Abstract
We have characterized and quantified a form of bacterial chemotaxis that manifests only as an emergent property by measuring symmetry breaking in a swarm of Myxococcus xanthus exposed to a two-dimensional nutrient gradient from within an agar substrate. M. xanthus chemotaxis requires cell-cell contact and coordinated motility, as individual motile cells exhibit only nonvectorial movement in the presence of a nutrient gradient. Genes that specifically affect M. xanthus chemotaxis include at least 10 of the 53 that express enhancer binding proteins of the NtrC-like class, an indication that this behavior is controlled through transcription, most likely by a complex signal transduction network.
Some of the conditions that define a bacterial swarm—genetically identical cells in close proximity—are theoretically both necessary and sufficient to shift the unit of selection from the individual to the group, i.e., a swarm evolves as a superorganism (5, 34). As a consequence, cellular autonomy diminishes, and each component cell becomes a superposable subunit with a genetic instruction set optimized for the swarm. This instruction set specifies both individual and group responses through interdependent signal transduction networks, which link inputs from neighboring cells and the environment to outputs that control each response (28). The behavior of the swarm evolves to become emergent, manifesting structures, patterns, and properties during the process of self-organization (8). Nature is rife with examples (3, 7).
A Myxococcus xanthus swarm is a model organism used to study prokaryotic multicellularity (4, 19). Its best-characterized behavior is the starvation stress response known as development, during which cells break symmetry and move to form dome-shaped aggregates called fruiting bodies (18). Each fruiting body is made up of approximately 1 × 105 cells, some of which differentiate into metabolically quiescent and environmentally resistant myxospores (20). From an experimental perspective, a spore-filled fruiting body provides a verifiable endpoint that can be used as a quantifiable metric; a development assay typically measures the number of viable myxospores produced by a swarm over a given period of time following the onset of starvation stress (9). Because these results are quantifiable, they can be used to make definitive statements about relative phenotypic differences among M. xanthus mutant strains, making possible both rank and cluster analysis.
Development represents only one response to nutrient limitation, and some researchers have reported a second: M. xanthus can also exhibit a chemotactic response to a gradient of nutrients (22, 30). These findings are controversial, however, since other reports claim there is no response (12, 35) (for a summary of the controversy, see Discussion). Unlike development, where sporulation efficiency functions as a quantifiable metric, all previous analyses of M. xanthus swarm chemotaxis have been qualitative in nature. Thus, genetic effects could not be ranked or clustered according to phenotype, and this has made a comparative analysis nearly impossible. A rigorous quantitative assay is required.
MATERIALS AND METHODS
Culture preparation.
M. xanthus strains were inoculated into flasks containing CTTYE broth (1.0% Casitone, 0.5% yeast extract, 10.0 mM Tris-HCl, 1.0 mM KH2PO4, and 8.0 mM MgSO4) and incubated in the dark at 32°C with vigorous swirling. Once the culture reached a density of 5 × 108 cells/ml, the cells were washed, pelleted, and resuspended in TPM buffer (10.0 mM Tris-HCl, 1.0 mM KH2PO4, and 8.0 mM MgSO4) to a density of 5 × 109 cells/ml.
Generation of mutants.
Plasmids containing internal fragments of target genes were electroporated into M. xanthus cells using standard techniques (29). Following electroporation, the cells were placed into flasks containing 1.5 ml of CTTYE broth and incubated at 32°C for 24 h with vigorous swirling. Aliquots (500 μl) of these cultures were then added to 5.0 ml of 50°C CTTYE soft agar and poured onto CTTYE plates containing kanamycin (40 μl/ml). To confirm the composition of the insertions, chromosomal DNA was isolated from kanamycin-resistant colonies and used in PCRs containing the appropriate gene-specific primers (37).
Nutritive-disk construction.
A sterile 0.5-mm-thick silicone rubber gasket (Grace Bio-Labs) was placed on top of a flame-sterilized glass microscope slide, forming a small well. Molten 1% agar in CTTYE broth was poured into this well and covered by a second flame-sterilized microscope slide, and the slides were clamped together to flatten the CTTYE agar. When the CTTYE agar had cooled and hardened, the clamps and one of the slides were removed. A 1-mm diameter nutritive disk was extracted from the well using a microsampling pipette with a 100-μl glass disposable tip (Fisher).
Tracking assay apparatus construction.
The nutritive disk was placed into a well created by a sterile gasket placed on a flame-sterilized coverslip. This well was then filled with molten 1% agar in TPM buffer that had been allowed to cool to 60°C to prevent the nutritive disk from melting. The gasket/coverslip was then covered by a flame-sterilized microscope slide, clamped to flatten the agar, and cooled to room temperature. Once the agar hardened, the clamp was removed and the slide was separated from the gasket/coverslip. This resulted in a smooth agar surface free of any visible defects or stress lines. A 0.5-μl aliquot of the resuspended (5 × 109 cells/ml) M. xanthus liquid culture was spotted so the swarm edge was 1.14 ± 0.04 mm away from the CTTYE disk (now embedded within the TPM buffer/agar) and allowed to dry for 1 min. A second gasket was placed over the first gasket to create an air space, and the entire apparatus was covered with a flame-sterilized microscope slide (Fig. 1A and B). The time interval between spotting and the initiation of image acquisition is no more than 5 min, and this time is taken into account when calculating the number of hours after the initiation of the tracking assay (T0 and Tn).
FIG. 1.
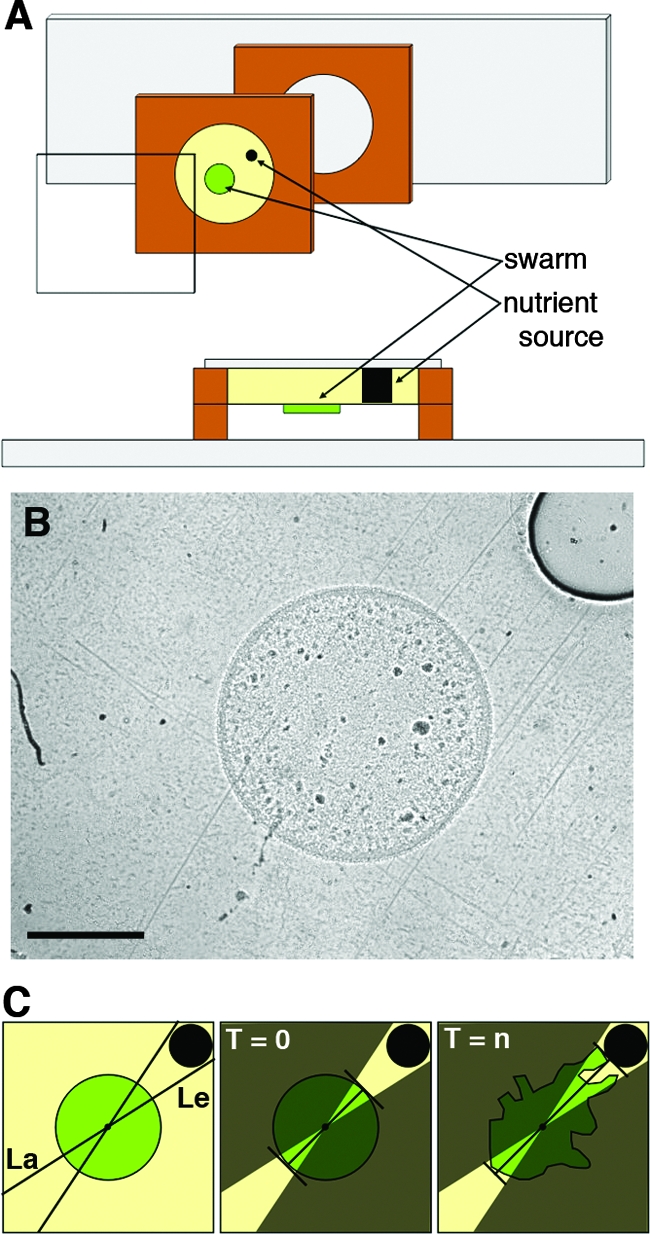
Tracking assay and quantification. A glass coverslip behind a silicon gasket creates a well that contains an agar substrate with an embedded nutritive disk. This is aligned with a second gasket that has been placed on a microscope slide. (A) Exploded view and cross section of the apparatus. (B) 20× bright-field image of tracking assay apparatus at T0. Scale bar, 1 mm. (C) Diagram of quantification protocol used to define and measure the leading (Le) and lagging (La) edges to determine the TR. See the text for an explanation of T0 and Tn.
Swarm image acquisition.
Completed tracking assay slides were placed on a heated stage (20/20 Technology, Inc.) and maintained at 32°C. Images were acquired every 60 s for a period of 24 h using a Nikon Eclipse E400 microscope equipped with an Insight Firewire camera (model no. 11.0 Monochrome; Diagnostic Images, Inc.) controlled through a computer (Apple, Inc.) running SPOT image acquisition software (Diagnostic Images, Inc.). The acquired images were automatically saved in a sequentially numbered tagged image file format (tiff). Once the assay was completed, the images were compiled into a time-lapse video using QuickTime (Apple, Inc.).
Initiation and quantification of the tracking assay.
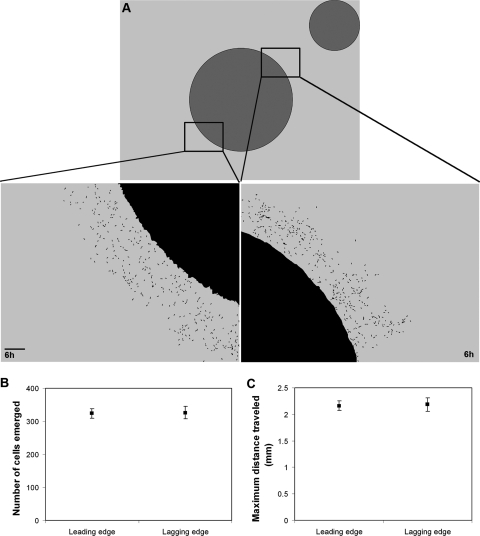
The tracking assay is initiated (T0) when the 0.5 μl of 3 × 106 M. xanthus cells dries (<1 min) (Fig. 1B). Reaching the nutritive disk, or an elapsed time of 24 h, represents a verifiable endpoint (Tn). We digitally marked the swarm and nutritive disk circumferences at T0 and used them to determine both the swarm and nutritive-disk centroids. We then used these two points to draw a straight line that transected both the T0 center (called the original centroid) of the Tn swarm and the center of the nutritive disk (called the center line). Next, we drew two straight lines tangential to the nutritive disk that transected the Tn swarm's original centroid (Fig. 1C). This defined two equal areas of the swarm that were geometrically opposite, one being closest to the nutritive disk (leading edge) and the other being furthest from the nutritive disk (lagging edge). The swarm expands during the assay through a series of group translocations called flares (13), and symmetry breaking is detected by measuring the ratio of the furthest distance traveled by a flare on the leading edge to the furthest distance traveled by a flare on the lagging edge from T0 to Tn (flare length line), where n is the number of hours required for the first flare of the leading edge to reach the nutritive disk, with a maximum Tn of 24 h (Fig. 1C). Perpendicular lines from the ends of both of the flare length lines to the center line were added to create normalized points on the center line. The distances from the original swarm edge along the center line to these normalized points were then used to generate tracking ratios (TR). The TRs were ranked and compared using the Duncan's multiple-range test procedure within the statistical analysis software package SAS (SAS Institute, Inc.). The images were processed using the ImageJ (http://rsb.info.nih.gov/ij) and Photoshop (Adobe Systems, Inc.) software packages. No filters were applied to any of the images or videos presented with the exception of Fig. 3A. The images of leading and lagging edges in Fig. 3A were processed to help clarify individual cells using Photoshop filters in the following order: auto levels, find edges, and threshold.
FIG. 3.
Chimeric swarm behavior. DK1622 (wt) cells diluted 1:100 into a DK11316 (A− S−) mutant background were subjected to the tracking assay for 6 h, and images were captured of the leading and the lagging edges. (A) The threshold images diagram leading and lagging edge reference points at T0 and show individual cells that have moved outside the nonmotile swarm at a Tn of 6. The number of (B) and maximum distance traveled by (C) individual cells that emerged from both the leading and lagging edges are shown. Scale bar in panel A, 0.1 mm.
Confirmation of nutritive gradient.
To prove that a gradient could be established and to measure the lower limit of diffusion in 1% agar, the CTTYE agar in the nutritive disk was replaced with Quantum dots (Q-dots) (Quantum Dot Corp.), which are inert fluorescent semiconductors that approximate the size of a large protein (∼10 nm in diameter). Fluorescent images were acquired as described above. Q-dots produced a visible fluorescent gradient that diffused 1 mm within the first 3 h and more than 2 mm within the first 6 h. These results confirm that nutrient molecules as large as a 10-nm diameter protein will be present in the diffusion front and will cross the leading edge of the expanding swarm within the first 2 h of the assay.
Elimination of elasticotaxis as a variable.
To determine if the asymmetry of the swarm expansion during a tracking assay was due to stress forces caused by the placement of the nutritive disk, we replaced it with a nonnutritive disk containing 1% agar in TPM buffer. The tracking assay was performed under these conditions and the results quantified as described above.
RESULTS
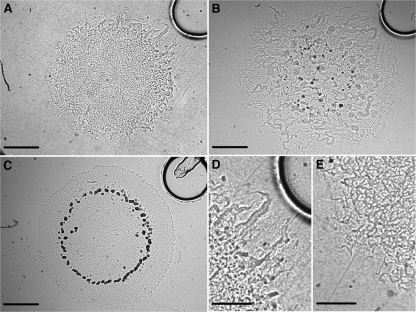
We have developed a tracking assay to quantify M. xanthus chemotaxis. In some ways the tracking assay is similar to the chemical-in-plug assay first described by Tso and Adler (36), but it has been modified and adapted for digital microcinematography and time-lapse analysis. The assay surface is nonnutritive agar in which a smaller nutritive-agar disk is embedded. The nutritive-agar and nonnutritive-agar disks are identical in composition, except for the presence of Casitone and yeast extract in the nutritive-agar disk; over several hours, a gradient extends out from the nutritive disk into the nonnutritive agar (see Fig. S1 in the supplemental material). Because the nutritive disk is covered by nonnutritive agar, the assay surface is smooth, thus eliminating any possible swarm response to surface irregularities, such as elasticotaxis (11, 13). To initiate a tracking assay, a swarm is created by spotting M. xanthus liquid culture onto the nonnutritive-agar surface approximately 1 mm from the edge of the nutritive disk. The swarm expands over several hours, and we report chemotaxis as its TR, which is a measure of expansion asymmetry. A TR of >1.0 indicates movement toward the nutrient source, a TR of <1.0 indicates movement away from the nutrient source, and a TR of 1.0 indicates symmetric movement. Wild-type (wt) cells have a TR of 1.94 ± 0.20 (Fig. 2A; see Video S1 in the supplemental material); in contrast, the same assay performed using a nonnutrient control disk results in a TR of 1.01 ± 0.03 (see Video S2 in the supplemental material).
FIG. 2.
Symmetry breaking during a tracking assay in M. xanthus. (A) A swarm of DK1622 (wt) (Tn = 6 h; TR = 1.94 ± 0.20). (B) A swarm of DK1253 (A+ S−; Tn = 18 h; TR = 1.23 ± 0.06). (C) A swarm of DK1218 (A− S+; Tn = 23 h; TR = 0.90 ± 0.15). (D and E) Higher magnification view of DK1622 leading and lagging edges, respectively. Scale bars in panels A to C, 1 mm; scale bars in panels D and E, 0.5 mm.
The asymmetry in swarm expansion indicates that cells are moving up the gradient and toward the nutritive disk; however, we must eliminate the possibility that this asymmetry is a growth response to differing nutrient levels across the swarm. Higher nutrient levels further up the gradient could accelerate the growth and division rates of cells at the leading edge, which is always closer to the nutritive disk, thus increasing the rate of expansion and resulting in a TR of >1.0. We used a genetic approach to explore this hypothesis, by performing a series of experiments with motility mutant strains DK1218 and DK1253 (16, 17). M. xanthus moves across agar via gliding motility, which can be genetically dissected into two distinct subdivisions called adventurous (A) and social (S) motility; DK1218 (genotype cglB2) is defective in A motility, while DK1253 (genotype tgl-1) is defective in S motility. Either A or S motility alone is sufficient for swarm expansion to occur. If the asymmetric expansion observed in wt swarms is caused by increased growth and division rates as a function of proximity to the nutritive disk, then the genetics of cell motility would be of no consequence, and both DK1218 and DK1253 swarms should produce TRs greater than 1.0. This is not observed: DK1253 swarms exhibit asymmetric expansion (TR = 1.23 ± 0.06) (Fig. 2B), but DK1218 swarms expand symmetrically (TR = 0.90 ± 0.15) (Fig. 2C). Therefore, the asymmetry of wt swarm expansion requires motility. In fact, it specifically requires A motility, which indicates that chemotaxis in M. xanthus is not a passive process. However, the data clearly show that the synergistic interactions of both A and S motility are required to produce a wt-level response.
An M. xanthus cell reverses direction by switching its leading pole (33), and isolated cells on an agar substrate have been shown to respond to a nutrient gradient by changing the frequency of their reversals (22, 30). This change lacks any observable polarity, however, so that it produces no net cell translocation that could cause the cell to move up a gradient. Therefore, the response of isolated cells to a gradient of nutrients has been more accurately described as a type of nondirected chemokinesis (38), rather than chemotaxis. Although these isolated cell data may seem to contradict the results of the tracking assay, they are at least partially consistent: if chemotaxis in M. xanthus is the result of all the cells in a swarm sensing and responding to a gradient autonomously by changing their reversal frequencies in a directed fashion to move up the gradient, then all cells and every flare within the gradient would move toward the nutritive disk. No swarm exhibits this phenotype at any time during the tracking assay. Instead, a swarm expands away from the center at all points during a tracking assay, and these observations are consistent with individual cell data that report a nondirected response. An observation that is not consistent with a nondirected response is that the flares on the leading edge of a wt swarm can be easily distinguished from flares on the lagging edge: leading-edge flares appear larger, thicker, and straighter (Fig. 2D) than lagging-edge flares, which appear more “spidery” (Fig. 2E). Perhaps a cell exhibits nondirected movement when isolated and chemotaxis when in a swarm, because a swarm provides the “context” within which M. xanthus cells interact with their environment?
Several physical properties are known to contribute to a swarm's context: the mass of a swarm that exerts stress on the agar substrate, which individual cells can sense and respond to through elasticotaxis (11, 13); the exopolysaccharide matrix generated by the swarm, which cells require for normal motility (21, 31); and the quorum sensing and cell-cell contact signals that require groups of cells in close proximity, which are known to alter individual cell behavior (21). Perhaps there is some subset of these properties that enables a single M. xanthus cell to direct its reversal frequency so that it moves up a gradient of nutrients. To investigate this hypothesis, we performed the tracking assay using a swarm in which a small number (1%) of motile wt cells were distributed throughout a swarm (99%) of DK11316 nonmotile mutant cells (genotype pilA::Tcr, ΔcglB) (Fig. 3A). This swarm chimera provides a significant subset of swarm context, such as the elasticotactic force, the exopolysaccharide matrix, and 3 × 106 living cells in close proximity for quorum sensing and cell-cell contact signaling. Although the cells in this swarm can communicate with each other, they cannot move together as a group because only 1% of the cells are capable of movement, and they are distributed throughout the swarm. Under these conditions, if the 1% of wt cells exhibit chemotaxis, by moving up the nutrient gradient from within the chimeric swarm, then more would emerge from the leading edge than from the lagging edge. This is not observed. Instead, equal numbers of cells emerge from both edges (Fig. 3B) and travel the same maximum distance away from the swarm center (Fig. 3C). These data confirm that individual M. xanthus cells do not exhibit chemotaxis, even within the context of a nonmotile living swarm. M. xanthus chemotaxis is a multicellular behavior that requires the movement of a swarm, and in this way, it is like development.
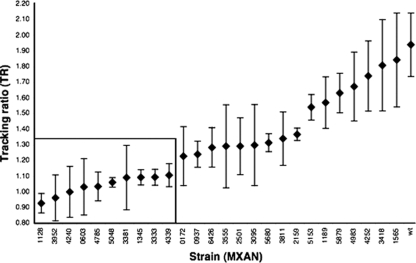
From a genomic perspective, development is a spatiotemporal cascade of transcription events controlled, at least in part, by σ54 enhancer binding proteins (EBPs) that exhibit significant homology to the NtrC-like class of activators (10, 23, 27). The M. xanthus genome contains 53 NtrC-like EBPs, an inordinate number compared to other prokaryotes, and approximately one-third of NtrC-like EBPs are involved in the control of development (6, 14). Some of these EBPs might also function in chemotaxis. Therefore, we examined the distribution of chemotaxis phenotypes among M. xanthus mutant strains containing one of the NtrC-like EBPs disrupted through homologous recombination. Mutant strains with no observable deviation from the wt phenotype were of particular interest because aberrant chemotaxis phenotypes would not be obscured by a more general motility defect. Using a set of standard assays (cell growth, swarm motility, and development) (6), we identified 26 out of the 53 mutant strains that appeared to have an overall wt phenotype. We then performed tracking assays on these 26 strains and ranked them by mean TR (Fig. 4). The overall distribution of TRs is continuous, with significantly different mutant classes, as determined by a one-way analysis of variance (F27,72 = 2.04; P = 0.0088). Using Duncan's multiple-range test, we identified 10 strains displaying TRs significantly different from the wt (Fig. 4).
FIG. 4.
Ranking NtrC-like EBPs by TR. Tracking assays were performed on each of the 26 NtrC-like EBP single gene disruption mutant strains that displayed no defect in growth rate, swarm expansion, or development. The TR results are displayed in the order of increasing mean (TR ± SE) (n = 3). NtrC-like EBPs found to be chemotaxis-specific are in the rectangle.
DISCUSSION
In this report, we have quantified M. xanthus swarm chemotaxis in response to a two-dimensional gradient of nutrients, and we have demonstrated that the response is the result of neither asymmetric growth nor elasticotaxis. Swarm motility is required for M. xanthus chemotaxis, as opposed to cell motility, and this distinction means that, like development, it is a multicellular response to an external stimulus. Also, like development, M. xanthus chemotaxis is affected by the disruption of genes that regulate transcription, specifically the NtrC-like EBPs, and a quantitative analysis reveals a continuous distribution of swarm phenotypes for both responses (A. G. Garza, personal communication). These data indicate that M. xanthus chemotaxis may have a complex and branching signal transduction schema, similar to the one proposed for the control of development (24).
Previous authors have referred to the movement of M. xanthus in response to chemical gradients as chemotaxis, but others avoid this term because it provokes a second and unrelated controversy regarding the exact definition of chemotaxis. We do not propose or imply any mechanistic connection between chemotaxis in M. xanthus and chemotaxis in flagellated bacteria (1). Because of the long and controversial history of chemotaxis in M. xanthus, here we provide a summary.
It was first hypothesized that chemotaxis directed cells toward aggregation points during development; in 1954, Lev reported that fruiting bodies produced diffusible substances that stimulated fruiting-body formation (25). McVittie and Zahler claimed to confirm the existence of this chemotactic substance in 1962 (26), but no substance was ever isolated. In 1981, Shimkets and Dworkin reported that development requires cells to be touching one another (32), and this finding meant that models for development no longer required cells to communicate with each other over long distances via a diffusible chemoattractant. An unintended effect of the Shimkets and Dworkin study was to shift the focus of research away from fruiting bodies as the source of a chemoattractant and toward external chemical signals from nutrients and prey.
In 1983, Dworkin concluded that directed motility in M. xanthus does occur but not as a response to chemical gradients (11). He surmised that what seemed to be chemotaxis was, in fact, elasticotaxis, the directed movement of cells perpendicular to stress lines in agar. In an accompanying report, Dworkin and Eide reported no swarm chemotaxis toward gradients of a variety of defined and complex materials, including the nutrients Casitone and yeast extract (12). Furthermore, they speculated that chemotaxis is neither “mechanistically appropriate nor developmentally or ecologically useful” for M. xanthus, due to the fact that the cells exist in a swarm and move too slowly to detect diffusing nutrients. This speculation assumed only the biased-random-walk model of individual cell chemotaxis.
In 1993, Shi et al. devised a chemotaxis assay that was based on a one-dimensional steep and stable chemical gradient (30) and reported that M. xanthus swarms do show chemotaxis toward nutrients, including Casitone and yeast extract. In 1996, Tieman et al. claimed to improve the assay but reported no chemotaxis toward the same nutrients (35). Both sets of contradictory findings were based on qualitative assessments of swarm expansion; we were able to quantify the figure in the Tiemen et al. manuscript that showed swarm expansion in the presence of a one-dimensional nutrient gradient by calculating the TR and determined it to be 1.3 to 1.4. In other words, although it was not clear to the authors, M. xanthus did exhibit chemotaxis, and therefore, the Tieman et al. results actually corroborated the results of Shi et al. More than anything else, these data clearly demonstrate the importance of a quantifiable metric when performing a comparative analysis of behavior. By providing such a metric, we were able to make an accurate comparison of asymmetric expansion among mutant M. xanthus strains and thus begin to characterize the behavioral genetics of chemotaxis. In particular, the broad effect of EBP disruption on swarm phenotype has profound implications regarding the complexity of the signal transduction network.
Other than the 1983 report by Dworkin and Eide and the 1996 report by Tieman et al., all studies on M. xanthus swarms have reported chemotaxis up a gradient of nutrients. In addition, the apparent contradiction between swarm chemotaxis and the nondirected movement of individual cells makes sense. M. xanthus chemotaxis satisfies all six requirements for emergence, as defined by Goldstein (15): (i) a swarm is an entity, rather than a population, in that it represents a global, or macro level (ii) that exhibits coherence or correlation, and (iii) chemotaxis is a dynamical process (iv) that is ostensive and (v) that exhibits both radical novelty and (vi) supervenience. Given the wide variety of other molecular mechanisms that control behavior in M. xanthus, including quorum sensing (2), localized cell-cell communication (21, 31), and stigmergy (11, 13), we hypothesize that the most plausible model for M. xanthus chemotaxis resembles a form of swarm intelligence rather than the canonical biased random walk indicative of chemotaxis in flagellated bacteria.
Supplementary Material
Acknowledgments
This research was made possible by a National Science Foundation Career award (MCB-0746066, Characterization of Transcriptional Activators that Regulate Emergent Behavior) to R.D.W. and a Syracuse University graduate research fellowship to R.G.T.
We thank A. G. Garza for providing plasmids and mutant strains and W. T. Starmer for comments and help with statistical analysis. We are grateful to L. J. Shimkets, B. S. Goldman, G. Suen, M. Singer, L. G. Welch, K. A. Murphy, H. G. Taylor, B. I. Arshinoff, and K. Longacker for helpful discussions and comments on the manuscript.
Footnotes
Published ahead of print on 22 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexandre, G., and I. B. Zhulin. 2001. More than one way to sense chemicals. J. Bacteriol. 1834681-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13273-286. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Jacob, E. 2003. Bacterial self-organization: co-enhancement of complexification and adaptability in a dynamic environment. Philos. Trans. A 3611283-1312. [DOI] [PubMed] [Google Scholar]
- 4.Bonner, J. T. 2000. First signals: the evolution of multicellular development. Princeton University Press, Princeton, NJ.
- 5.Buss, L. W. 1987. The evolution of individuality. Princeton University Press, Princeton, NJ.
- 6.Caberoy, N. B., R. D. Welch, J. S. Jakobsen, S. C. Slater, and A. G. Garza. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 1856083-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camazine, S., J. L. Deneubourg, N. R. Franks, J. Sneyd, G. Theraylaz, and E. Bonabeau. 2001. Self-organization in biological systems. Princeton University Press, Princeton, NJ.
- 8.Corning, P. A. 2005. Holistic Darwinism: synergy, cybernetics, and the bioeconomics of evolution. University of Chicago Press, Chicago, IL.
- 9.Curtis, P. D., R. G. Taylor, R. D. Welch, and L. J. Shimkets. 2007. Spatial organization of Myxococcus xanthus during fruiting body formation. J. Bacteriol. 1899126-9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diodati, M. E., F. Ossa, N. B. Caberoy, I. R. Jose, W. Hiraiwa, M. M. Igo, M. Singer, and A. G. Garza. 2006. Nla18, a key regulatory protein required for normal growth and development of Myxococcus xanthus. J. Bacteriol. 1881733-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dworkin, M. 1983. Tactic behavior of Myxococcus xanthus. J. Bacteriol. 154452-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dworkin, M., and D. Eide. 1983. Myxococcus xanthus does not respond chemotactically to moderate concentration gradients. J. Bacteriol. 154437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontes, M., and D. Kaiser. 1999. Myxococcus cells respond to elastic forces in their substrate. Proc. Natl. Acad. Sci. USA 968052-8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 10315200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein, J. 1999. Emergence as a construct: history and issues. Emergence 149-72. [Google Scholar]
- 16.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): genes controlling movement of single cells. Mol. Gen. Genet. 171167-176. [Google Scholar]
- 17.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol. Gen. Genet. 171177-191. [Google Scholar]
- 18.Igoshin, O. A., D. Kaiser, and G. Oster. 2004. Breaking symmetry in myxobacteria. Curr. Biol. 14R459-R462. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser, D. 2001. Building a multicellular organism. Annu. Rev. Genet. 35103-123. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 145-54. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser, D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 5875-98. [DOI] [PubMed] [Google Scholar]
- 22.Kearns, D. B., and L. J. Shimkets. 1998. Chemotaxis in a gliding bacterium. Proc. Natl. Acad. Sci. USA 9511957-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keseler, I. M., and D. Kaiser. 1997. σ54, a vital protein for Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 941979-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kroos, L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 4113-39. [DOI] [PubMed] [Google Scholar]
- 25.Lev, M. 1954. Demonstration of a diffusible fruiting factor in Myxobacteria. Nature 173501.13144794 [Google Scholar]
- 26.McVittie, A., and S. A. Zahler. 1962. Chemotaxis in Myxococcus. Nature 1941299-1300. [Google Scholar]
- 27.Morett, E., and L. Segovia. 1993. The σ54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 1756067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkinson, J. S., P. Ames, and C. A. Studdert. 2005. Collaborative signaling by bacterial chemoreceptors. Curr. Opin. Microbiol. 8116-121. [DOI] [PubMed] [Google Scholar]
- 29.Plamann, L., J. M. Davis, B. Cantwell, and J. Mayor. 1994. Evidence that asgB encodes a DNA-binding protein essential for growth and development of Myxococcus xanthus. J. Bacteriol. 1762013-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi, W., T. Köhler, and D. R. Zusman. 1993. Chemotaxis plays a role in the social behaviour of Myxococcus xanthus. Mol. Microbiol. 9601-611. [DOI] [PubMed] [Google Scholar]
- 31.Shimkets, L. J. 1990. Social and developmental biology of the myxobacteria. Microbiol. Rev. 54473-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimkets, L. J., and M. Dworkin. 1981. Excreted adenosine is a cell density signal for the initiation of fruiting body formation in Myxococcus xanthus. Dev. Biol. 8451-60. [DOI] [PubMed] [Google Scholar]
- 33.Spormann, A. M., and A. D. Kaiser. 1995. Gliding movements in Myxococcus xanthus. J. Bacteriol. 1775846-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szathmáry, E., and J. M. Smith. 1995. The major evolutionary transitions. Nature 374227-232. [DOI] [PubMed] [Google Scholar]
- 35.Tieman, S., A. Koch, and D. White. 1996. Gliding motility in slide cultures of Myxococcus xanthus in stable and steep chemical gradients. J. Bacteriol. 1783480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tso, W. W., and J. Adler. 1974. Negative chemotaxis in Escherichia coli. J. Bacteriol. 118560-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viswanathan, P., T. Ueki, S. Inouye, and L. Kroos. 2007. Combinatorial regulation of genes essential for Myxococcus xanthus development involves a response regulator and a LysR-type regulator. Proc. Natl. Acad. Sci. USA 1047969-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward, M. J., K. C. Mok, and D. R. Zusman. 1998. Myxococcus xanthus displays Frz-dependent chemokinetic behavior during vegetative swarming. J. Bacteriol. 180440-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.