliA-dependent genes in C. jejuni was surprisingly much larger (roughly 140 genes downregulated in fliA mutants) (23) than the regulons found in both H. hepaticus and H. pylori (of which H. pylori had the smallest set of 5 clearly FliA-dependent genes (21). Part of this divergence in the number of FliA-dependent genes may be due to differences in methodology, since the microarray analyses of C. jejuni (4, 23) and H. hepaticus (present study) were performed later during the growth phase than for H. pylori (21). FliA-dependent transcriptional activity is presumed to be most active in late exponential growth (35). Regulation of flagellar modification genes by FliA was more obvious in C. jejuni than in the two other species. Roughly one-fifth of the genes whose transcript abundance was influenced in C. jejuni fliA mutants (4, 23) were associated with posttranslational glycosylation functions and included genes of the known peb protein glycosylation gene cluster, which comprises approximately 27 genes (32). In the intestinal pathogen C. jejuni, in addition, several novel FliA-regulated genes of unknown function were found to be involved in host interaction, two of which were secreted by the flagellar type III apparatus (FspA, FlaC, and nonsecreted protein Cj0977) (17, 38).
The major flagellin genes flaA_1 and flaA_2 were the dominant genes controlled by H. hepaticus FliA. Previously, a duplication of the flaA gene in H. hepaticus was described (46). The two flaA copies are located in entirely different regions of the H. hepaticus genome with different neighboring genes of unknown function and appear to be monocistronic operons in both cases. Since the two flaA gene copies, including their noncoding upstream regions, are 100% identical and contain no point mutations, we speculate that the gene duplication occurred quite recently. This duplication was present in all of the isolates in our strain collection. However, since all of those strains were derived from laboratory-housed mouse colonies, which are currently the only source of H. hepaticus isolates, the duplication may be restricted to a particular strain of H. hepaticus. A role for this gene duplication in vivo is not clear, since H. hepaticus isolates from wild rodents are not available and niche requirements in laboratory-kept mice are not well defined. Inactivation of either one of the two flaA copies appeared to have a minor impact on flagellar morphology and FlaA protein expression; inactivation of flaA_1 seemed to have a larger impact on motility than the mutation of flaA_2. Possibly, the influence of either flaA copy on other motility-relevant genes differs. One role for the duplicated flaA genes could be to provide a complement for functional replacement and also intergenic recombination, in case one flaA gene is accidentally inactivated.
Interestingly, the inactivation of both flaA genes in the double mutant or, under some conditions, lack of FliA seemed to affect the posttranslational modification of the FlaB protein and possibly other proteins, as assessed by Western immunoblotting with antibodies of different specificities and by migration in SDS-polyacrylamide gel electrophoresis. Very likely, the proposed flagellin modification in H. hepaticus is, at least in part, associated with glycosylation events, as has been reported for the flagellin proteins of H. pylori (21, 40), H. felis (19), and Campylobacter sp. (47, 48). FlaB appeared to undergo increased posttranslational modification in particular in doubly FlaA-deficient mutants. Potentially, the almost complete lack of the very abundant, probably glycan-modified, FlaA protein in double flaA mutants might lead to hypermodification of other posttranslationally modified proteins such as FlaB.
In some bacterial species, flagella are crucial for successful colonization of the host. Mutants of H. pylori (7, 24, 36), C. jejuni (33), or C. coli (37) deficient in flagella and motility were deficient in persistent colonization. Lack of flagella and motility appeared to impair gastrointestinal colonization less drastically in some infection models by some other intestinal pathogenic bacteria (22) which are less restricted to a specific host or niche, such as V. cholerae (12, 39) or Salmonella enterica serovar Typhimurium (28). Thus, we asked whether the presence of flagella or motility is required for successful colonization of H. hepaticus in the intestinal tracts of mice and found that mutants deficient in FliA were not able to colonize mice. Even at very early time points (3 days after inoculation), fliA mutant bacteria could not be detected in the feces, while the wild-type strain was detected in all of the infected mice, suggesting that the fliA mutant is not only deficient in long-term colonization but lacks the capacity of initial colonization. In order to address the issue of whether the colonization deficiency of the FliA mutant could be due to functions missing in fliA mutants other than only the almost complete lack of flagella or impaired motility, we challenged mice with either a flaA_1 flaA_2 flagellin double mutant that did not express any FlaA or with a flaA_1 single mutant with morphologically normal flagella but strongly diminished motility. flaA_1 flaA_2 double mutants were not detectably colonizing, demonstrating that either the presence of flagella or motility is crucial for the infection. Also, mice challenged with the H. hepaticus flaA_1 single mutant were not colonized, indicating that reduced motility rather than structural impairment of flagella was the crucial factor which prevented the successful colonization of mice. flaA_2 mutants, which displayed only slightly reduced motility, colonized mice at 4 weeks postinoculation (T. Sterzenbach and C. Josenhans, unpublished data); however, colonization was slightly reduced in comparison with the wild type. For H. pylori, it was reported that a deletion mutant in the flagellar motor protein MotB that affects motility but not the assembly of complete flagellar structures leads to a deficiency in murine colonization (36). Hence, our present study revealed that FliA and FlaA expression and motility are essential prerequisites for intestinal colonization of H. hepaticus, whereas the presence of flagella was not sufficient to ensure even initial colonization. Whether other factors regulated by FliA are crucial for the ability to colonization could not been shown.
Flagella are major antigenic targets both for the innate and adaptive immune systems. Infection with various bacterial species, including H. pylori (31), P. aeruginosa (1), C. jejuni (50), and C. coli (30), produces host immunoglobulins that are directed against flagellar proteins. Furthermore, for H. hepaticus, the flagellar hook protein FlgE represents a dominant T-cell antigen (25). It was therefore surprising to find that in the intestinal tract, where immune functions are highly active, intact flagella and motility are absolutely required for colonization by H. hepaticus. Flagella of H. pylori and H. hepaticus are encased by a sheath (16), which probably offers some protection from the action of host immune functions. In addition, flagellins and lipopolysaccharide of both species possess low activation potential for Toll-like receptors TLR5 and TLR4 (27, 44), which minimizes the common innate responses of the host to flagella and flagellar sheath. Both of these evasion strategies could compensate in vivo for the necessity of maintaining flagella and motility, which appear to be very important during H. hepaticus infection. In addition to motility, cytolethal distending toxin was the only other factor that has been shown so far to play an important role in the persistent colonization of Swiss Webster or A/JCr mice by H. hepaticus (13, 15). Whether H. hepaticus motility and related functions are necessary only for initial colonization or also for long-term colonization could not be determined in this study. Motility and chemotaxis could very likely play dominant roles in enabling the bacteria to find their appropriate intestinal habitat, in particular during the initial steps of colonization.
Acknowledgments
We are grateful to Sophie Borchert for carefully reading the manuscript and helpful discussions and to Hilde de Reuse and Agnès Labigne for generously sharing the gm cassette. Hans-J. Hedrich, André Bleich, Martina Dorsch, and the staff members of the Central Animal Facility of Hannover Medical School are gratefully acknowledged for continuous support and discussions. We thank the Hannover Medical School Microarray Facility, in particular Oliver Dittrich-Breiholz, Heike Schneider, and Michael Kracht, for excellent technical support.
The German Research Council (SFB621) is acknowledged for funding to S.S. and the U.S. National Institute of Health for support to J.G.F. (grant R01 CA067529) and D.B.S. (grant PHS R01 DK052413).
Footnotes
Published ahead of print on 8 August 2008.
REFERENCES
- 1.Anderson, T. R., T. C. Montie, M. D. Murphy, and V. P. McCarthy. 1989. Pseudomonas aeruginosa flagellar antibodies in patients with cystic fibrosis. J. Clin. Microbiol. 272789-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrutis, K. A., J. G. Fox, D. B. Schauer, R. P. Marini, X. Li, L. Yan, C. Josenhans, and S. Suerbaum. 1997. Infection of the ferret stomach by isogenic flagellar mutant strains of Helicobacter mustelae. Infect. Immun. 651962-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahill, R. J., C. J. Foltz, J. G. Fox, C. A. Dangler, F. Powrie, and D. B. Schauer. 1997. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect. Immun. 653126-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrillo, C. D., E. Taboada, J. H. Nash, P. Lanthier, J. Kelly, P. C. Lau, R. Verhulp, O. Mykytczuk, J. Sy, W. A. Findlay, K. Amoako, S. Gomis, P. Willson, J. W. Austin, A. Potter, L. Babiuk, B. Allan, and C. M. Szymanski. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 27920327-20338. [DOI] [PubMed] [Google Scholar]
- 5.Contreras, M., J. M. Thiberge, M. A. Mandrand-Berthelot, and A. Labigne. 2003. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 49947-963. [DOI] [PubMed] [Google Scholar]
- 6.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50809-824. [DOI] [PubMed] [Google Scholar]
- 7.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 642445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eppinger, M., C. Baar, G. Raddatz, D. H. Huson, and S. C. Schuster. 2004. Comparative analysis of four Campylobacterales. Nat. Rev. Microbiol. 2872-885. [DOI] [PubMed] [Google Scholar]
- 9.Fox, J. G. 1998. Review article: Helicobacter species and in vivo models of gastrointestinal cancer. Aliment. Pharmacol. Ther. 12(Suppl. 1)37-60. [DOI] [PubMed] [Google Scholar]
- 10.Fox, J. G., F. E. Dewhirst, J. G. Tully, B. J. Paster, L. Yan, N. S. Taylor, M. J. J. Collins, P. L. Gorelick, and J. M. Ward. 1994. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J. Clin. Microbiol. 321238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, J. G., and A. Lee. 1997. The role of Helicobacter species in newly recognized gastrointestinal tract diseases of animals. Lab. Anim. Sci. 47222-255. [PubMed] [Google Scholar]
- 12.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 642246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge, Z., Y. Feng, M. T. Whary, P. R. Nambiar, S. Xu, V. Ng, N. S. Taylor, and J. G. Fox. 2005. Cytolethal distending toxin is essential for Helicobacter hepaticus colonization in outbred Swiss Webster mice. Infect. Immun. 733559-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge, Z., K. Hiratsuka, and D. E. Taylor. 1995. Nucleotide sequence and mutational analysis indicate that two Helicobacter pylori genes encode a P-type ATPase and a cation-binding protein associated with copper transport. Mol. Microbiol. 1597-106. [DOI] [PubMed] [Google Scholar]
- 15.Ge, Z., A. B. Rogers, Y. Feng, A. Lee, S. Xu, N. S. Taylor, and J. G. Fox. 2007. Bacterial cytolethal distending toxin promotes the development of dysplasia in a model of microbially induced hepatocarcinogenesis. Cell. Microbiol. 92070-2080. [DOI] [PubMed] [Google Scholar]
- 16.Geis, G., S. Suerbaum, B. Forsthoff, H. Leying, and W. Opferkuch. 1993. Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J. Med. Microbiol. 38371-377. [DOI] [PubMed] [Google Scholar]
- 17.Goon, S., C. P. Ewing, M. Lorenzo, D. Pattarini, G. Majam, and P. Guerry. 2006. A σ28-regulated nonflagella gene contributes to virulence of Campylobacter jejuni 81-176. Infect. Immun. 74769-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 1832937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Josenhans, C., R. L. Ferrero, A. Labigne, and S. Suerbaum. 1999. Cloning and allelic exchange mutagenesis of two flagellin genes from Helicobacter felis. Mol. Microbiol. 33350-362. [DOI] [PubMed] [Google Scholar]
- 20.Josenhans, C., A. Labigne, and S. Suerbaum. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 1773010-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josenhans, C., E. Niehus, S. Amersbach, A. Horster, C. Betz, B. Drescher, K. T. Hughes, and S. Suerbaum. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43307-322. [DOI] [PubMed] [Google Scholar]
- 22.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291605-614. [DOI] [PubMed] [Google Scholar]
- 23.Kamal, N., N. Dorrell, A. Jagannathan, S. M. Turner, C. Constantinidou, D. J. Studholme, G. Marsden, J. Hinds, K. G. Laing, B. W. Wren, and C. W. Penn. 2007. Deletion of a previously uncharacterized flagellar-hook-length control gene fliK modulates the σ54-dependent regulon in Campylobacter jejuni. Microbiology 1533099-3111. [DOI] [PubMed] [Google Scholar]
- 24.Kavermann, H., B. P. Burns, K. Angermuller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kullberg, M. C., J. F. Andersen, P. L. Gorelick, P. Caspar, S. Suerbaum, J. G. Fox, A. W. Cheever, D. Jankovic, and A. Sher. 2003. Induction of colitis by a CD4+ T cell clone specific for a bacterial epitope. Proc. Natl. Acad. Sci. USA 10015830-15835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kullberg, M. C., D. Jankovic, C. G. Feng, S. Hue, P. L. Gorelick, B. S. McKenzie, D. J. Cua, F. Powrie, A. W. Cheever, K. J. Maloy, and A. Sher. 2006. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J. Exp. Med. 2032485-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, S. K., A. Stack, E. Katzowitsch, S. I. Aizawa, S. Suerbaum, and C. Josenhans. 2003. Helicobacter pylori flagellins have very low intrinsic activity to stimulate human gastric epithelial cells via TLR5. Microbes Infect. 51345-1356. [DOI] [PubMed] [Google Scholar]
- 28.Lockman, H. A., and R. Curtiss. 1990. Salmonella typhimurium mutants lacking flagella or motility remain virulent in BALB/c mice. Infect. Immun. 58137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marani, P., S. Wagner, L. Baars, P. Genevaux, J. W. de Gier, I. Nilsson, R. Casadio, and G. von Heijne. 2006. New Escherichia coli outer membrane proteins identified through prediction and experimental verification. Protein Sci. 15884-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, P. M., J. Mathiot, J. Ipero, A. J. Georges, and M. C. Georges-Courbot. 1988. Antibody response to Campylobacter coli in children during intestinal infection and carriage. J. Clin. Microbiol. 261421-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattsson, A., M. Quiding-Jarbrink, H. Lonroth, A. Hamlet, I. Ahlstedt, and A. Svennerholm. 1998. Antibody-secreting cells in the stomachs of symptomatic and asymptomatic Helicobacter pylori-infected subjects. Infect. Immun. 662705-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNally, D. J., J. P. Hui, A. J. Aubry, K. K. Mui, P. Guerry, J. R. Brisson, S. M. Logan, and E. C. Soo. 2006. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J. Biol. Chem. 28118489-18498. [DOI] [PubMed] [Google Scholar]
- 33.Nachamkin, I., X. H. Yang, and N. J. Stern. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl. Environ. Microbiol. 591269-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52947-961. [DOI] [PubMed] [Google Scholar]
- 35.Niehus, E., F. Ye, S. Suerbaum, and C. Josenhans. 2002. Growth phase-dependent and differential transcriptional control of flagellar genes in Helicobacter pylori. Microbiology 1483827-3837. [DOI] [PubMed] [Google Scholar]
- 36.Ottemann, K. M., and A. C. Lowenthal. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 701984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavlovskis, O. R., D. M. Rollins, R. L. Harberberger, A. E. Green, L. Habash, S. Strocko, and R. I. Walker. 1991. Significance of flagella in colonization resistance of rabbits immunized with Campylobacter spp. Infect. Immun. 592259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poly, F., C. Ewing, S. Goon, T. E. Hickey, D. Rockabrand, G. Majam, L. Lee, J. Phan, N. J. Savarino, and P. Guerry. 2007. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect. Immun. 753859-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson, K. 1991. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect. Immun. 592727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schirm, M., E. C. Soo, A. J. Aubry, J. Austin, P. Thibault, and S. M. Logan. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 481579-1592. [DOI] [PubMed] [Google Scholar]
- 41.Shames, B., J. G. Fox, F. Dewhirst, L. Yan, Z. Shen, and N. S. Taylor. 1995. Identification of widespread Helicobacter hepaticus infection in feces in commercial mouse colonies by culture and PCR assay. J. Clin. Microbiol. 332968-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solnick, J. V., and D. B. Schauer. 2001. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 1459-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sterzenbach, T., S. K. Lee, B. Brenneke, F. von Goetz, D. B. Schauer, J. G. Fox, S. Suerbaum, and C. Josenhans. 2007. Inhibitory effect of enterohepatic Helicobacter hepaticus on innate immune responses of mouse intestinal epithelial cells. Infect. Immun. 752717-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suerbaum, S., T. Sterzenbach, and C. Josenhans. 2004. Comparative genome analysis of Helicobacter hepaticus, Helicobacter pylori and Campylobacter jejuni. Nova Acta Leopoldina NF 88. 333151-160. [Google Scholar]
- 46.Suerbaum, S., C. Josenhans, T. Sterzenbach, B. Drescher, P. Brandt, M. Bell, M. Droege, B. Fartmann, H.-P. Fischer, Z. Ge, A. Hörster, R. Holland, K. Klein, J. König, L. Macko, G. L. Mendz, G. Nyakatura, D. B. Schauer, Z. Shen, J. Weber, M. Frosch, and J. G. Fox. 2003. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 1007901-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol. 11233-238. [DOI] [PubMed] [Google Scholar]
- 48.Thibault, P., S. M. Logan, J. F. Kelly, J. R. Brisson, C. P. Ewing, T. J. Trust, and P. Guerry. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 27634862-34870. [DOI] [PubMed] [Google Scholar]
- 49.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wenman, W. M., J. Chai, T. J. Louie, C. Goudreau, H. Lior, D. G. Newell, A. D. Pearson, and D. E. Taylor. 1985. Antigenic analysis of Campylobacter flagellar protein and other proteins. J. Clin. Microbiol. 21108-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whary, M. T., and J. G. Fox. 2004. Natural and experimental Helicobacter infections. Comp. Med. 54128-158. [PubMed] [Google Scholar]
- 52.Ye, F., T. Brauer, E. Niehus, K. Drlica, C. Josenhans, and S. Suerbaum. 2007. Flagellar and global gene regulation in Helicobacter pylori modulated by changes in DNA supercoiling. Int. J. Med. Microbiol. 29765-81. [DOI] [PubMed] [Google Scholar]
- 53.Young, V. B., K. A. Knox, and D. B. Schauer. 2000. Cytolethal distending toxin sequence and activity in the enterohepatic pathogen Helicobacter hepaticus. Infect. Immun. 68184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]