Abstract
IS6110 restriction fragment length polymorphism (RFLP) genotyping is the most widely used genotyping method to study the epidemiology of Mycobacterium tuberculosis. However, due to the complexity of the IS6110 RFLP genotyping technique, and the interpretation of RFLP data, mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) genotyping has been proposed as the new genotyping standard. This study aimed to determine the discriminatory power of different MIRU-VNTR locus combinations relative to IS6110 RFLP genotyping, using a collection of Beijing genotype M. tuberculosis strains with a well-established phylogenetic history. Clustering, diversity index, clustering concordance, concordance among unique genotypes, and divergent and convergent evolution were calculated for seven combinations of 27 different MIRU-VNTR loci and compared to IS6110 RFLP results. Our results confirmed previous findings that MIRU-VNTR genotyping can be used to estimate the extent of recent or ongoing transmission. However, molecular epidemiological linking of cases varied significantly depending on the genotyping method used. We conclude that IS6110 RFLP and MIRU-VNTR loci evolve independently and at different rates, which leads to discordance between transmission chains predicted by the respective genotyping methods. Concordance between the two genotyping methods could be improved by the inclusion of genetic distance (GD) into the clustering formulae for some of the MIRU-VNTR loci combinations. In summary, our findings differ from previous reports, which may be explained by the fact that in settings of low tuberculosis incidence, the genetic distance between epidemiologically unrelated isolates was sufficient to define a strain using either marker, whereas in settings of high incidence, continuous evolution and persistence of strains revealed the weaknesses inherent to these markers.
Over the past 2 decades, molecular genotyping methods have enhanced our understanding of the epidemiology of tuberculosis (TB) in numerous geographical settings. These methods have enabled geo-temporal tracking of Mycobacterium tuberculosis strains with the view to identifying source cases responsible for TB outbreaks (3), tracking of recent and ongoing disease transmission (31), distinguishing between reinfection and relapse (28), evaluating the effectiveness of direct observed therapy short-course-based TB control programs (5, 16), and identifying global genetic lineages (7). Ideally, molecular genotyping tools should be inexpensive, highly discriminative, deliver rapid results, be straightforward to perform, and produce easily interpretable results that allow for accurate interlaboratory comparisons (universally comparable databases).
Three genotyping methods are currently widely used in molecular epidemiological studies of TB: IS6110 restriction fragment length polymorphism (RFLP) genotyping (27), spoligotyping (14), and mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) genotyping (21, 22). Currently, IS6110 RFLP genotyping is the most widely used genotyping method (27). However, this method is time-consuming, laborious, and complex. Furthermore, differences in application can make interlaboratory comparisons difficult, and the data generated may have limitations (i.e., comparison of strains with high versus low IS6110 copy numbers). More recently, the validity of the calculation of IS6110 RFLP clustering, as a surrogate for transmission, has been questioned, as the IS6110 banding pattern may change during transmission (33, 35). A nearest genetic distance model has been evaluated to incorporate IS6110 banding changes into the calculation of ongoing transmission (24). The term “cluster” has also been questioned in studies which have compared contact tracing data with IS6110 RFLP data (4, 26). In response, numerous studies have been conducted to try to identify alternative methods that have the ability to accurately describe epidemiological events in different settings at a similar discriminatory level to that of IS6110 RFLP genotyping. One of the most promising methods is MIRU-VNTR genotyping, a PCR-based method for detecting the number of tandem repeats at a given genetic locus. Supply et al. (21) defined a set of 15 MIRU-VNTR loci for molecular epidemiological investigations and a set of 24 MIRU-VNTR loci for phylogenetic analysis of M. tuberculosis strains worldwide. In support of this, another study concluded that this “real-time” MIRU-VNTR genotyping approach was highly applicable for population-based studies (18). This view was reinforced by a study conducted in the Brussels region, where the authors concluded that a standardized MIRU-VNTR genotyping method could be a new reference for epidemiological and phylogenetic screening of M. tuberculosis strains (2).
A study from Japan (10) investigated the differentiation power of the proposed 15- and 24-loci MIRU-VNTR genotyping methods for strains with the Beijing genotype and concluded that the analyses of these loci were of limited use for discriminating strains of this genotype. In their study they showed that VNTR loci 3820, 3232, and 4120 were highly polymorphic in Beijing genotype strains and thus proposed the use of these loci to enhance the discriminatory power of the proposed 15-MIRU-VNTR genotyping method. However, other studies have excluded these loci due to difficulties associated with the reproducibility of PCR amplification (15, 21, 36).
Subsequently, a study in Hong Kong, which also examined strains of the Beijing genotype, showed that a different combination of 12 VNTR and QUB (Queen's University of Belfast) loci gave a Hunter-Gaston discriminatory index value which was almost equal to that obtained in IS6110 RFLP genotyping (12, 13). However, this was refuted by a more recent study from China which suggested that MIRU-VNTR genotyping may overestimate transmission in isolates with the Beijing genotype (11). Collectively, these findings suggest that the selection of MIRU-VNTR loci for optimal differentiation of M. tuberculosis requires further validation in different geographical settings. To date, the performance of the MIRU-VNTR genotyping method has not been evaluated in an epidemic setting, nor has it been tested within the context of a robust M. tuberculosis phylogeny.
In this study the discriminatory power of different MIRU-VNTR locus combinations was determined as previously described (8, 10, 21, 22) and compared to the IS6110 RFLP genotyping method by using a collection of Beijing genotype M. tuberculosis strains with a well-established phylogenetic history (9). The results are discussed in the context of concordance between the different genotyping methods in their abilities to define a strain and to accurately describe the epidemiology of TB in a high-incidence setting.
MATERIALS AND METHODS
Study population.
Sputum samples were collected during the period from January 1993 to December 2004 from new and retreatment TB patients who were resident and attending health care clinics in an epidemiological field site in Cape Town, South Africa (31). This study is part of a larger, long-term molecular epidemiological project which has been approved by the ethics committee of Stellenbosch University.
IS6110 RFLP genotyping.
M. tuberculosis isolates were cultured on MGIT (Becton Dickinson) or Löwenstein-Jensen medium, and DNA was extracted as previously described (32). Each isolate was classified by IS6110 RFLP genotyping (27) and spoligotyping (14) using internationally standardized protocols. IS6110 RFLP patterns were analyzed using Gelcompar II (Applied-Maths, Sint-Martens-latem, Belgium) with tolerance settings allowing a 5% shift in lane position and a 0.6% variation in individual band position to compensate for minor technical errors. Isolates were assigned as members of the Beijing genotype if they had the characteristic Beijing spoligotype (30). Only the first M. tuberculosis isolate from each case was included for subsequent analysis. Each Beijing isolate was grouped into one of seven phylogenetic sublineages according to 40 different genetic markers, as previously described (9).
DNA sequencing.
The DNA sequence of the katG, rpoB, embB, and rrs genes of isolates classified as members of the Beijing sublineage 5 were determined as previously described (19, 25).
MIRU-VNTR typing.
Twenty-seven MIRU-VNTR loci were amplified by PCR as described previously (8, 10, 21, 22). The number of repeats at each genomic locus was calculated according to the electrophoretic mobility of the corresponding PCR product (23). Alleles were assigned numerical values according to the number of repeats present in that genomic locus. Isolates were genotypically classified according to seven different MIRU-VNTR locus combinations (Table 1).
TABLE 1.
MIRU-VNTR locus combinations
| Locus | Locus included in MIRU-VNTR combinationa
|
||||||
|---|---|---|---|---|---|---|---|
| 12-MIRUb | 12-MIRU + ETR A, B, Cc | 12-MIRU + hypervariable locid | 15-MIRU-VNTRe | 15-MIRU-VNTR + hypervariable locif | 24-MIRU-VNTRe | 24-MIRU-VNTR + hypervariable locid | |
| MIRU02 | • | • | • | • | • | ||
| MIRU04 | • | • | • | • | • | • | • |
| MIRU10 | • | • | • | • | • | • | • |
| MIRU16 | • | • | • | • | • | • | • |
| MIRU20 | • | • | • | • | • | ||
| MIRU23 | • | • | • | • | • | ||
| MIRU24 | • | • | • | • | • | ||
| MIRU26 | • | • | • | • | • | • | • |
| MIRU27 | • | • | • | • | • | ||
| MIRU31 | • | • | • | • | • | • | • |
| MIRU39 | • | • | • | • | • | ||
| MIRU40 | • | • | • | • | • | • | • |
| VNTR1955 | • | • | • | • | |||
| VNTR2165/ETR-A | • | • | • | • | • | ||
| QUB11b | • | • | • | • | |||
| QUB26b | • | • | • | • | |||
| VNTR0424 | • | • | • | • | |||
| VNTR2401 | • | • | • | • | |||
| VNTR4156 | • | • | • | • | |||
| VNTR3690 | • | • | • | • | |||
| ETR-C | • | • | • | • | • | ||
| VNTR2347 | • | • | |||||
| ETR-B | • | • | • | ||||
| Mtub 34 | • | • | |||||
| QUB3232 | • | • | • | ||||
| VNTR3820 | • | • | • | ||||
| VNTR4120 | • | • | • | ||||
Analytical calculations. (i) Estimation of clustering.
A cluster (representing either recent or ongoing transmission or a <2-year interval) was defined as a series of isolates having the same genotype (IS6110 RFLP or MIRU-VNTR), while isolates with unique IS6110 RFLP or MIRU-VNTR genotypes were considered to represent reactivation or influx of disease into the study community (20). Secondary analyses which incorporated the concept of evolution during transmission were done using data sets (genotypes according to IS6110 RFLP or a particular MIRU-VNTR locus combination) in which isolates separated by a single evolutionary event were combined into transmission chains with a genetic distance of 1 (24).
(ii) Estimation of genetic diversity.
The genetic diversity for each individual MIRU-VNTR locus, each of the seven MIRU-VNTR locus combinations (Table 1), and the IS6110 RFLP fingerprints was calculated as h=1−∑xi2[n/(n−1)], where xi is the frequency of the ith allele at the locus, n is the number of isolates in the sample, and the term n/(n − 1) is a correction for bias in small samples (17).
(iii) Estimation of matching and mismatching concordance.
Concordance between the IS6110 RFLP genotypes and the respective MIRU-VNTR genotypes was calculated as follows: each isolate was paired with every other isolate in the data set, and their genotypes (IS6110 RFLP and MIRU-VNTR) were scored as either a match (identical) or mismatch (nonidentical). Matching concordance between the respective genotyping methods was calculated according to the number of paired isolates having a match for both of the methods as a proportion of the total number of pairs having matching IS6110 RFLP genotypes. This is a measure of agreement between two methods as to whether any two isolates form part of the same transmission chain. Mismatching concordance was calculated as the number of paired isolates having nonmatching genotypes for both of the methods as a proportion of the total number of pairs having nonmatching IS6110 RFLP genotypes. This is a measure of agreement between two methods for any two isolates that do not form part of the same transmission chain.
(iv) Estimation of concordance among unique genotypes.
Concordance between uniquely occurring IS6110 RFLP genotypes and the MIRU-VNTR genotypes was calculated as the proportion of isolates having unique IS6110 RFLP genotypes that also had unique MIRU-VNTR genotypes.
(v) Estimation of number of convergent events.
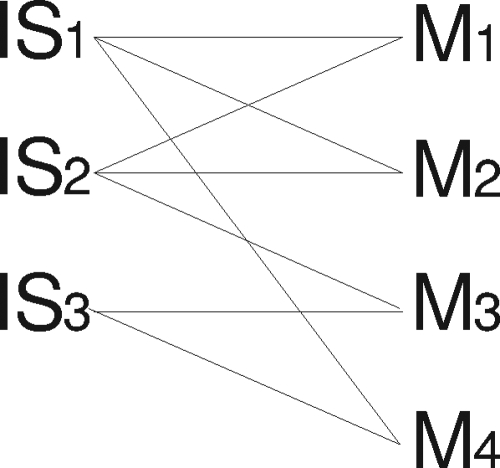
Convergent evolution was identified by drawing connecting lines between each IS6110 RFLP genotype and each MIRU-VNTR genotype for which isolates were found to have that particular genotype combination (Fig. 1). Convergent evolution was defined, conservatively, as the existence of isolates representing each of the four possible combinations of two IS6110 RFLP genotypes (e.g., IS1 and IS2) and two MIRU-VNTR genotypes (e.g., M1 and M2) (Fig. 1). This scenario would only be possible if one of the MIRU-VNTR genotypes had evolved more than once, assuming that the chance of IS6110 RFLP genotype convergence was significantly lower than that of MIRU-VNTR genotype convergence. The validity of this method was confirmed by plotting the IS6110 RFLP genotypes onto a phylogenetic tree constructed using the MIRU-VNTR data in combination with a neighbor-joining algorithm (data not shown) (34).
FIG. 1.
An example of MIRU-VNTR (Mx) and IS6110 RFLP (ISx) genotypes. The connecting lines represent the MIRU-VNTR and IS6110 RFLP genotype combinations observed in M. tuberculosis isolates in the study setting. M1 and M2 are both linked to IS1 and IS2 and therefore represent a convergent event. Neither M3 nor M4 share common connections to more than one ISx with any other Mx. Their connecting lines therefore indicate simple, linear evolution.
(vi) Estimation of number of divergent events.
A divergent evolutionary event was scored for each MIRU-VNTR genotype which existed in combination with only one IS6110 RFLP genotype and where this IS6110 RFLP genotype was found in combination with more than one MIRU-VNTR genotype (Fig. 2). This implies that the MIRU-VNTR genotype arose subsequent to the IS6110 RFLP genotype. A divergent event was also added for each convergent event, since a convergent event implies a prior divergent event.
FIG. 2.
IS6110 RFLP banding patterns of Beijing sublineage 5 isolates sharing identical katG, rpoB, embB, and rrs gene mutations. The isolate numbers are indicated in bold, while the IS6110-RFLP cluster numbers are indicated in standard text (these numbers correspond to the numbers given in data set S2 of the supplemental material).
(vii) Sensitivity and specificity calculations.
The sensitivity and specificity (and positive and negative predictive values) of the IS6110 RFLP and respective MIRU-VNTR genotyping methods were calculated using GraphPad Prism 5 software (La Jolla, CA) based on their ability to correctly identify an independently genotyped drug-resistant cluster.
RESULTS
IS6110 RFLP genotyping identified 74 different strains among the 321 isolates with the Beijing spoligotype collected over a 12-year period (Table 2). Of these strains, 272 were grouped into 25 clusters (containing between 2 and 100 isolates), and 49 were unique strains. The overall percent clustering was calculated to be 84.7% using the n/T formula (1). Each isolate was subsequently genotyped with 27 MIRU-VNTR loci and analyzed according to seven different MIRU-VNTR locus combinations (Table 1; see also data sets in the supplemental material). The performance of these locus combinations, in relation to the IS6110 RFLP genotyping method, was determined either over a 12-year period (Table 2) or over six consecutive 2-year periods (Table 3). In both analyses the traditional 12-MIRU loci genotyping method underestimated the number of genotypes (strains) identified and thereby overestimated the percentage of clustering (Tables 2 and 3). The inclusion of exact tandem repeat (ETR) alleles A, B, and C to the 12-MIRU loci set did not significantly improve the number of strains detected or the estimate of clustering (Tables 2 and 3). Analysis of the isolates using the newly proposed 15- and 24-MIRU-VNTR locus combinations increased the number of strains identified; however, the discriminatory power of these locus combinations remained lower than that observed using IS6110 RFLP genotyping (Tables 2 and 3). Consequently, these locus combinations overestimated clustering. The addition of the VNTR loci 3232, 4120, and 3820 to the 12-, 15-, and 24-MIRU-VNTR locus combinations increased the number of strains detected and thereby produced clustering estimates similar to or slightly lower than that of IS6110 RFLP genotyping (Tables 2 and 3). This implies that some MIRU-VNTR locus combinations could be selected as epidemiological markers to estimate the extent of both recent (<2-year interval) and ongoing (unrestricted interval) transmission in settings with a high incidence of strains with the Beijing genotype.
TABLE 2.
Comparison between molecular epidemiological data generated over a 12-year interval using IS6110 RFLP and MIRU-VNTR genotyping methods
| Method | Genetic distance of 0
|
Genetic distance of 0
|
Genetic distance of 1
|
% Beijing sublineage discrimination
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of genotypes | % Clustering | Unique IS6110 genotype (n = 49) with:
|
Clustered IS6110 genotype (n = 272) with:
|
% Pair-wise matching concordance | % Pair-wise mismatching concordance | % Concordance between unique strains | No. of converged genotypes | No. of diverged genotypes | Diversity indexf | No. of genotypes | % Pair-wise matching concordance | % Pair-wise mismatching concordance | % Concordance between unique strains | |||||
| Unique MIRU-VNTR genotype (n) | Clustered MIRU-VNTR genotype (n) | Unique MIRU-VNTR genotype (n) | Clustered MIRU-VNTR genotype (n) | GD = 0 | GD = 1 | |||||||||||||
| IS6110 RFLP | 74 | 84.7 | NAg | NA | NA | NA | 100 | NA | 100 | NA | NA | 0.85 | 40 | 100 | NA | 100 | 100 | 99 |
| MIRU-VNTR locus combinations | ||||||||||||||||||
| 12-MIRUa | 27 | 95.0 | 6 | 43 | 10 | 262 | 68 | 69 | 20.4 | 3 | 14 | 0.63 | 3 | 99 | 2 | 0 | 96 | 1 |
| 12-MIRU + ETR A, B, Cb | 39 | 91.2 | 8 | 41 | 18 | 254 | 60 | 72 | 22.4 | 5 | 26 | 0.67 | 4 | 99 | 3 | 4 | 96 | 1 |
| 12-MIRU + hypervariable locic | 67 | 84.1 | 14 | 35 | 38 | 234 | 53 | 71 | 28.6 | 5 | 47 | 0.7 | 11 | 96 | 8 | 4 | 99 | 3 |
| 15-MIRU-VNTRd | 47 | 89.4 | 12 | 37 | 22 | 250 | 67 | 68 | 34.7 | 3 | 27 | 0.63 | 11 | 99 | 80 | 15 | 99 | 99 |
| 15-MIRU-VNTR + hypervariable locie | 83 | 78.8 | 19 | 30 | 49 | 223 | 51 | 71 | 40.8 | 5 | 58 | 0.7 | 20 | 96 | 82 | 30 | 99 | 99 |
| 24-MIRU-VNTRd | 57 | 87.2 | 12 | 37 | 28 | 244 | 52 | 73 | 40.8 | 7 | 38 | 0.72 | 15 | 99 | 83 | 22 | 99 | 99 |
| 24-MIRU-VNTR + hypervariable locic | 91 | 77.3 | 19 | 30 | 54 | 218 | 39 | 81 | 42.9 | 9 | 68 | 0.78 | 27 | 96 | 88 | 41 | 99 | 99 |
TABLE 3.
Comparison between molecular epidemiological data generated over six consecutive 2-year intervals by IS6110 RFLP and MIRU-VNTR genotyping methods
| Method | Avg value (range)
|
|||||
|---|---|---|---|---|---|---|
| No. of strains | No. of clusters | % Clustering | % Pair-wise matching concordance | % Pair-wise mismatching concordance | % Concordance between unique strains | |
| IS6110 RFLP | 17.9 (7-20) | 6.1 (4-10) | 74.5 (55.4-86.7) | 100 | NAf | 100 |
| MIRU-VNTR locus combinations | ||||||
| 12-MIRUa | 9.9 (5-14) | 4.6 (2-7) | 85.1 (60.0-93.1) | 69.7 (60-80) | 92.8 (72-99) | 19.1 (6.1-60.0) |
| 12-MIRU + ETR A, B, Cb | 10.9 (7-14) | 4.9 (3-7) | 83.1 (60.0-91.7) | 63.7 (44-72) | 95.0 (84-99) | 25.7 (15.6-60.0) |
| 12-MIRU + hypervariable locic | 15.4 (8-22) | 4.0 (2-6) | 73.1 (60.0-77.8) | 52.7 (40-58) | 94.0 (77-99) | 42.2 (27.0-66.7) |
| 15-MIRU-VNTRd | 12.0 (5-17) | 3.7 (2-5) | 80.3 (66.7-93.1) | 68.2 (45-78) | 92.3 (70-99) | 31.3 (16.7-60.0) |
| 15-MIRU-VNTR + hypervariable locie | 16.6 (7-23) | 3.6 (2-6) | 70.7 (57.6-76.4) | 51.0 (22-63) | 94.0 (76-100) | 50.9 (32.0-66.7) |
| 24-MIRU-VNTRd | 13.6 (7-19) | 4.6 (3-6) | 77.8 (60.0-89.7) | 55.0 (44-69) | 95.2 (84-99) | 35.4 (24.0-60.0) |
| 24-MIRU-VNTR + hypervariable locic | 18.1 (9-25) | 4.3 (3-7) | 67.7 (54.5-75.0) | 40.2 (22-51) | 95.0 (84-99) | 51.3 (32.0-66.7) |
To determine whether a correlation existed between the definitions of a strain according to IS6110 RFLP or MIRU-VNTR genotyping methods, the respective genotypes were compared. From the results shown in Table 2 it is evident that a strain classified as a cluster according to IS6110 RFLP genotyping may in some instances be classified as unique according to the different MIRU-VNTR locus combinations, or vice versa. Using a pair-wise analysis, we estimated the degree of matching concordance between the IS6110 RFLP and MIRU-VNTR genotyping methods to range between 39% and 68% depending on the locus combinations used (Tables 2 and 3). The inclusion of additional MIRU-VNTR loci decreased the degree of matching concordance, as a result of an increased rate of divergence caused by more rapid evolution, with the hypervariable loci having the greatest effect. Conversely, the inclusion of additional loci increased the degree of mismatching concordance, as well as concordance between strains identified as having unique genotypes according to both genotyping methods (IS6110 RFLP and MIRU-VNTR). A consequence of more rapid evolution was the increased risk of convergent evolutionary events (Table 2).
To determine whether concordance between the respective genotyping methods could be improved, the analysis was repeated to allow for a genetic distance of 1, i.e., evolution of single MIRU-VNTR loci or single-band changes in the IS6110 pattern within the definition of a cluster. The results showed that the inclusion of genetic distance had a significant influence on the MIRU-VNTR definition of a cluster, collapsing many of the genotypes (Table 2; see also data set S1 in the supplemental material). This was less pronounced for IS6110 RFLP analysis (Table 2; see also data set S1 in the supplemental material). Matching concordance was improved by allowing for evolution of the MIRU-VNTR genotypes; however, mismatching concordance was concomitantly reduced for genotypes based on the 12-MIRU loci combinations. This may be explained by the loss of discriminatory power as a result of the collapsing of genotypes, which is associated with a low rate of evolution. In contrast, mismatching concordance was improved for 15- and 24-MIRU-VNTR combinations due to the higher evolutionary rates of these markers. However, the concordance among unique genotypes remained low (Table 2).
To establish which of the genotyping methods provided the most accurate description of ongoing transmission in the study setting, the largest group of drug-resistant isolates (found within sublineage 5) was selected, based on identical mutations conferring resistance to isoniazid, rifampin, ethambutol, and streptomycin (see data set S2 in the supplemental material). These isolates represent the continuing spread of a previously described multidrug-resistant TB outbreak (29). A total of 35 isolates were identified with the katG315 AGC to ACC, rpoB531 TCG to TTG, embB306 ATG to ATA, and rrs513 CAG to CCG mutations, forming a single drug resistance-based cluster (Fig. 2). The sensitivity, specificity, and positive and negative predictive values related to the abilities of the different markers to identify the drug-resistant cluster are given in Table 4. While the sensitivities of all the markers were high, with some of those based on MIRU-VNTR loci outperforming IS6110 RFLP, the specificity of all MIRU-VNTR markers was substantially lower than that of IS6110 RFLP. The inclusion of genetic distance (single events) within the definition of a cluster appeared to improve the sensitivity of most of the markers but concomitantly decreased the specificity of the MIRU-VNTR markers. The specificity of IS6110 was not affected by the inclusion of genetic distance. Positive predictive values were not significantly affected by allowing for evolution of the markers; however, with the exceptions of IS6110, which increased, and the 24 MIRU and three hypervariable loci, which remained unchanged, the negative predictive values for all markers were reduced to zero.
TABLE 4.
Sensitivity, specificity, PPV, and NPV values for IS6110 RFLP and MIRU-VNTR genotyping methods based on correct identification of an independently genotyped drug-resistant cluster characterized by unique mutations in the katG, rpoB, embB, and rrs genes
| Method | Value (95% CI) when GD = 0f
|
Value (95% CI) when= 1f
|
||||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | Sensitivity | Specificity | PPV | NPV | |
| IS6110 RFLP | 0.83 (0.66-0.93) | 1.00 (0.48-1.00) | 1.00 (0.88-1.00) | 0.45 (0.17-0.77) | 0.94 (0.81-0.99) | 1.00 (0.48-1.00) | 1.00 (0.89-1.00) | 0.71 (0.29-0.96) |
| MIRU-VNTR locus combinations | ||||||||
| 12-MIRUa | 1.00 (0.90-1.00) | 0.20 (0.01-0.72) | 0.90 (0.76-0.97) | 1.00 (0.03-1.00) | 1.00 (0.72-1.00) | 0.00 (0.00-0.54) | 0.88 (0.72-0.95) | ND |
| 12-MIRU + ETR A, B, Cb | 0.97 (0.85-1.00) | 0.20 (0.01-0.72) | 0.89 (0.75-0.97) | 0.50 (0.01-0.99) | 1.00 (0.72-1.00) | 0.00 (0.00-0.54) | 0.88 (0.72-0.95) | ND |
| 12-MIRU + hypervariable locic | 0.80 (0.63-0.92) | 0.20 (0.01-0.72) | 0.88 (0.71-0.96) | 0.13 (0.00-0.53) | 0.97 (0.85-1.00) | 0.00 (0.00-0.52) | 0.87 (0.73-0.96) | 0.00 (0.00-0.98) |
| 15-MIRU-VNTRd | 0.91 (0.77-0.98) | 0.40 (0.05-0.85) | 0.91 (0.77-0.98) | 0.40 (0.05-0.85) | 1.00 (0.72-1.00) | 0.00 (0.00-0.537) | 0.88 (0.72-0.95) | ND |
| 15-MIRU-VNTR + hypervariable locie | 0.74 (0.57-0.88) | 0.40 (0.05-0.85) | 0.90 (0.73-0.98) | 0.18 (0.02-0.52) | 0.94 (0.81-0.99) | 0.00 (0.00-0.52) | 0.87 (0.72-0.96) | 0.00 (0.00-0.84) |
| 24-MIRU-VNTRd | 0.91 (0.77-0.98) | 0.40 (0.05-0.85) | 0.91 (0.77-0.98) | 0.40 (0.05-0.85) | 1.00 (0.72-1.00) | 0.00 (0.00-0.54) | 0.88 (0.72-0.95) | ND |
| 24-MIRU-VNTR + hypervariable locic | 0.74 (0.57-0.88) | 0.40 (0.05-0.85) | 0.90 (0.73-0.98) | 0.18 (0.02-0.52) | 0.74 (0.57-0.88) | 0.40 (0.05-0.85) | 0.90 (0.73-0.98) | 0.18 (0.02-0.52) |
To determine whether MIRU-VNTR genotyping could be used as a method to phylogenetically group strains with the Beijing genotype, the correlation between MIRU-VNTR genotype and Beijing sublineage was quantified. As sublineages 3 and 4 and sublineages 5 and 6 were distinguished solely on the basis of IS6110 in our data set, these two pairs of sublineages were combined for the purposes of this analysis. Table 2 shows that the respective MIRU-VNTR locus combinations correctly grouped >96% of the isolates according to their sublineage designation, in comparison to 100% with IS6110 RFLP genotyping. The incorporation of genetic distance reduced the ability of genotyping methods based on the 12-MIRU locus combinations to correctly group isolates (Table 2).
DISCUSSION
IS6110 RFLP genotyping is the most widely used genotyping method for investigating and understanding the epidemiology of M. tuberculosis (27). However, studies comparing IS6110 RFLP molecular epidemiological and contact tracing data have questioned the validity of the definition of transmission (4, 26). In order to address these concerns MIRU-VNTR genotyping using either 15- or 24-MIRU-VNTR loci combinations have been extensively evaluated as the new genotyping standard for molecular epidemiological studies of M. tuberculosis (21). Concordance between MIRU-VNTR genotyping and contact tracing data was found to be superior to that of IS6110 RFLP in settings of low incidence (2, 18). However, these MIRU-VNTR locus combinations have not been fully tested in geographical regions of TB endemicity or within a robust M. tuberculosis phylogeny. Our results confirm previous findings (2, 10, 18, 21) which suggested that MIRU-VNTR genotyping, using carefully selected locus combinations, could be used to estimate the extent of recent or ongoing transmission. The inclusion of the three hypervariable loci improved the discriminatory power of the MIRU-VNTR genotyping method in this Beijing lineage, thereby supporting a previous suggestion for their inclusion (10). However, the use of these loci needs further evaluation in other evolutionary lineages, as difficulties associated with amplification reproducibility have been reported (15, 21, 36).
We conclude that the PCR-based MIRU-VNTR genotyping method could be applied as an epidemiological tool to measure the performance of a TB control program over time in a defined geographical setting. However, the observed concordance in the estimate of recent and ongoing transmission when using the IS6110 RFLP or MIRU-VNTR genotyping methods was only coincidental. A subsequent analysis of the MIRU-VNTR data, in comparison to the IS6110 RFLP genotyping data, revealed that the classification of a strain according to its genotype differed significantly depending on the genotyping method used. Accordingly, our study showed that the degree of matching and mismatching concordance as well as concordance among unique strains was low. This led to discordance between the transmission chains predicted by the respective genotyping methods. Matching concordance increased when genetic distance was incorporated into the clustering calculation for all of the MIRU-VNTR combinations. However, this effect was offset in the case of 12-MIRU-based markers by the concomitant reduction in mismatching concordance, which was not the case for the 15- and 24-MIRU-VNTR combinations. From this, it is apparent that the additional loci included in the 15- and 24-MIRU-VNTR combinations (with or without the addition of the hypervariable loci) improved the overall concordance of MIRU-VNTR with respect to IS6110 RFLP. This may be due to these loci being inherently less stable and therefore more informative. However, a caveat to the inclusion of genetic distance in the clustering formula is that epidemiologically unrelated cases may be incorrectly linked within a transmission chain.
Our analysis of the drug-resistant cluster to elucidate which of the genotyping methods provided the most accurate reflection of the epidemiology highlighted shortcomings of both the IS6110 RFLP and MIRU-VNTR genotyping methods. This analysis supported a previous study which demonstrated that ongoing transmission was characterized by the evolution of variant IS6110 RFLP genotypes while simultaneously preserving existing genotypes (33). A similar observation was found when using the different MIRU-VNTR locus combinations. This could be explained by the fact that the evolution of different loci could take place both convergently and divergently. Together, these results substantiate previous findings which have suggested that the definition of ongoing transmission according to IS6110 RFLP or MIRU-VNTR genotyping should include closely related genotypes (18, 24, 35). However, when allowing for single MIRU-VNTR changes within the definition of a cluster, the MIRU-VNTR genotyping method collapsed many of the sublineage 5 isolates into a limited number of clusters. As a result, most of the isolates were grouped as resistant, giving the method a high sensitivity, but in doing so, compromising specificity. In contrast, the identification of isolates within the drug-resistant cluster was largely retained by IS6110 RFLP analysis despite the inclusion of genetic distance. This suggests that IS6110 RFLP analysis in combination with genetic distance provides a more accurate reflection of ongoing transmission of this multidrug-resistant TB outbreak in this setting. This finding is important for the interpretation of molecular epidemiological data in settings where contact tracing is extremely difficult. However, we acknowledge that the concordance between IS6110 RFLP findings and transmission needs further investigation in different settings and in M. tuberculosis strains with different genetic backgrounds.
Our results differ from previous studies (2, 18), which demonstrated a close correlation between IS6110 RFLP and MIRU-VNTR genotyping. These studies were conducted in settings in western Europe with a low incidence of TB and where the TB epidemic is primarily driven by reactivation and immigration (6). In these settings, efficient TB control programs would largely prevent recent and ongoing transmission and the subsequent generation of closely related clonal variants. Thus, genetic diversity is predicted to be preserved. In most instances, this would imply that the strains cultured from TB cases would be genetically distantly related and thus would not share either IS6110 RFLP banding patterns or MIRU-VNTR genotypes. Accordingly, MIRU-VNTR genotyping would discriminate strains at a level similar to that of IS6110 RFLP genotyping. In contrast, our setting of high TB incidence has promoted the evolution of a large number of genetically closely related strains which are maintained within the host population. The genetic distance between these strains is often of such a nature that strains either have identical IS6110 RFLP genotypes and variant MIRU-VNTR genotypes or vice versa. Accordingly, we hypothesize that the degree of discordance between IS6110 RFLP and MIRU-VNTR genotyping is dependent on the genetic distance between isolates. This is supported by the observation that distantly related isolates from the different Beijing sublineages have evolved distinct IS6110 RFLP and MIRU-VNTR genotypes.
In summary, we conclude that both IS6110 RFLP and MIRU-VNTR genotyping methods have limitations in defining chains of transmission of Beijing genotype M. tuberculosis strains in this setting of high incidence.
Supplementary Material
Acknowledgments
The Harry Crossley Foundation and the European Commission 6th Framework Program on Research Technological Development Demonstration (project no. 037919) are thanked for financially supporting this project.
Footnotes
Published ahead of print on 20 August 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 3301710-1716. [DOI] [PubMed] [Google Scholar]
- 2.Allix-Beguec, C., M. Fauville-Dufaux, and P. Supply. 2008. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 461398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. B. Plikaytis, V. Kapur, K. Stockbauer, X. Pan, M. L. Lutfey, S. L. Moghazeh, W. Eisner, T. M. Daniel, M. H. Kaplan, J. T. Crawford, J. M. Musser, and B. N. Kreiswirth. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275452-457. [PubMed] [Google Scholar]
- 4.Cowan, L. S., L. Diem, T. Monson, P. Wand, D. Temporado, T. V. Oemig, and J. T. Crawford. 2005. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J. Clin. Microbiol. 43688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz-Ferro, E., and E. Fernandez-Nogueira. 2007. Epidemiology of tuberculosis in Galicia, Spain, 1996-2005. Int. J. Tuberc. Lung Dis. 111073-1079. [PubMed] [Google Scholar]
- 6.Dahle, U. R., V. Eldholm, B. A. Winje, T. Mannsaker, and E. Heldal. 2007. Impact of immigration on the molecular epidemiology of Mycobacterium tuberculosis in a low-incidence country. Am. J. Respir. Crit. Care Med. 176930-935. [DOI] [PubMed] [Google Scholar]
- 7.Gagneux, S., K. DeRiemer, T. Van, M. Kato-Maeda, B. C. de Jong, S. Narayanan, M. Nicol, S. Niemann, K. Kremer, M. C. Gutierrez, M. Hilty, P. C. Hopewell, and P. M. Small. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1032869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson, A., T. Brown, L. Baker, and F. Drobniewski. 2005. Can 15-locus mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis provide insight into the evolution of Mycobacterium tuberculosis? Appl. Environ. Microbiol. 718207-8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanekom, M., G. D. van der Spuy, E. Streicher, S. L. Ndabambi, C. R. McEvoy, M. Kidd, N. Beyers, T. C. Victor, P. D. van Helden, and R. M. Warren. 2007. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J. Clin. Microbiol. 451483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwamoto, T., S. Yoshida, K. Suzuki, M. Tomita, R. Fujiyama, N. Tanaka, Y. Kawakami, and M. Ito. 2007. Hypervariable loci that enhance the discriminatory ability of newly proposed 15-loci and 24-loci variable-number tandem repeat typing method on Mycobacterium tuberculosis strains predominated by the Beijing family. FEMS Microbiol. Lett. 27067-74. [DOI] [PubMed] [Google Scholar]
- 11.Jiao, W. W., I. Mokrousov, G. Z. Sun, Y. J. Guo, A. Vyazovaya, O. Narvskaya, and A. D. Shen. 2008. Evaluation of new variable-number tandem-repeat systems for typing Mycobacterium tuberculosis with Beijing genotype isolates from Beijing, China. J. Clin. Microbiol. 461045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kam, K. M., C. W. Yip, L. W. Tse, K. L. Leung, K. L. Wong, W. M. Ko, and W. S. Wong. 2006. Optimization of variable number tandem repeat typing set for differentiating Mycobacterium tuberculosis strains in the Beijing family. FEMS Microbiol. Lett. 256258-265. [DOI] [PubMed] [Google Scholar]
- 13.Kam, K. M., C. W. Yip, L. W. Tse, K. L. Wong, T. K. Lam, K. Kremer, B. K. Au, and D. van Soolingen. 2005. Utility of mycobacterial interspersed repetitive unit typing for differentiating multidrug-resistant Mycobacterium tuberculosis isolates of the Beijing family. J. Clin. Microbiol. 43306-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. Van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kremer, K., C. Arnold, A. Cataldi, M. C. Gutierrez, W. H. Haas, S. Panaiotov, R. A. Skuce, P. Supply, A. G. van der Zanden, and D. van Soolingen. 2005. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J. Clin. Microbiol. 435628-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Calleja, A. I., M. A. Lezcano, M. A. Vitoria, M. J. Iglesias, A. Cebollada, C. Lafoz, P. Gavin, L. Aristimuno, M. J. Revillo, C. Martin, and S. Samper. 2007. Genotyping of Mycobacterium tuberculosis over two periods: a changing scenario for tuberculosis transmission. Int. J. Tuberc. Lung Dis. 111080-1086. [PubMed] [Google Scholar]
- 17.Nei, M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oelemann, M. C., R. Diel, V. Vatin, W. Haas, S. Rusch-Gerdes, C. Locht, S. Niemann, and P. Supply. 2007. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J. Clin. Microbiol. 45691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pretorius, G. S., P. D. van Helden, F. Sirgel, K. D. Eisenach, and T. C. Victor. 1995. Mutations in katG gene sequences in isoniazid-resistant clinical isolates of Mycobacterium tuberculosis are rare. Antimicrob. Agents Chemother. 392276-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 3301703-1709. [DOI] [PubMed] [Google Scholar]
- 21.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, H. P. de, D. H. van, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 444498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 393563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36762-771. [DOI] [PubMed] [Google Scholar]
- 24.van der Spuy, G. D., R. M. Warren, M. Richardson, N. Beyers, M. A. Behr, and P. D. van Helden. 2003. Use of genetic distance as a measure of ongoing transmission of Mycobacterium tuberculosis. J. Clin. Microbiol. 415640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Zanden, A. G., E. M. Te Koppele-Vije, B. N. Vijaya, D. van Soolingen, and L. M. Schouls. 2003. Use of DNA extracts from Ziehl-Neelsen-stained slides for molecular detection of rifampin resistance and spoligotyping of Mycobacterium tuberculosis. J. Clin. Microbiol. 411101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Deutekom, H., P. Supply, P. E. de Haas, E. Willery, S. P. Hoijng, C. Locht, R. A. Coutinho, and D. van Soolingen. 2005. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J. Clin. Microbiol. 434473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Rie, A., R. Warren, M. Richardson, T. C. Victor, R. P. Gie, D. A. Enarson, N. Beyers, and P. D. van Helden. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 3411174-1179. [DOI] [PubMed] [Google Scholar]
- 29.van Rie, A., R. M. Warren, N. Beyers, R. P. Gie, C. N. Classen, M. Richardson, S. L. Sampson, T. C. Victor, and P. D. van Helden. 1999. Transmission of a multidrug-resistant Mycobacterium tuberculosis strain resembling “strain W” among noninstitutionalized, human immunodeficiency virus-seronegative patients. J. Infect. Dis. 1801608-1615. [DOI] [PubMed] [Google Scholar]
- 30.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 333234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verver, S., R. M. Warren, Z. Munch, E. Vynnycky, P. D. van Helden, M. Richardson, G. D. van der Spuy, D. A. Enarson, M. W. Borgdorff, M. A. Behr, and N. Beyers. 2004. Transmission of tuberculosis in a high incidence urban community in South Africa. Int. J. Epidemiol. 33351-357. [DOI] [PubMed] [Google Scholar]
- 32.Warren, R., M. de Kock, E. Engelke, R. Myburgh, N. Gey van Pittius, T. Victor, and P. van Helden. 2006. Safe Mycobacterium tuberculosis DNA extraction method that does not compromise integrity. J. Clin. Microbiol. 44254-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warren, R. M., G. D. van der Spuy, M. Richardson, N. Beyers, C. Booysen, M. A. Behr, and P. D. van Helden. 2002. Evolution of the IS6110-based restriction fragment length polymorphism pattern during the transmission of Mycobacterium tuberculosis. J. Clin. Microbiol. 401277-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warren, R. M., T. C. Victor, E. M. Streicher, M. Richardson, G. D. van der Spuy, R. Johnson, V. N. Chihota, C. Locht, P. Supply, and P. D. van Helden. 2004. Clonal expansion of a globally disseminated lineage of Mycobacterium tuberculosis with low IS6110 copy numbers. J. Clin. Microbiol. 425774-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh, R. W., A. Ponce de Leon, C. B. Agasino, J. A. Hahn, C. L. Daley, P. C. Hopewell, and P. M. Small. 1998. Stability of Mycobacterium tuberculosis DNA genotypes. J. Infect. Dis. 1771107-1111. [DOI] [PubMed] [Google Scholar]
- 36.Yokoyama, E., K. Kishida, M. Uchimura, and S. Ichinohe. 2007. Improved differentiation of Mycobacterium tuberculosis strains, including many Beijing genotype strains, using a new combination of variable number of tandem repeats loci. Infect. Genet. Evol. 7499-508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.