Abstract
Cell cycle progression and polar differentiation are temporally coordinated in Caulobacter crescentus. This oligotrophic bacterium divides asymmetrically to produce a motile swarmer cell that represses DNA replication and a sessile stalked cell that replicates its DNA. The initiation of DNA replication coincides with the proteolysis of the CtrA replication inhibitor and the accumulation of DnaA, the replication initiator, upon differentiation of the swarmer cell into a stalked cell. We analyzed the adaptive response of C. crescentus swarmer cells to carbon starvation and found that there was a block in both the swarmer-to-stalked cell polar differentiation program and the initiation of DNA replication. SpoT is a bifunctional synthase/hydrolase that controls the steady-state level of the stress-signaling nucleotide (p)ppGpp, and carbon starvation caused a SpoT-dependent increase in (p)ppGpp concentration. Carbon starvation activates DnaA proteolysis (B. Gorbatyuk and G. T. Marczynski, Mol. Microbiol. 55:1233-1245, 2005). We observed that SpoT is required for this phenomenon in swarmer cells, and in the absence of SpoT, carbon-starved swarmer cells inappropriately initiated DNA replication. Since SpoT controls (p)ppGpp abundance, we propose that this nucleotide relays carbon starvation signals to the cellular factors responsible for activating DnaA proteolysis, thereby inhibiting the initiation of DNA replication. SpoT, however, was not required for the carbon starvation block of the swarmer-to-stalked cell polar differentiation program. Thus, swarmer cells utilize at least two independent signaling pathways to relay carbon starvation signals: a SpoT-dependent pathway mediating the inhibition of DNA replication initiation, and a SpoT-independent pathway(s) that blocks morphological differentiation.
The initiation of DNA replication in Caulobacter crescentus is one event in an ordered sequence of cell cycle events that are temporally coordinated with asymmetric polar morphogenesis (27, 45). Upon differentiation of the motile swarmer cell into a sessile stalked cell, chromosome replication is initiated (8, 22, 31). The swarmer-to-stalked cell transition includes the release of the swarmer cell's single polar flagellum (43), retraction of polar pili (25, 46), proteolytic destruction of the polar chemosensory apparatus (1), and the growth of a stalk at the site previously occupied by the flagellum (40).
Control of DNA replication is maintained through the opposing activities of two essential regulators of cell cycle progression: CtrA, which binds to and silences the origin of replication (38), and DnaA, which activates replication initiation (13). In late-stage swarmer cells, at the beginning of the swarmer-to-stalked cell transition, CtrA is cleared from the cell by ClpXP-mediated proteolysis (9, 19). Coincident with CtrA proteolysis, and the morphological transition of swarmer cells into stalked cells, there is an approximate twofold increase in DnaA abundance (6). DnaA both binds to the origin, allowing replisome formation, and serves as transcription factor for multiple genes involved in the DNA replication process (6, 13, 18).
C. crescentus, an oligotroph, is able to adapt to and survive in nutrient-poor environments. Relatively little is known about the molecular mechanisms that facilitate communication between extracellular signals, such as the deprivation of specific nutrients, and the regulatory molecules that control cell cycle progression. Carbon starvation destabilizes the DnaA protein, possibly through the activation of ClpP-mediated proteolysis (14), suggesting that DnaA clearance prevents the initiation of DnaA replication when cells are deprived of the resources necessary to complete replication (14). Identification of the factors that sense and transduce carbon starvation signals to activate DnaA proteolysis is central to our understanding of the cell's response to nutrient deprivation.
Many bacteria respond to nutrient deprivation by producing the stress-signaling nucleotides guanosine pentaphosphate (pppGpp) and guanosine tetraphosphate (ppGpp), referred to here as (p)ppGpp, which link the sensation of nutrient deprivation to global alterations in transcription profiles (48). In Escherichia coli, where (p)ppGpp was first discovered, its abundance is regulated by two gene products, RelA and SpoT (48). RelA is a (p)ppGpp synthase that associates with the ribosome. In response to amino acid starvation, ribosomes stall due to uncharged tRNA molecules occupying the ribosome's A site. This event activates an idling reaction by RelA in which (p)ppGpp is synthesized from GTP/GDP and ATP (15, 58). The second source of (p)ppGpp, SpoT, is a bifunctional enzyme that has the capacity to both hydrolyze and synthesize (p)ppGpp (61). SpoT in E. coli mediates an increase in (p)ppGpp concentration during carbon starvation, phosphate starvation, fatty acid starvation, and iron starvation (3, 47, 53, 61). Unlike E. coli, C. crescentus and other members of the alphaproteobacteria possess a single bifunctional RelA/SpoT homolog capable of synthesizing and hydrolyzing (p)ppGpp (57).
In E. coli, (p)ppGpp mediates adaptation to nutrient deprivation, in part, through global alterations in transcription that involve direct and indirect effects on RNA polymerase (48). (p)ppGpp also facilitates adaptation to nutrient deprivation through its regulation of DNA replication. An elevated intracellular concentration of (p)ppGpp inhibits the initiation of E. coli DNA replication (28, 41). Although the mechanism for this inhibition is not known, it has been hypothesized to involve inhibition of dnaA transcription (63). In Bacillus subtilis, (p)ppGpp stops DNA replication by blocking elongation through a direct inhibitory interaction with DNA primase (56).
Since it has been reported that carbon starvation blocks the swarmer-to-stalked cell transition (14) and that (p)ppGpp accumulates in C. crescentus cells starved of carbon (5), we explored the possibility that the (p)ppGpp stress-signaling molecule could provide a means to relay carbon starvation signals to downstream effectors responsible for blocking polar morphogenesis and/or the initiation of DNA replication. To test this hypothesis, we examined whether carbon starvation of swarmer cells blocks the initiation of DNA replication and stalk biogenesis. We also measured the accumulation of polar organelle proteins. We deleted the C. crescentus ortholog of spoT and found that SpoT is required for the activation of (p)ppGpp production that occurs during carbon starvation. While SpoT was not required to block polar morphogenesis during carbon starvation, it was required to block DNA replication initiation. In swarmer cells deprived of carbon, DnaA was rapidly degraded and this proteolysis was dependent on SpoT. Thus, DnaA proteolysis prevents the inappropriate initiation of DNA replication when resources are limiting, and we propose that this event is mediated by a SpoT-dependent increase in (p)ppGpp concentration. Further, SpoT-mediated (p)ppGpp accumulation was necessary to maintain viability during carbon starvation. Collectively, these observations suggest that C. crescentus swarmer cells use at least two independent signaling pathways to coordinate the simultaneous inactivation of polar morphogenesis and DNA replication initiation during carbon starvation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used for this study are listed in Table 1. All experiments were performed using derivatives of the synchronizable Caulobacter crescentus strain CB15N (NA1000) (12). CB15N strains were grown in peptone-yeast extract (PYE), M2 minimal medium supplemented with 0.2% glucose as a carbon source (M2G) (10), or M5 minimal low-phosphate medium supplemented with 1 mM glutamate and 0.2% glucose as a carbon source (M5G) (9) and incubated at 28°C. When indicated, xylose (0.03%) was added to the medium to induce expression from the xylose-inducible promoter. Escherichia coli strain TOP10 (Invitrogen) was used for cloning. TOP10 was grown in Luria-Bertani medium at 37°C. When appropriate, C. crescentus media were supplemented with antibiotics at the following concentrations (for liquid/solid media, in μg/ml): spectinomycin (25/50), gentamicin (0.5/5), kanamycin (5/20).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| NA1000 | syn-1000; previously called CB15N, a synchronizable derivative of CB15 | 12 |
| LS4427 | NA1000 ΔspoT | This study |
| MT16 | NA1000 ori::(tetO)nxylX::pHPV472 | 55 |
| LS4428 | MT16 ΔspoT | This study |
| SC993 | CB15 hisB::Tn5 proA103 str-140 | 11 |
| LS2596 | NA1000 hisB::Tn5 | R. Roberts |
| Plasmids | ||
| pNPTS138 | Nonreplicating vector used for integration and allelic replacement | C. Mohr, R. Roberts, and L. Shapiro |
| pJL23-9 | pNPTS138-derived vector used for ΔspoT allelic replacement | This study |
| pJL37 | pNPTS138-derived vector harboring 550-bp fragment upstream of translational start for spoT | This study |
| pJL38 | pNPTS138-derived vector harboring the full-length spoT and 550 bp upstream of the spoT translational start site | This study |
Culture synchronization.
Swarmer cells were isolated from a mixed culture through a modified version of the percoll density centrifugation protocol (52). A PYE overnight culture was grown to an optical density at 660 nm (OD660) of 0.4 and back-diluted to an OD660 of 0.025 in M2G. This culture was then grown to an OD660 of 0.4 and back-diluted a second time to an OD660 of 0.01 in M2G and grown to an OD660 of 0.5 in order to dilute remnants of the PYE. The culture was cooled on ice for 10 min, and a 13-ml aliquot of the mixed culture was pelleted at 4,500 × g at 4°C and washed once with 600 μl of ice-cold M2. The washed cells were pelleted at 6,000 × g for 3 min at 4°C and resuspended in 600 μl of ice-cold M2 and 750 μl of ice-cold percoll. The percoll mixture was centrifuged at 11,000 × g for 20 min at 4°C in order to isolate the lower swarmer band. Swarmer cells were then washed four times in ice-cold M2 using 3-min 6,000 × g pelleting steps. The washed swarmer cells were finally diluted into prewarmed (28°C) M2G medium, and the swarmer cells were able to progress synchronously through the cell cycle. When larger amounts of swarmer cells were required, multiple 13-ml aliquots of mixed culture were synchronized in parallel and swarmer bands were pooled prior to the final four washing steps with M2 medium.
Generation of ΔspoT mutant and complemented strains.
A stable in-frame deletion of spoT was generated by homologous recombination and allelic replacement using a two-step selection procedure with the nonreplicating vector pNPTS138 as previously described (16, 49, 51). To generate the ΔspoT allelic replacement plasmid, 500-bp DNA fragments located upstream and downstream of the spoT open reading frame were PCR amplified and cloned into pNPTS138. The upstream DNA fragment was amplified using primers OJL65 (5′ ACA TGC ATG CCG AGG AAC TCA TGG CCC GCGT 3′) and OJL66 (5′ CGG AAT TCC GGC GCT TCG GTC ACA GCGA 3′). This fragment was framed by SphI and EcoRI sites and it included the first 13 codons of spoT. The downstream DNA fragment was PCR amplified using primers OJL68 (5′ CGG AAT TCT CGG TGG AGA CGG TGG ACC GGA 3′) and OJL69 (5′ ACA TGC ATG CCT CTT GCT GGT CAA GCG GGT CAAG 3′). This fragment was framed by EcoRI and SphI sites and it included the final 11 codons of spoT. Triple ligation of the upstream and downstream DNA fragments into the SphI site of pNPTS138 generated plasmid pJL23-9, which harbored a ΔspoT allele in which codons 14 to 732 were deleted. Confirmation of the desired plasmid construction was obtained by DNA sequencing. To create a complementing plasmid for the ΔspoT strain, a 2.9-kb fragment of DNA encompassing spoT as well as 0.55 kb of DNA upstream of its translational start site was PCR amplified using primers OJL70 (5′ GGA CTA GTG CTC GCG AAA GCA GGA TGC TCGT 3′) and OJL71 (5′ GGA CTA GTG GCA CGC CGT CAG ACT GCG TAA 3′). This DNA fragment, framed by SpeI sites, was cloned into a SpeI-digested pNPTS138 to generate plasmid pJL38. A negative control vector was created by PCR amplifying only the 0.55 kb of DNA upstream of the spoT translational start site using primers OJL70 and OJL102 (5′ TTT ACT AGT GCG CCC GAT TCG GTC CCC GT 3′). This DNA fragment, framed by SpeI sites, was cloned into the SpeI site of pNPTS138 to generate plasmid pJL37. pJL38 and pJL37 were electroporated into the ΔspoT strain as described previously (55). Only pJL37 was electroporated into the wild-type strain. Transformants with pJL37 or pJL38 integrated upstream of the spoT locus were obtained by selecting for kanamycin resistance.
Fluorescence time-lapse microscopy.
Swarmer cells from the fluorescent repressor-operator system (FROS) strains MT16 (55) and MT16 ΔspoT, generated in this study, were obtained by culture synchronization as described above with the exception that 60 min prior to the start of the synchrony protocol, synthesis of TetR-yellow fluorescent protein (YFP) was induced by the addition of 0.03% xylose to the culture (55). M2 washed swarmer cells were spotted onto 1% (wt/vol) agarose pads prepared using either M2G if cells were to be imaged during growth in the presence of a carbon source or M2 if cells were to be imaged during carbon starvation. A coverslip was added and sealed to the slide with wax (1:1:1, vaseline-lanolin-paraffin), and images were obtained as a function of time using both Nomarski differential interference contrast (DIC) and the YFP fluorescence channel. Images were obtained using a Nikon E800 microscope with a 5-MHz Micromax 5600 cooled charge-coupled-device camera controlled through Metamorph verson 4.5. DIC and YFP fluorescence images were overlaid using Photoshop (Adobe).
Flow cytometry.
Swarmer cells were isolated as described above and inoculated into either M2 or M2G. At various time points, aliquots were removed and treated with a modified version of the flow cytometry preparation protocol previously described (59) in order to analyze chromosome content. Briefly, aliquots were removed from the cultures at various time points, pelleted at 16,000 × g for 1 min, and resuspended in M2G with 15 μg/ml rifampin to block new rounds of DNA replication initiation. Samples were then incubated for an additional 3 h at 28°C, fixed with 70% ethanol, and stored overnight at 4°C. Fixed cells were pelleted at 4,000 × g and gently resuspended in TMS buffer (10 mM Tris-HCl [pH 7.2], 1.5 mM MgCl2, 150 mM NaCl) and stained with 10 μM Vybrant DyeCycle Orange (Invitrogen). Fixed cells were analyzed on a Becton Dickinson FACScan flow cytometer. Flow cytometry data were analyzed using FlowJo software (Tree Star).
DAPI staining.
Swarmer cells were isolated as described above and inoculated into either M2 or M2G. At 0 and 120 min after inoculation, aliquots were removed and fixed with a solution consisting of 30 mM sodium phosphate buffer (pH 7.5) and 2.5% formaldehyde. After 48 h of storage at 4°C, cells were pelleted at 4,000 × g and gently resuspended in 4′,6-diamidino-2-phenylindole (DAPI) stain (1.5 μg/ml) for 2 h at 22°C. Fixed and stained cells were mounted on 1% (wt/vol) agarose pads and imaged using a UV filter.
Immunoblot analysis.
Immunoblotting to monitor the steady-state levels of FliF, PilA, McpA, DnaA, and CtrA was performed as described elsewhere (6). In brief, CtrA, PilA, McpA, and FliF were separated from other proteins in total cell extracts using a 15% (CtrA and PilA) or 10% (McpA and FliF) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, while DnaA was separated using an 8% SDS-PAGE gel. Immunodetection was performed with polyclonal antibodies recognizing FliF and McpA (1:30,000 dilution) as well as PilA, DnaA, and CtrA (1:10,000 dilution), a goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (1:35,000 dilution), chemiluminescence reagent (Perkin-Elmer), and BioMax MR film. The developed film was scanned and processed with Photoshop (Adobe), and relative band intensities were determined using ImageQuant (Molecular Dynamics).
DnaA turnover.
Swarmer cells were isolated as described above, suspended in M2 medium, and grown at 28°C with aeration for 10 min. Rifampin (10 μg/ml) was added to the culture to inhibit transcription and ultimately protein synthesis. Aliquots of cells were removed as a function of time after rifampin addition, and Western blotting was utilized to monitor DnaA levels in these samples. In order to adjust for the nonlinearity of chemiluminescent signal versus protein quantity in Western blots analyzed by chemiluminescent detection, a standard curve was generated to determine the relationship between chemiluminescent DnaA signal and actual DnaA abundance. DnaA levels quantified from the Western blot assay were fit to a single exponential decay curve in order to calculate the half-life.
Determination of cell viability.
Viability of C. crescentus swarmer cells during carbon starvation was determined by inoculating M2 medium with synchronized and washed swarmer cells isolated as described above. M2 cultures were incubated with aeration (250 rpm) at 28°C, and aliquots of the cultures were removed followed by serial dilution and plating onto PYE agar to achieve approximately 500 to 50 colonies/plate. The plates were grown for 2 to 3 days at 28°C. Colonies were counted and multiplied by the appropriate dilution factor to calculate CFU/ml.
(p)ppGpp assays.
(p)ppGpp production was assayed as described previously (57). C. crescentus strains were grown overnight in PYE to an OD660 of 0.4, back-diluted to OD660 0.05 in M5G supplemented with 1 mM glutamate, grown to OD660 0.4, back-diluted a second time to OD660 0.01, and grown to OD660 0.4 to dilute remnants of PYE. A 1.0-ml aliquot was removed from each culture, pelleted at 16,000 × g, washed two times in M5 with or without glucose and glutamate, and resuspended in 250 μl M5 with or without glucose and glutamate. [32P]phosphate was added at 100 μCi/ml, and cultures were labeled for 1 h with shaking (200 rpm) at 28°C. Following the labeling, samples were removed and mixed with an equal volume of 2 M formic acid, placed on ice for 30 min, and stored at −20°C overnight. Samples were pelleted for 5 min at 16,000 × g in a microcentrifuge, and 2 μl of supernatant was spotted directly onto polyethyleneimine (PEI) cellulose thin-layer chromatography (TLC) plates and developed in 1.5 M KH2PO4 for 1 h. Nucleotides were visualized by phosphorimaging.
RESULTS
Carbon starvation blocks the swarmer-to-stalked cell polar morphogenesis program.
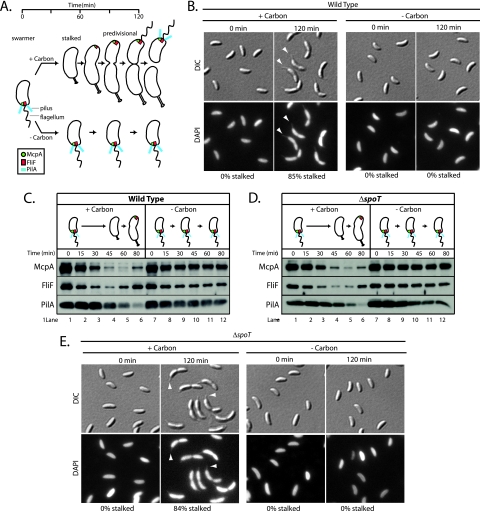
The hallmark feature of the swarmer-to-stalked cell transition is the biogenesis of a stalk at the site previously occupied by the flagellum (Fig. 1A). To assess the effect of carbon starvation on stalk formation, swarmer cells were isolated and inoculated into minimal medium that either lacked or possessed a carbon source (0.2% glucose). At 0 and 120 min after the start of incubation, aliquots were removed and cells were fixed with formaldehyde, stained with the fluorescent dye DAPI, and imaged using both DIC and fluorescence microscopy in order to identify stalked cells. Just as DAPI staining has been used to visualize the C. crescentus flagellum (26), we have found that DAPI staining also enhances the visualization of the stalk. Carbon starvation completely blocked stalk formation (Fig. 1B). No stalked cells were observed in the 0- and 120-min incubation samples. In contrast, after 120 min of incubation in the presence of a carbon source, 85% of cells possessed visible stalks (Fig. 1B). The swarmer-to-stalked cell transition is a multifaceted process. In addition to stalk biogenesis, the transition is also marked by significant changes in polar protein complexes (Fig. 1A). Pili, which consist of PilA polymers, are retracted and the PilA steady-state level declines (25, 46, 54). The flagellum is released, and the membrane-embedded components of the flagellar apparatus, such as the inner membrane protein FliF, are degraded (20, 43). The chemosensory apparatus, which includes many copies of the McpA chemoreceptor, is also degraded as swarmer cells become stalked cells (1) (Fig. 1A). To determine the effect of carbon starvation on each of these processes, swarmer cells were isolated and inoculated into minimal medium that either lacked or possessed a carbon source. Samples were removed from incubated cultures at 15-min intervals. Western blot assays of the protein lysates derived from these samples using antibodies directed against McpA, FliF, and PilA revealed that these proteins remained abundant during incubation of swarmer cells in the absence of carbon (Fig. 1C, lanes 7 to 12). In the presence of glucose, McpA levels in swarmer cells declined markedly by 60 min, at which point swarmer cells had developed into stalked cells (Fig. 1C, lanes 1 to 5). McpA levels then began to slowly increase as stalked cells transitioned to early predivisional cells (Fig. 1C, lanes 5 and 6). By contrast, swarmer cells starved of carbon maintained a constant level of McpA (Fig. 1C, lanes 7 to 12). In the presence of a carbon source, FliF and PilA exhibited a decline in abundance during the swarmer-to-stalked cell transition. For FliF, this drop in abundance occurred during the first 45 min of incubation (Fig. 1C, lanes 1 to 4), and then FliF increased as the cell cycle progressed (Fig. 1C, lanes 5 and 6). For PilA, this drop occurred by 80 min of incubation (Fig. 1C, lanes 1 to 6). The reaccummulation of PilA occurs just before cell division (54), which was beyond the time span of this experiment. In contrast, during carbon starvation, FliF and PilA remained abundant over the course of the experiment (Fig. 1C, lanes 7 to 12). Thus, carbon starvation inhibited the cell cycle programmed reduction in the steady-state levels of McpA, FliF, and PilA. We have not ruled out the possibility that carbon-starved swarmer cells are able to shed their flagellum and retract their pili despite the effect of carbon starvation on FliF and PilA steady-state levels.
FIG. 1.
Carbon starvation blocks swarmer-to-stalked cell polar differentiation. (A) Schematic depicting polar changes that occur over the course of the cell cycle. Complexes of the McpA chemoreceptor (green), the FliF flagellar basal body protein (red), and the PilA pilus subunit (blue) are shown. (B) DIC and DAPI fluorescence images of wild-type swarmer cells inoculated into M2 medium possessing or lacking a carbon source (0.2% glucose). Arrowheads point to stalks on synchronized cells grown in the presence of glucose. The percentage of stalked cells was determined by counting at least 100 cells. (C) Immunoblots of cell extracts from a synchronized wild-type culture incubated in minimal medium in the presence (M2G) or absence (M2) of glucose. McpA, FliF, and PilA antibodies were used to probe each protein's accumulation at the indicated time points. (D) Immunoblots of cell extracts from a synchronized ΔspoT strain grown in either the presence or absence of glucose. McpA, FliF, and PilA antibodies were used to probe each protein's accumulation at the indicated time points. (E) DIC and DAPI fluorescence images of ΔspoT swarmer cells inoculated into M2 medium possessing or lacking a carbon source (0.2% glucose). Arrowheads point to stalks on synchronized ΔspoT cells grown in the presence of glucose. The percentage of stalked cells was determined by counting at least 100 cells.
Carbon starvation inhibits the initiation of DNA replication.
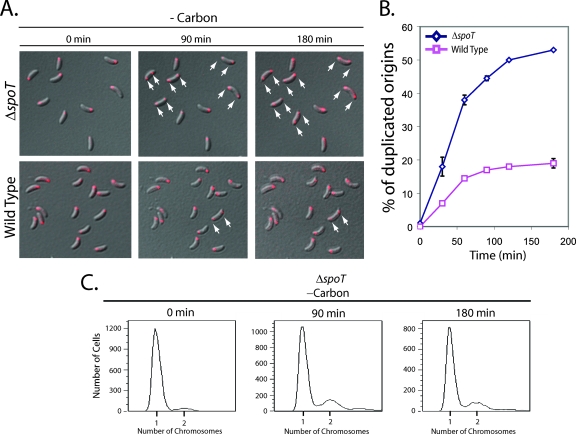
To assess the effect of carbon starvation on DNA replication, we employed two assays: origin replication and separation was visualized by the fluorescent repressor-operator system (FROS) (55), and chromosome duplication was assessed by fluorescence-activated cell sorting (FACS). FROS uses an inducible Tet repressor (TetR)-YFP fusion protein and a TetR operator array (tetO)n integrated next to the origin of replication. When the TetR-YFP proteins bind to (tetO)n, the localization of the origin can be visualized by fluorescence microscopy of live cells. In swarmer cells, the chromosomal origin of DNA replication is positioned at the flagellar pole. Upon replication initiation, one of the duplicated origin regions traverses the length of the cell and localizes to the opposite cell pole, thereby yielding a cell with a bipolar distribution of chromosomal origins (21, 55). Thus, origin duplication could be used as a readout for the initiation of DNA replication.
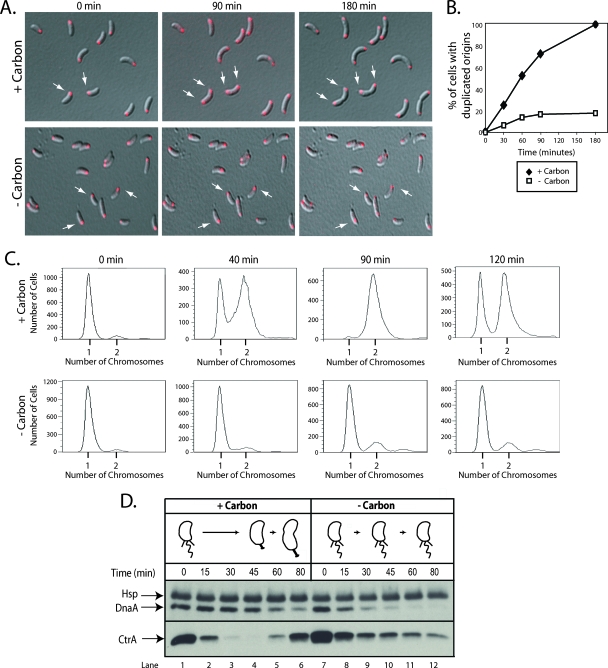
Isolated populations of swarmer cells with (tetO)n next to the origin locus were washed in minimal medium lacking a carbon source and spotted onto minimal medium agarose pads that either lacked or possessed a carbon source (0.2% glucose). Fluorescence microscopy images were taken every 30 min for the first 90 min and then again after 180 min. The percentage of cells with bipolar origins, indicating that replication had initiated, was then determined as a function of time (Fig. 2A and B). By 90 min after cells were spotted onto agarose pads containing a carbon source, 75% had bipolar origins. In contrast, origin duplication was a rare event in cells spotted onto agarose pads that lacked a carbon source. By 90 min, only 18% of the cells had duplicated their origins, and by 180 min, this number had not increased. These results indicate that either the initiation of DNA replication was inhibited in these cells or that replication initiated normally but carbon starvation disrupted DNA segregation. To differentiate between these two possibilities, we used FACS to examine both the timing of DNA replication and the DNA content of synchronized swarmer cells grown either with or without a carbon source (Fig. 2C). At the indicated times, samples were removed, and treated with rifampin to block new rounds of replication initiation. Based on the distribution of cells with one or two chromosomes, we observed that over 50% of synchronized cells grown in the presence of a carbon source had initiated replication by 40 min of incubation, and nearly 100% had initiated replication by 90 min (Fig. 2C). In contrast, the majority of synchronized cells starved for carbon retained only a single chromosome even after 2 h of incubation, indicating that the initiation of DNA replication was blocked (Fig. 2C).
FIG. 2.
Carbon starvation inhibits the initiation of DNA replication and promotes a reduction in the steady-state level of DnaA in swarmer cells. (A) FROS images of the chromosomal replication origin in synchronized swarmer cells grown on minimal medium agarose pads that either possessed (+carbon) or lacked (-carbon) 0.2% glucose. The origin is shown in red, and white arrows point to examples of cells with either a single or a duplicated origin. (B) Quantification of origin duplication as a function of incubation time. The origin duplication status of at least 150 cells was determined at each time point, and the number that possessed a duplicated origin was expressed as a percentage of the total number of cells counted. The experiment to determine the origin duplication status of carbon-starved swarmer cells was performed two times, and the plotted values are the means of the two experiments. (C) FACS analysis of wild-type swarmer cells grown in minimal medium either with (+carbon) or without (-carbon) 0.2% glucose. At the indicated times, aliquots were incubated at 28°C for 3 h with rifampin (15 μg/ml), which allowed cells that had initiated DNA replication prior to rifampin addition to finish chromosome duplication but blocked new rounds of replication initiation. Cells were fixed, stained with Vybrant DyeCycle Orange, and analyzed by flow cytometry. (D) Immunoblots of cell extracts from a synchronized wild-type culture incubated in medium that either possessed (+carbon) or lacked (-carbon) 0.2% glucose. DnaA and CtrA antibodies were used to probe each protein's steady-state levels at the indicated time points. The DnaA antibody cross-reacts with a heat shock protein (Hsp) thought to be GroEL (14) and which runs at a slightly larger apparent molecular weight during SDS-PAGE.
In C. crescentus, the initiation of DNA replication is directly controlled by two master regulators of the cell cycle, CtrA and DnaA. CtrA silences replication initiation by binding to five sites in the origin sequence (38), while DnaA activates initiation, after CtrA is cleared from the cell, by enabling replisome formation at the origin (13). To investigate the mechanism by which carbon starvation blocks the initiation of DNA replication, the levels of DnaA and CtrA were compared in synchronized cells incubated either with or without a carbon source. A previous study demonstrated that carbon starvation of mixed cell populations increased the rate of DnaA turnover, resulting in a decrease in DnaA accumulation (14). In swarmer cells deprived of carbon, we observed a rapid and pronounced decrease in the steady-state level of DnaA (Fig. 2D, lanes 7 to 12). Whereas DnaA remains abundant for the first 45 min of the cell cycle in cells incubated with a carbon source (Fig. 2D, lanes 1 to 4), in cells lacking a carbon source, nearly all DnaA was eliminated during that time frame (Fig. 2D, lanes 7 to 10). Carbon starvation also influenced the steady-state level of CtrA (Fig. 2D). Synchronized swarmer cells grown in the presence of a carbon source exhibited a rapid and pronounced decrease in CtrA abundance during the swarmer-to-stalked cell transition (Fig. 2D, lanes 1 to 3). Synchronized swarmer cells subjected to carbon starvation exhibited a slower decline in the steady-state level of CtrA (Fig. 2D, lanes 7 to 12). These results suggest that the persistence of CtrA and the significant decline in the steady-state level of DnaA are contributing factors to the inhibition of DNA replication initiation caused by carbon starvation.
C. crescentus spoT.
In many bacteria, adaptive responses to carbon starvation are mediated by activating the production of the intracellular signaling molecule (p)ppGpp (36, 61). In C. crescentus, carbon starvation stimulates (p)ppGpp accumulation (5). In alphaproteobacteria like C. crescentus, the synthesis and degradation of (p)ppGpp is mediated by a single member of the RelA/SpoT family of (p)ppGpp synthases and (p)ppGpp hydrolases (57). A BLAST search of the C. crescentus genome using the sequence of the RelA/SpoT family member from Sinorhizobium meliloti identified a single orthologous gene, CC1553, which was 37% identical and 55% similar. We named the gene spoT.
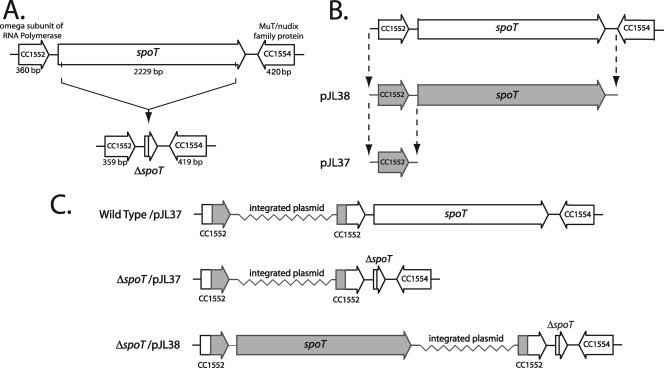
To evaluate the function of C. crescentus SpoT upon carbon starvation, we generated an in-frame spoT deletion strain (Fig. 3A). We also generated a complementing vector with the goal of rescuing mutant phenotypes. The complementing vector was created by cloning spoT along with 550 bp of sequence upstream of the predicted translational start site into a nonreplicating plasmid for chromosomal integration (Fig. 3B). The vector was then integrated in the ΔspoT strain to create the complemented strain ΔspoT/pJL38 (Fig. 3C). A negative control vector, pJL37, was also created by cloning only the 550 bp upstream of the translational start site for spoT (Fig. 3B). This control vector was integrated into both the ΔspoT strain and wild-type strain to create ΔspoT/pJL37 and wild-type/pJL37 (Fig. 3C).
FIG. 3.
Construction of a C. crescentus ΔspoT strain and a complemented ΔspoT strain. (A) Schematic representation of the spoT locus and the ΔspoT strain used to characterize SpoT function. (B) Construction of the ΔspoT-complementing vector pJL38 and the negative control vector, pJL37. (C) Schematic representation of the complemented ΔspoT strain, ΔspoT/pJL38, and the control strains harboring the empty vector ΔspoT/pJL37 and wild-type/pJL37.
SpoT is required for rapid adaptation to nutritional downshift.
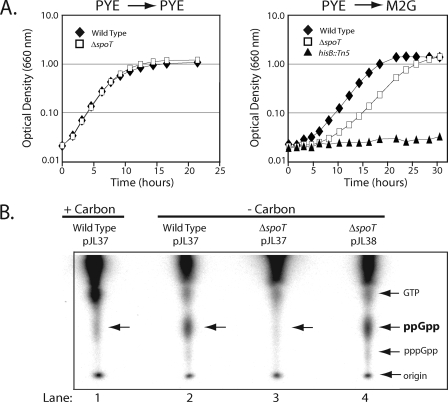
The C. crescentus ΔspoT strain did not exhibit a growth defect when grown in rich (PYE) medium (Fig. 4A). Inactivation of relA and spoT, the genes required for (p)ppGpp synthesis in enteric bacteria, renders these bacteria auxotrophic for several different amino acids (61). This auxotrophy stems from (p)ppGpp's role in activating transcription of amino acid biosynthetic genes (2, 37). Since C. crescentus SpoT appeared to be the cell's only source of (p)ppGpp production, we asked if the ΔspoT strain exhibited amino acid auxotrophy when grown in minimal medium. To test this possibility, the wild type, the ΔspoT strain, and a histidine auxotroph (hisB::Tn5) (11) were grown in PYE medium overnight, washed with minimal medium, and inoculated into minimal medium. As shown in Fig. 4A, while the ΔspoT strain exhibited an extended lag phase relative to the wild-type strain, suggesting a role for SpoT in adaptation to sudden nutritional downshift, it clearly did not exhibit the auxotrophy evident in the hisB::Tn5 mutant, which as expected failed to exhibit any growth in minimal medium (Fig. 4A).
FIG. 4.
SpoT is required for rapid adaptation to nutrient downshift from rich medium (PYE) to minimal medium (M2G) and for (p)ppGpp accumulation during carbon starvation. (A) Growth curves of wild-type, ΔspoT, and hisB::Tn5 strains grown in PYE medium, washed in M2G, and then either resuspended in PYE (left panel) or M2G (right panel). (B) One-dimensional thin-layer chromatography analysis of total intracellular nucleotides extracted from wild-type/pJL37 grown in minimal medium with 0.2% glucose (+carbon) as well as total intracellular nucleotides extracted from wild-type/pJL37, ΔspoT/pJL37 (uncomplemented), and ΔspoT/pJL38 (complemented) grown in minimal medium lacking 0.2% glucose (-carbon).
SpoT is required for carbon starvation-induced production of (p)ppGpp.
To determine if the stimulation of (p)ppGpp production upon carbon starvation was dependent on SpoT, we used one-dimensional thin-layer chromatography (TLC) to assess (p)ppGpp levels in a ΔspoT strain and the isogenic wild-type strain (Fig. 4B). The wild-type strain bearing an empty integrated vector, wild-type/pJL37, was grown to mid-logarithmic phase in minimal medium, washed with minimal medium either lacking or possessing a carbon source, and then inoculated into the same medium. After 60 min of incubation along with [32P]phosphate, nucleotides were extracted and resolved on a TLC plate. As shown in Fig. 4B, lanes 1 and 2, carbon starvation stimulated the accumulation of (p)ppGpp. This phenomenon was entirely SpoT dependent. When the carbon starvation protocol was repeated using a ΔspoT strain harboring an empty vector, ΔspoT/pJL37, no (p)ppGpp was observed (Fig. 4B, lane 3). Complementation of the ΔspoT strain with vector pJL38 rescued (p)ppGpp production (Fig. 4B, lane 4).
SpoT is not required to block polar development in carbon-starved swarmer cells.
When wild-type swarmer cells were starved of carbon, the block in morphological differentiation from a swarmer cell into a stalked cell (Fig. 1B) was reflected in the stable maintenance of the polar organelle proteins McpA, FliF, and PilA (Fig. 1C, compare lanes 7 to 12 and 1 to 6). If SpoT-mediated (p)ppGpp production were required to inhibit the degradation of McpA, FliF, and PilA during carbon starvation, then these proteins would no longer be maintained in carbon-starved ΔspoT swarmer cells. To test this hypothesis, ΔspoT swarmer cells were inoculated into minimal medium that either lacked or possessed a carbon source. Samples were removed approximately every 15 min, and Western blot assays were used to monitor changes in the abundance of McpA, FliF, and PilA. As shown in Fig. 1D, the steady-state levels of McpA, FliF, and PilA exhibited greater stability in carbon-starved ΔspoT swarmer cells than they did in unstarved ΔspoT swarmer cells (Fig. 1D, compare lanes 7 to 12 with lanes 1 to 6). Therefore, SpoT is not required to inhibit the degradation of McpA, FliF, and PilA during carbon starvation. In addition, ΔspoT swarmer cells starved for carbon failed to exhibit stalk formation after 120 min of incubation (Fig. 1E). Thus, SpoT-mediated (p)ppGpp production is not required to block the swarmer-to-stalked cell transition during carbon starvation.
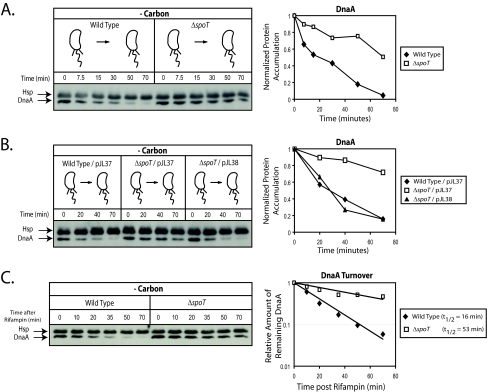
SpoT is required to activate DnaA depletion in carbon-starved swarmer cells.
To determine if SpoT contributes to the depletion of DnaA upon carbon starvation, swarmer cells were isolated from wild-type and ΔspoT strains, washed in minimal medium lacking a carbon source, and then incubated in the same medium. Samples were removed as a function of time, and Western blot assays were used to monitor changes in DnaA levels. The ΔspoT strain exhibited a marked stabilization in DnaA levels (Fig. 5A). When wild-type strains were deprived of carbon, 50% of the DnaA was depleted within 18 min and nearly all DnaA was eliminated by 70 min. However, in ΔspoT swarmer cells, DnaA levels decreased modestly and gradually over the course of the experiment such that approximately 50% remained after 70 min of carbon starvation (Fig. 5A). To determine if the effect on DnaA in the ΔspoT swarmer cells could be rescued by a complementing copy of the spoT gene, the experiment was repeated using wild-type/pJL37, ΔspoT/pJL37, and ΔspoT/pJL38. As shown in Fig. 5B, complementing the ΔspoT swarmer cells fully restored the rate of DnaA loss in the absence of carbon to wild-type levels.
FIG. 5.
Carbon starvation-induced reduction in the steady-state level of DnaA requires a SpoT-dependent activation of DnaA proteolysis. (A) Immunoblots of cell extracts from synchronized swarmer cells of the wild type or a ΔspoT strain which were grown in medium lacking glucose (-carbon). DnaA antibody was used to probe the DnaA steady-state level at the indicated time points. (B) Immunoblots of cell extracts from synchronized swarmer cells of wild-type/pJL37, ΔspoT/pJL37 (uncomplemented), and ΔspoT/pJL38 (complemented) which were grown in medium lacking glucose (-carbon). DnaA antibody was used to probe the protein's steady-state level at the indicated time points. (C) SpoT promotes DnaA proteolysis during carbon starvation. The half-life of DnaA was measured by Western blot analysis of samples taken at the indicated times after the addition of rifampin (10 μg/ml) to block further dnaA transcription and ultimately DnaA synthesis. Samples were taken from synchronized swarmer cells of wild-type and ΔspoT strains that were starved for carbon for 10 min prior to the addition of rifampin. In order to adjust for the nonlinearity of chemiluminescent signal versus protein quantity in Western blots analyzed by chemiluminescent detection, a standard curve was generated to determine the relationship between chemiluminescent DnaA signal and actual DnaA abundance. DnaA levels quantified from the Western blot assay were fit to a single exponential decay curve in order to calculate the half-life.
Rapid loss of DnaA in carbon-starved swarmer cells is due to a SpoT-dependent activation of proteolysis.
In mixed populations of C. crescentus, it is known that carbon starvation leads to a decrease in DnaA levels by increasing the rate of proteolysis (14). To determine if SpoT affects DnaA proteolysis during carbon starvation of swarmer cells, we compared the rates of DnaA turnover in wild-type and ΔspoT swarmer cells starved for carbon. As shown in Fig. 5C, upon carbon starvation, the DnaA turnover rate was significantly reduced in a ΔspoT mutant compared to the wild type. Carbon-starved ΔspoT swarmer cells exhibited a 53-min half-life, while carbon-starved wild-type swarmer cells exhibited a 16-min half-life. Therefore, SpoT mediates the depletion of DnaA during carbon starvation by enhancing proteolysis.
SpoT is required to block the initiation of DNA replication during carbon starvation.
We have presented experiments (Fig. 2A, B, and C) demonstrating that carbon starvation blocks the initiation of DNA replication. Experiments that monitored the level of DnaA upon carbon depletion suggested that by promoting the degradation of the DnaA activator of initiation, the cell is unable to initiate replication. The identification of SpoT as a factor that is required for DnaA degradation allowed us to directly test the proposition that changes in DnaA abundance during carbon starvation contribute to the cell's inhibition of replication initiation.
To test this hypothesis, we utilized the FROS strain with (tetO)n integrated near the origin of replication, thereby enabling origin duplication to serve as a readout for the initiation of DNA replication. A deletion of spoT was then generated in this FROS strain. Swarmer cells were isolated from wild-type and ΔspoT FROS strains grown in minimal medium, and the cells were washed with minimal medium lacking a carbon source and spotted onto M2 agarose pads lacking a carbon source. Origin duplication was measured as a function of time (Fig. 6A and B). Consistent with the hypothesis that SpoT-dependent depletion of DnaA is necessary for efficient inhibition of DNA replication initiation during carbon starvation, the ΔspoT FROS strain exhibited a 2.5- to 3-fold elevated frequency of origin duplication (Fig. 6B). Since approximately 50% of carbon-starved ΔspoT swarmer cells exhibited inappropriate replication initiation, we asked if these initiated chromosomes completed replication. FACS analysis showed that the chromosome complement remained at 1 per cell (Fig. 6C). Therefore, the chromosomes that initiated replication under these conditions were unable to complete duplication.
FIG. 6.
SpoT is required for the carbon starvation-induced block in the initiation of DNA replication. (A) FROS images of the chromosomal origin of replication in swarmer cells of the wild type and a ΔspoT strain incubated on M2 minimal medium agarose pads in the absence of a carbon source. The origin region is shown in red, and white arrows point to examples of cells that have undergone origin duplication. (B) Quantification of the origin duplication status of carbon-starved swarmer cells from wild-type and ΔspoT strains is presented as a function of time. At each time point, at least 150 cells were counted, and the plotted values are the means of two independent experiments. Bars represent 1 standard deviation from the mean. (C) FACS analysis of synchronized ΔspoT swarmer cells grown in M2 minimal medium without (-carbon) 0.2% glucose. At the indicated times, aliquots were incubated at 28°C for 3 h with rifampin (15 μg/ml), which allowed cells that had initiated replication prior to rifampin addition to finish chromosome duplication but blocked new rounds of replication initiation. Cells were fixed, stained with Vybrant DyeCycle Orange, and analyzed by flow cytometry.
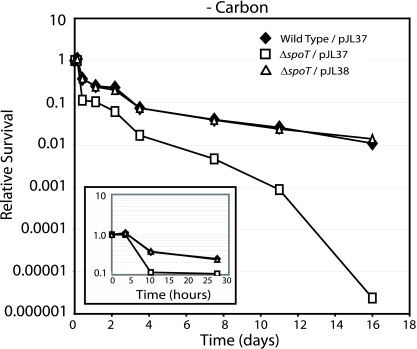
SpoT is required for swarmer cells to survive carbon starvation.
In many bacteria, the absence of a functional spoT allele causes a marked defect in survival when these bacteria are subjected to carbon starvation (24, 36). Because SpoT was required for C. crescentus swarmer cells to efficiently block the initiation of DNA replication during carbon starvation (Fig. 6), we asked if swarmer cells from a ΔspoT strain exhibit a carbon starvation survival defect. Accordingly, swarmer cells from a ΔspoT strain bearing an empty vector (ΔspoT/pJL37), an isogenic wild-type strain (wild-type/pJL37), and a complemented ΔspoT strain (ΔspoT/pJL38) were subjected to carbon starvation by washing them in minimal medium lacking a carbon source followed by incubation in the same medium. Survival was assayed by monitoring CFU per milliliter of medium as a function of time. Over the course of 2 weeks of carbon starvation, the presence of a functional spoT allele strongly influenced survival (Fig. 7). Over this time frame, the wild-type and complemented ΔspoT strains' viabilities declined approximately 2 orders of magnitude, while the ΔspoT strain's viability declined approximately 6 orders of magnitude. The loss of survival occurred in distinct stages. The first stage occurred during the first 10 h of carbon starvation, during which the wild-type strain and the complemented ΔspoT strain exhibited a reproducible 3-fold decline in survival while the ΔspoT with the vector control exhibited a reproducible 10-fold decline in survival (Fig. 7). After this initial decline in viability, the survival difference between wild-type and complemented ΔspoT strain versus the ΔspoT strain with the vector control expanded from 2.5-fold to 30-fold. After 10 days of carbon starvation, the ΔspoT vector control exhibited a pronounced loss of viability (Fig. 7).
FIG. 7.
SpoT is required for swarmer cells to survive carbon starvation. Synchronized wild-type/pJL37, ΔspoT/pJL37 (uncomplemented), and ΔspoT/pJL38 (complemented) swarmer cells were washed in minimal medium lacking a carbon source, suspended in the same medium, and incubated at 28°C. Cell viability was measured as CFU/ml of culture medium and was determined at the indicated time points and normalized to viability measured at time zero. (Insert) A blown-up depiction of carbon starvation survival data from the first 30 h of the experiment. The cell viability values are the means from three independent experiments. Standard deviations were less than 25% of the means.
DISCUSSION
In this report, we demonstrate that SpoT-mediated (p)ppGpp production is necessary for mediating the C. crescentus adaptive response to carbon starvation. The depletion of glucose as the sole carbon source blocks both the swarmer-to-stalked cell polar morphogenesis and the initiation of DNA replication. Although the SpoT-mediated production of (p)ppGpp is not required to block polar morphogenesis, it is required to prevent the initiation of DNA replication. The capacity of the swarmer cell to block the initiation of DNA replication during carbon starvation stems in part from its ability to regulate the abundance of DnaA. During favorable growth conditions DNA replication is initiated upon CtrA proteolysis and the simultaneous accumulation of DnaA. During carbon starvation, the initiation of DNA replication is blocked by the SpoT-dependent degradation of DnaA. SpoT-mediated (p)ppGpp production was also found to be necessary for survival upon carbon starvation.
Carbon starvation simultaneously inhibits the swarmer-to-stalked cell differentiation program and the initiation of DNA replication.
Since C. crescentus is an oligotroph that thrives in nutrient-poor aquatic environments, its ability to control the swarmer-to-stalked cell differentiation program as well as the initiation of DNA replication may be essential for it to out-compete other microorganisms within its environmental niche. Each C. crescentus cell division is asymmetric, producing a motile swarmer cell and a sessile stalked cell. The swarmer cell's ability to block differentiation into a nonmotile stalked cell during carbon starvation provides it the opportunity to escape its unfavorable environment. Once it reaches a new environment where the carbon source is sufficient to support survival, the swarmer to stalked cell differentiation block can be removed. The swarmer cell's ability to block the initiation of DNA replication during carbon starvation also increases its prospects for survival. If DNA replication initiation is not tightly regulated during carbon starvation, stalled replication forks could arise and lead to both replication fork collapse and cell death. In E. coli, control of replication initiation is an important factor in enabling survival during replication fork arrest (50).
SpoT-dependent control of DnaA proteolysis regulates the initiation of DNA replication during carbon starvation.
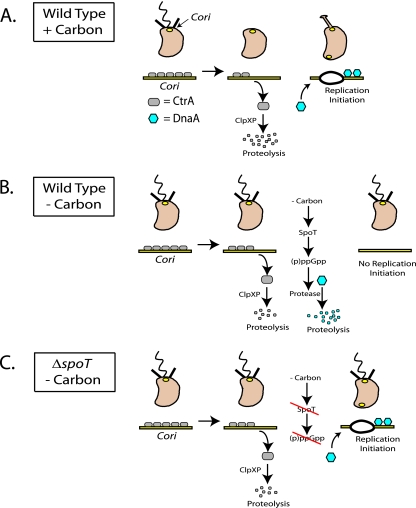
C. crescentus replicates its DNA once per cell cycle (30, 35). It achieves this control by tightly regulating the initiation event (30), which occurs upon the swarmer-to-stalked cell transition (8). DNA replication initiation is controlled by two master regulators of cell cycle progression: CtrA, which binds to and silences the origin of replication (38), and DnaA, which activates initiation (13). The precise timing of DNA replication initiation is established by tightly controlling both the activity and the abundance of CtrA and DnaA. CtrA is a member of the response regulator family of DNA binding transcription factors, and phosphorylation increases its affinity for DNA (44). In swarmer cells where CtrA is both abundant and phosphorylated, it binds to five sites within the C. crescentus origin of replication (Cori) (Fig. 8A) (32). During the swarmer-to-stalked cell transition, when CtrA is both dephosphorylated and degraded (9), the DnaA protein accumulates (6). DnaA then binds directly to the Cori sequence and stimulates DNA strand unwinding and replisome loading (Fig. 8A). During carbon starvation, the cell cycle-regulated oscillations in CtrA and DnaA abundance that allow faithful initiation of DNA replication are perturbed (Fig. 2D). Steady-state levels of DnaA in carbon-starved swarmer cells decrease due to a SpoT- and (p)ppGpp-dependent activation of proteolysis. In the absence of DnaA, the replisome is unable to load at the Cori sequence and initiate replication (Fig. 8B). In a ΔspoT strain subjected to carbon starvation, (p)ppGpp levels fail to rise, and the carbon starvation signal is not transmitted to the cellular factors responsible for activating the proteolysis of DnaA. As a result, the steady-state level of DnaA remains stable, and DNA replication can initiate (Fig. 8C), despite the fact that the cell is starving and initiation at that time could be lethal due to the generation of stalled replication forks that eventually collapse. In fact, carbon-starved ΔspoT swarmer cells that inappropriately initiate DNA replication (Fig. 6A and B) are unable to complete chromosome duplication (Fig. 6C), suggesting that their replication forks have stalled. Carbon-starved ΔspoT swarmer cells initiate DNA replication 2.5-fold more often than wild-type carbon-starved swarmer cells (Fig. 6B). We speculate that replication fork stalling and fork collapse are reflected in the threefold-higher viability loss observed among ΔspoT swarmer cells starved of carbon for 10 h (Fig. 7B).
FIG. 8.
Model for SpoT-dependent control of DnaA proteolysis and the inhibition of DNA replication initiation in carbon-starved swarmer cells. A detailed description of the figure is given in the Discussion section.
Additional mechanisms to regulate the initiation of DNA replication during carbon starvation.
While the experiments reported here indicate that SpoT and (p)ppGpp play important roles in the inhibition of DNA replication upon carbon starvation (Fig. 6A and B), our observations suggest that swarmer cells utilize additional control mechanisms to prevent the initiation of DNA replication during carbon starvation. For example, after 3 h of incubation, only 50% of carbon-starved ΔspoT swarmer cells inappropriately initiated DNA replication (Fig. 6B). While this value is greater than the proportion of carbon-starved wild-type swarmer cells that initiate DNA replication (18%), it is less than the approximately 100% of wild-type swarmer cells that initiate replication when they are grown in the presence of a carbon source for 3 h (Fig. 2B). Thus, the persistence of DnaA in carbon-starved ΔspoT swarmer cells is not sufficient to cause all cells to initiate replication. Several factors could explain this observation. First, although DnaA persists in carbon-starved ΔspoT swarmer cells during carbon starvation, it does not exhibit the approximate twofold increase in DnaA abundance that occurs in wild-type cells during the swarmer-to-stalked cell transition (6). This increase in the steady-state level of DnaA may be important for initiating DNA replication. Second, in wild-type swarmer cells starved for carbon, CtrA persists at a higher steady-state level than it does when cells undergo the swarmer-to-stalked cell transition in the presence of carbon (Fig. 2D, compare lanes 9 to 12 with lanes 3 to 4). This CtrA persistence is SpoT independent (data not shown), and the failure to completely proteolyze CtrA during carbon starvation may contribute to the suppression of DNA replication initiation. Finally, carbon starvation may not only influence the abundance of DnaA in the cell but also its activity. In E. coli, ATP-bound DnaA is more active in the initiation of DNA replication than ADP-bound DnaA (42, 62). During carbon starvation, when the swarmer cell's intracellular ATP level declines, the amount of ATP-bound DnaA may also decline and thereby limit the initiation of DNA replication.
Additional components of the SpoT-dependent pathway for DnaA proteolysis.
While it is clear that SpoT and (p)ppGpp are components of the signaling pathway that links carbon starvation to the activation of DnaA proteolysis, additional components of this signaling pathway remain to be discovered. For example, we do not yet understand how the carbon starvation signal is relayed to SpoT. Since SpoT is a bifunctional enzyme capable of both synthesizing and hydrolyzing (p)ppGpp, the key parameter that determines the cell's steady-state level of (p)ppGpp is the ratio of those two catalytic activities (34). The crystal structure of SpoT's catalytic domain from Streptococcus equisimilis revealed that the protein possesses two distinct active sites for (p)ppGpp synthesis and (p)ppGpp hydrolysis (17). These active sites are connected by a common bundle of α-helices that participate in allosteric conformational transitions to coordinate the catalytic activities of the active sites and avoid a futile cycle of synthesis and degradation (17). Therefore, it is possible that the carbon starvation signal could be propagated to SpoT by a factor that interacts with SpoT to alter its conformation, thus changing the ratio of (p)ppGpp synthesis to (p)ppGpp hydrolysis so that it tips in favor of greater net synthesis. Several recent reports of SpoT-interacting proteins suggest potential regulatory components acting upstream of SpoT. In E. coli, the acyl carrier protein, which is required for fatty acid biosynthesis, has been found to interact with SpoT (3). It has also been hypothesized that carbon starvation could disrupt fatty acid biosynthesis by decreasing the cell's pool of intracellular precursor molecules, thereby altering the acylation status of acyl carrier protein and enabling an interaction with SpoT (3). In E. coli and V. cholerae, the essential GTP-binding protein CgtA (also known as Obg) has also been shown to interact with SpoT (39, 60). In addition, in both organisms, disruption of CgtA/Obg function either by producing a mutant CgtA (23) or by depleting Obg (39) led to an increase in intracellular (p)ppGpp concentration, suggesting that CgtA's interaction with SpoT may be involved in activating (p)ppGpp hydrolysis (23, 39). C. crescentus possesses an ortholog of cgtA (cgtAc), and a temperature-sensitive allele (cgtAc G80E) has been isolated (7, 29). cgtAc G80E swarmer cells are unable to replicate their DNA during growth at the nonpermissive temperature, and it has been speculated that this G1-S-phase block in cell cycle progression is SpoT mediated through an alteration in the level of (p)ppGpp (7).
In C. crescentus, the regulatory components that function downstream of SpoT and its (p)ppGpp product are not known. During normal cell cycle progression in the presence of all requisite nutrients, the turnover of C. crescentus DnaA is largely ClpP dependent, but no evidence has been found to support a requirement for the ClpP protease chaperones ClpX and ClpA (14). It has been speculated that ClpP could be required for the rapid clearance of DnaA during carbon starvation (14). Since (p)ppGpp interacts directly and/or indirectly with RNA polymerase and leads to global alterations in transcription profiles (48), it is possible that a DnaA protease chaperone could be a member of the SpoT-dependent regulon of genes whose transcription is activated during carbon starvation, thereby targeting DnaA for ClpP-mediated proteolysis. Alternatively, the turnover of DnaA during carbon starvation may involve a protease other than ClpP, and a SpoT-mediated increase in (p)ppGpp could be directly or indirectly involved in the transcriptional activation of that starvation-induced protease. Finally, (p)ppGpp has recently been shown to interact with GTPases involved in both ribosome assembly (4) and translation initiation (33). Therefore, it is possible that a SpoT-mediated increase in (p)ppGpp could influence the translation of a factor controlling DnaA proteolysis. The identification of factors functioning both upstream and downstream of SpoT will enable us to clarify how carbon starvation signals are sensed and integrated by critical regulators of cell cycle progression like DnaA.
Acknowledgments
This work was supported by NIH grant GM51426 and by DOE grants DE-FG03ER63219-A001 and DE-FG02ER63219 to L.S. J.A.L. was supported by the Stanford NIH Genome Training Program.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Alley, M. R., J. R. Maddock, and L. Shapiro. 1993. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science 2591754-1757. [DOI] [PubMed] [Google Scholar]
- 2.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305673-688. [DOI] [PubMed] [Google Scholar]
- 3.Battesti, A., and E. Bouveret. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 621048-1063. [DOI] [PubMed] [Google Scholar]
- 4.Buglino, J., V. Shen, P. Hakimian, and C. D. Lima. 2002. Structural and biochemical analysis of the Obg GTP binding protein. Structure 101581-1592. [DOI] [PubMed] [Google Scholar]
- 5.Chiaverotti, T. A., G. Parker, J. Gallant, and N. Agabian. 1981. Conditions that trigger guanosine tetraphosphate accumulation in Caulobacter crescentus. J. Bacteriol. 1451463-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collier, J., S. R. Murray, and L. Shapiro. 2006. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J. 25346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta, K., J. M. Skidmore, K. Pu, and J. R. Maddock. 2004. The Caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal subunit levels. Mol. Microbiol. 541379-1392. [DOI] [PubMed] [Google Scholar]
- 8.Degnen, S. T., and A. Newton. 1972. Chromosome replication during development in Caulobacter crescentus. J. Mol. Biol. 64671-680. [DOI] [PubMed] [Google Scholar]
- 9.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90415-424. [DOI] [PubMed] [Google Scholar]
- 10.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204372-384. [DOI] [PubMed] [Google Scholar]
- 11.Ely, B., and R. H. Croft. 1982. Transposon mutagenesis in Caulobacter crescentus. J. Bacteriol. 149620-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbatyuk, B., and G. T. Marczynski. 2001. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol. Microbiol. 40485-497. [DOI] [PubMed] [Google Scholar]
- 14.Gorbatyuk, B., and G. T. Marczynski. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol. Microbiol. 551233-1245. [DOI] [PubMed] [Google Scholar]
- 15.Haseltine, W. A., and R. Block. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. USA 701564-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinz, A. J., D. E. Larson, C. S. Smith, and Y. V. Brun. 2003. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol. Microbiol. 47929-941. [DOI] [PubMed] [Google Scholar]
- 17.Hogg, T., U. Mechold, H. Malke, M. Cashel, and R. Hilgenfeld. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p) ppGpp metabolism during the stringent response. Cell 11757-68. [DOI] [PubMed] [Google Scholar]
- 18.Hottes, A. K., L. Shapiro, and H. H. McAdams. 2005. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 581340-1353. [DOI] [PubMed] [Google Scholar]
- 19.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 175658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenal, U., and L. Shapiro. 1996. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 152393-2406. [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen, R. B., and L. Shapiro. 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. USA 9610661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, R. B., S. C. Wang, and L. Shapiro. 2001. A moving DNA replication factory in Caulobacter crescentus. EMBO J. 204952-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, M., S. M. Sullivan, P. K. Wout, and J. R. Maddock. 2007. G-protein control of the ribosome-associated stress response protein SpoT. J. Bacteriol. 1896140-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, S., K. Watanabe, H. Suzuki, and M. Watarai. 2005. Roles of Brucella abortus SpoT in morphological differentiation and intramacrophagic replication. Microbiology 1511607-1617. [DOI] [PubMed] [Google Scholar]
- 25.Lagenaur, C., and N. Agabian. 1977. Caulobacter crescentus pili: structure and stage-specific expression. J. Bacteriol. 131340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam, H., W. B. Schofield, and C. Jacobs-Wagner. 2006. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 1241011-1023. [DOI] [PubMed] [Google Scholar]
- 27.Laub, M. T., L. Shapiro, and H. H. McAdams. 2007. Systems biology of Caulobacter. Annu. Rev. Genet. 41429-441. [DOI] [PubMed] [Google Scholar]
- 28.Levine, A., F. Vannier, M. Dehbi, G. Henckes, and S. J. Seror. 1991. The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J. Mol. Biol. 219605-613. [DOI] [PubMed] [Google Scholar]
- 29.Maddock, J., A. Bhatt, M. Koch, and J. Skidmore. 1997. Identification of an essential Caulobacter crescentus gene encoding a member of the Obg family of GTP-binding proteins. J. Bacteriol. 1796426-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marczynski, G. T. 1999. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J. Bacteriol. 1811984-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marczynski, G. T., A. Dingwall, and L. Shapiro. 1990. Plasmid and chromosomal DNA replication and partitioning during the Caulobacter crescentus cell cycle. J. Mol. Biol. 212709-722. [DOI] [PubMed] [Google Scholar]
- 32.Marczynski, G. T., and L. Shapiro. 2002. Control of chromosome replication in Caulobacter crescentus. Annu. Rev. Microbiol. 56625-656. [DOI] [PubMed] [Google Scholar]
- 33.Milon, P., E. Tischenko, J. Tomsic, E. Caserta, G. Folkers, A. La Teana, M. V. Rodnina, C. L. Pon, R. Boelens, and C. O. Gualerzi. 2006. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl. Acad. Sci. USA 10313962-13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray, K. D., and H. Bremer. 1996. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J. Mol. Biol. 25941-57. [DOI] [PubMed] [Google Scholar]
- 35.Nathan, P., M. A. Osley, and A. Newton. 1982. Circular organization of the DNA synthetic pathway in Caulobacter crescentus. J. Bacteriol. 151503-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostling, J., K. Flardh, and S. Kjelleberg. 1995. Isolation of a carbon starvation regulatory mutant in a marine Vibrio strain. J. Bacteriol. 1776978-6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul, B. J., M. B. Berkmen, and R. L. Gourse. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. USA 1027823-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA 95120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raskin, D. M., N. Judson, and J. J. Mekalanos. 2007. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 1044636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt, J. M., and R. Y. Stanier. 1966. The development of cellular stalks in bacteria. J. Cell Biol. 28423-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreiber, G., E. Z. Ron, and G. Glaser. 1995. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr. Microbiol. 3027-32. [DOI] [PubMed] [Google Scholar]
- 42.Sekimizu, K., D. Bramhill, and A. Kornberg. 1987. ATP activates DnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50259-265. [DOI] [PubMed] [Google Scholar]
- 43.Shapiro, L., and J. V. Maizel, Jr. 1973. Synthesis and structure of Caulobacter crescentus flagella. J. Bacteriol. 113478-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siam, R., and G. T. Marczynski. 2000. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 191138-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skerker, J. M., and M. T. Laub. 2004. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2325-337. [DOI] [PubMed] [Google Scholar]
- 46.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 193223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spira, B., N. Silberstein, and E. Yagil. 1995. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J. Bacteriol. 1774053-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srivatsan, A., and J. D. Wang. 2008. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11100-105. [DOI] [PubMed] [Google Scholar]
- 49.Stephens, C., A. Reisenauer, R. Wright, and L. Shapiro. 1996. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc. Natl. Acad. Sci. USA 931210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sutera, V. A., Jr., and S. T. Lovett. 2006. The role of replication initiation control in promoting survival of replication fork damage. Mol. Microbiol. 60229-239. [DOI] [PubMed] [Google Scholar]
- 51.Thanbichler, M., and L. Shapiro. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126147-162. [DOI] [PubMed] [Google Scholar]
- 52.Tsai, J. W., and M. R. Alley. 2001. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J. Bacteriol. 1835001-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinella, D., C. Albrecht, M. Cashel, and R. D'Ari. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56958-970. [DOI] [PubMed] [Google Scholar]
- 54.Viollier, P. H., N. Sternheim, and L. Shapiro. 2002. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 214420-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viollier, P. H., M. Thanbichler, P. T. McGrath, L. West, M. Meewan, H. H. McAdams, and L. Shapiro. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. USA 1019257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, J. D., G. M. Sanders, and A. D. Grossman. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wells, D. H., and S. R. Long. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 431115-1127. [DOI] [PubMed] [Google Scholar]
- 58.Wendrich, T. M., G. Blaha, D. N. Wilson, M. A. Marahiel, and K. H. Nierhaus. 2002. Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10779-788. [DOI] [PubMed] [Google Scholar]
- 59.Winzeler, E., and L. Shapiro. 1995. Use of flow cytometry to identify a Caulobacter 4.5S RNA temperature-sensitive mutant defective in the cell cycle. J. Mol. Biol. 251346-365. [DOI] [PubMed] [Google Scholar]
- 60.Wout, P., K. Pu, S. M. Sullivan, V. Reese, S. Zhou, B. Lin, and J. R. Maddock. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 1865249-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 2665980-5990. [PubMed] [Google Scholar]
- 62.Yung, B. Y., E. Crooke, and A. Kornberg. 1990. Fate of the DnaA initiator protein in replication at the origin of the Escherichia coli chromosome in vitro. J. Biol. Chem. 2651282-1285. [PubMed] [Google Scholar]
- 63.Zyskind, J. W., and D. W. Smith. 1992. DNA replication, the bacterial cell cycle, and cell growth. Cell 695-8. [DOI] [PubMed] [Google Scholar]