Abstract
Polyomavirus and papillomavirus (papovavirus) capsids are composed of 72 capsomeres of their major capsid proteins, VP1 and L1, respectively. After translation in the cytoplasm, L1 and VP1 pentamerize into capsomeres and are then imported into the nucleus using the cellular α and β karyopherins. Virion assembly only occurs in the nucleus, and cellular mechanisms exist to prevent premature capsid assembly in the cytosol. We have identified the karyopherin family of nuclear import factors as possible “chaperones” in preventing the cytoplasmic assembly of papovavirus capsomeres. Recombinant murine polyomavirus (mPy) VP1 and human papillomavirus type 11 (HPV11) L1 capsomeres bound the karyopherin heterodimer α2β1 in vitro in a nuclear localization signal (NLS)-dependent manner. Because the amino acid sequence comprising the NLS of VP1 and L1 overlaps the previously identified DNA binding domain, we examined the relationship between karyopherin and DNA binding of both mPy VP1 and HPV11 L1. Capsomeres of L1, but not VP1, bound by karyopherin α2β1 or β1 alone were unable to bind DNA. VP1 and L1 capsomeres could bind both karyopherin α2 and DNA simultaneously. Both VP1 and L1 capsomeres bound by karyopherin α2β1 were unable to assemble into capsids, as shown by in vitro assembly reactions. These results support a role for karyopherins as chaperones in the in vivo regulation of viral capsid assembly.
The virus families Polyomaviridae and Papillomaviridae, collectively referred to as papovaviruses, have a nonenveloped icosahedral capsid surrounding a double-stranded circular DNA genome. The structural proteins of polyomavirus (VP1, VP2, and VP3) and papillomavirus (L1 and L2) are synthesized late in viral infection and imported into the nucleus, where they assemble around newly synthesized viral genomes. Viral capsids are constructed from 72 capsomeres (pentamers) of L1 or VP1, arranged on a T=7 icosahedral lattice (1). The carboxyl terminus of VP1 or L1 mediates contacts between the pentamers in the assembled capsid (14, 26, 27, 32). Disulfide bonds stabilize the interpentamer contacts for L1 (25, 30, 42), while both disulfide bonds and calcium bridges stabilize these contacts for VP1 (2, 4, 21, 28, 44, 45).
The papillomavirus L1 and polyomavirus VP1 capsid proteins each have a well-defined, canonical nuclear localization signal (NLS) (3, 12, 20, 31, 35, 38, 40, 49). In the cytoplasm, NLS-containing proteins bind the adaptor protein karyopherin α, which has at its amino terminus a karyopherin β-binding domain and at its carboxyl terminus the NLS binding domain (8). Karyopherin α binds karyopherin β, which then docks the protein complex at the nuclear pore (19, 36). After translocation through the nuclear pore, RanGTP dissociates karyopherin β from the complex, resulting in the accumulation of karyopherin α plus NLS protein in the nucleus (15, 36). Once in the nucleus, factors such as nucleoporin Nup2 and the export receptor for karyopherin α, CAS, may facilitate the dissociation of karyopherin α from the NLS (24, 29). An alternative model for the dissociation of karyopherin α from an NLS involves the “NLS-like” sequences at the amino terminus of karyopherin α forming an intramolecular bond with the karyopherin α NLS binding domain, thus competing α from the VP1 or L1 NLS (17, 18).
The subcellular location of papovavirus assembly is controlled such that capsids are assembled in the cell nucleus and not in the cytoplasm (13, 33, 41). One such control mechanism may involve the hsc70 family of cellular chaperone proteins. For example, in mouse cells infected with polyomavirus, hsp70 binds polyomavirus VP1 in the cytoplasm and colocalizes with VP1 after transport into the nucleus (9). Moreover, in vitro capsid assembly of polyomavirus VP1 capsomeres is inhibited by bound hsc70 (7). We have investigated a potential role for karyopherins as chaperones of mouse polyomavirus (mPy) VP1 and human papillomavirus type 11 (HPV11) L1 capsid assembly. We show that the NLS sequences of both mPy VP1 and HPV11 L1 are required for binding karyopherin α2β1. Karyopherins affected the DNA binding properties of HPV11 L1 but not mPy VP1. In vitro, capsid assembly of both HPV11 L1 and mPy VP1 was inhibited in the presence of karyopherin α2β1. We propose that karyopherins play a chaperone function in the papovavirus life cycle by inhibiting capsid formation in the cytoplasm while importing L1 and VP1 capsomeres into the nucleus.
MATERIALS AND METHODS
Preparation of recombinant proteins.
Recombinant full-length HPV11 L1, mPy VP1, truncated HPV11 L1 (CΔ29), and truncated mPy VP1 (NΔ6) were expressed as glutathione S-transferase (GST) fusion protein in Escherichia coli BL21(DE3) cells. Cells were grown at 37°C in 2× YT (yeast extract-tryptone) medium plus ampicillin to an optical density at 600 nm of ∼0.2 and then cooled to 25°C and induced with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 15 h. Cells were harvested by centrifugation followed by resuspension in buffer L (40 mM Tris-HCl [pH 8.0], 0.2 M NaCl, 5% glycerol, 1 mM EDTA, 5 mM dithiothreitol [DTT]) with protease inhibitors pepstatin (1.25 μg/ml), leupeptin (2 μg/ml) leupeptin, and phenylmethylsulfonyl fluoride (1 mM [PMSF]). Cells were lysed by adding 0.5 mg/ml lysozyme and 0.1% deoxycholic acid and incubated at room temperature for 1 h. Lysate was treated with 10/ml DNase I and 5 mM MgCl2 and incubated at 4°C with gentle shaking for 4 to 5 h. The lysates were centrifuged at 25,000 × g for 1 h, and the protein-containing supernatant was applied to a glutathione-Sepharose column (Amersham) equilibrated with buffer L. The column was then washed with 20 bed volumes of buffer L, and the recombinant proteins were eluted by thrombin cleavage. Protein was concentrated in a Millipore/BIOMAX concentrator. Protein was further purified using a fast protein liquid chromatography gel filtration chromatograph (Superdex 200; Amersham).
Recombinant full-length His-tagged karyopherin α2 (48) and GST-tagged and His-tagged karyopherin β1 (5) were prepared as previously described. The purified proteins were dialyzed in buffer A (20 mM HEPES-KOH [pH 7.3], 110 mM potassium acetate, 2 mM magnesium acetate, 2 mM DTT) containing protease inhibitors.
Karyopherin β1 binding to karyopherin α2 and capsid protein VP1 or L1.
Glutathione-Sepharose 4B beads (Amersham) were preincubated with buffer A (20 mM HEPES-KOH [pH 7.0], 100 mM potassium acetate, 2 mM magnesium acetate, 0.1% Triton X-100, 2 mM EGTA, 1 mM DTT) supplemented with 1% bovine serum albumin (BSA) for 1 h at room temperature (RT) followed by three washes in buffer A. Recombinant GST-β1 protein was immobilized on BSA-blocked glutathione-Sepharose 4B beads (1.5 μg of protein/10 μg of beads) and incubated for 30 min at RT with 1.5 μg mPy VP1 or HPV11 L1 with karyopherin α2 in buffer A. To control for nonspecific binding of karyopherins to the GST protein moiety or the glutathione-Sepharose beads, 1.5 μg GST protein was immobilized on BSA-blocked glutathione-Sepharose 4B beads and incubated with 1.5 μg mPy VP1 or HPV11 L1 with karyopherin α2 in buffer A. GST-L1 binding to karyopherins α2 and β1 was also performed as described above for GST-β1. Bound protein was washed in buffer A with 0.3% Triton X-100. The bound proteins were eluted from the Sepharose beads by boiling in Laemmli buffer (2% [wt/vol] sodium dodecyl sulfate [SDS], 10% glycerol, 60 mM Tris-HCl [pH 6.8], 5% β-mercaptoethanol, 0.01% [wt/vol] bromophenol blue) for 10 min followed by SDS-polyacrylamide gel electrophoresis (PAGE). Protein bands were visualized using GelCode blue stain reagent (Pierce).
DNA binding measured by EMSA.
Purified recombinant HPV11 L1 or mPy VP1 (2.6 μM) with or without karyopherins (0.5 μM) was incubated with a 6-kb linearized plasmid DNA (0.5 μg/reaction) for 30 min at RT in buffer B (20 mM HEPES-KOH [pH 7.0], 110 mM KCl, 2 mM MgCl2, 1 mM DTT). Linear DNA was used in the reactions instead of circular plasmid to avoid the appearance of supercoiled, relaxed-circular, and linear forms of plasmid DNA that made the interpretation of L1- or VP1-bound DNA difficult. A 10× concentration of DNA loading buffer (50% glycerol, 0.4% bromophenol blue) was added to the sample to a final concentration of 1×, and the DNA-protein complexes were resolved on a 0.8% agarose gel containing 3 μg/ml ethidium bromide. Samples were electrophoresed at 150 V for 1 h in Tris-acetate-EDTA buffer (40 mM Tris, 20 mM glacial acetic acid, 1 mM EDTA [pH 8.0]). DNA bands were imaged using a UV light source. After electrophoretic mobility shift assay (EMSA) of L1 or VP1 DNA-protein complexes in 0.8% agarose gels as described above, the gels were soaked for 15 min in transfer buffer (25 mM Tris, 200 mM glycine, 10% methanol). Proteins were transferred to Immobilon-P (Millipore) using a semidry transfer apparatus at 150 V/150 mA for 1 h. Western blots were blocked in Tris-buffered saline (TBS) containing 3% BSA for 1 h at RT. Blots were hybridized overnight with the primary antibody anti-HPV11 L1 rabbit polyclonal antibody (R8363), mouse anti-karyopherin α2 (Rch-1 [BD Transduction Laboratories), anti-karyopherin β1 goat polyclonal antibody (Sc1919 [Santa Cruz Biotechnology]), or anti-VP1 rabbit polyclonal antibody (I58) in TBS containing 1% BSA. Blots were washed twice in a mixture of TBS, 1% BSA, and 0.3% Tween 20 for 5 min followed by 1 h of hybridization with horseradish peroxidase (HRP)-coupled goat anti-rabbit (Bio-Rad), goat anti-mouse (Bio-Rad), or donkey anti-goat (Bio-Rad) secondary antibody. The blot was washed twice in TBS, 1% BSA, and 0.3% Tween 20 followed by a final 10 min wash in TBS, 1% BSA, and 0.1% Tween 20. The HRP signal was detected with SuperSignal West Pico chemiluminescent substrate (Pierce) according to the manufacturer's protocol.
In vitro assembly reactions.
In vitro assembly reactions were performed using recombinant mPy VP1 or HPV11 L1 capsid proteins at 150 μg/ml. Fifty microliters of VP1 or L1, diluted to 150 μg/ml in a mixture of 20 mM HEPES-KOH (pH 7.0), 110 mM KCl, and 2 mM MgCl2, was dialyzed in mini-dialysis units (molecular weight cutoff, 3,500 [Pierce]) against assembly buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5 mM CaCl2, 5% glycerol [pH 7.2]) at RT overnight. Assembly reaction mixtures were concentrated to 50 μl using a Microcon 0.5-ml concentrator (Millipore). Fifty-microliter drops were spotted onto parafilm and then absorbed to glow-discharged, carbon- and formvar-coated copper transmission electron microscopy (TEM) grids (G400 copper; EM Science). Grids were blotted dry on Kimwipe (Kimberly-Clark), followed by staining with uranyl acetate (2% [wt/vol]) and TEM (Philips CM10) at 80 kV. The number of 50-nm particles was determined by direct count of the indicated number (n = 6) of grid squares (1,369 μm2) at ×4,800 magnification. To test assembly inhibition by karyopherin α2, β1, or α2β1, karyopherin proteins were added to the capsid proteins at a 1:3 molar concentration relative to the VP1 or L1 protein concentration followed by dialysis and capsid formation assessed by TEM as described above.
RESULTS
Karyopherin α2β1 binds to the NLS sequences of both mPy VP1 and HPV11 L1.
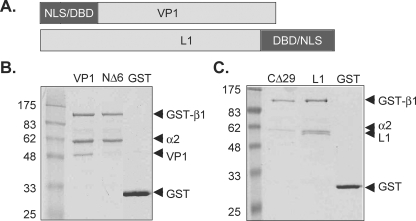
Previous studies have shown that HPV16, HPV45, HPV11 L1, and simian virus 40 (SV40) VP1 proteins are bound by a heterodimer of αβ karyopherins (31, 37-39). The NLS of mPy VP1 is located at its amino terminus, in contrast to HPV11 L1, where the NLS is situated at the carboxyl terminus (Fig. 1A). To determine whether karyopherins specifically interact with the NLS of mPy VP1 pentamers, both the full-length recombinant VP1 protein and a recombinant VP1 protein lacking the NLS (VP1NΔ6) were incubated with karyopherins α2 and GST-β1. Proteins bound to GST-β1 were detected by glutathione-Sepharose chromatography followed by SDS-PAGE. As shown in Fig. 1B, GST-β1 bound both karyopherin α2 and full-length VP1 pentamers, but did not bind VP1NΔ6 pentamers (Fig. 1B, NΔ6 lane). Recombinant full-length HPV11 L1, previously shown to bind the karyopherin heterodimer α2β1 (38), was used as a positive control for the GST-β1 interaction experiments. Glutathione-Sepharose chromatography was performed using GST-β1 incubated with full-length HPV11 L1 pentamers or recombinant HPV11 L1 pentamers lacking the NLS (L1CΔ29) and karyopherin α2 (Fig. 1C). GST-β1 bound karyopherin α2 and full-length L1, but not L1CΔ29 lacking an intact NLS (Fig. 1C, compare lanes L1 and CΔ29). These results demonstrate that the karyopherin heterodimer α2β1 binds both mPy VP1 and HPV11 L1 pentamers in an NLS-dependent manner.
FIG. 1.
The karyopherin heterodimer α2β1 binds VP1 and L1 in an NLS-dependent manner. (A) Schematic representation of the mPy VP1 and HPV11 L1 proteins depicting the location of the NLS and the DNA binding domain (DBD). (B) GST-β1 or GST bound to glutathione-Sepharose incubated with VP1 or VP1NΔ6 in the presence of karyopherin α2. (C) L1 or L1CΔ29 in the presence of karyopherin α2. Bound protein complexes were identified by SDS-PAGE and Coomassie staining.
Karyopherins affect HPV11 L1, but not mPy VP1, DNA binding.
During translocation of classical NLS-containing proteins through the nuclear pore, RanGTP dissociates karyopherin β1 from the karyopherin α2β1 heterodimer bound to the NLS, resulting in accumulation of the karyopherin α2-cargo protein complex in the nucleoplasm (8). After import into the nucleus, VP1 or L1 capsomeres assemble around the viral genome through interactions between the DNA binding domain of VP1 or L1 and the viral DNA genome. Because the NLSs of both mPy VP1 and HPV11 L1 overlap their DNA binding domains, we determined whether karyopherin α2, β1, or α2β1 bound to the NLS of VP1 or L1 would affect capsomere binding to DNA.
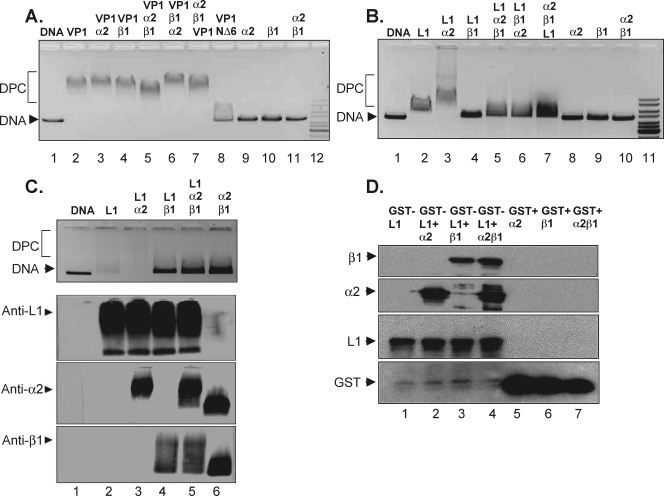
To determine the effect of karyopherins on the binding of VP1 or L1 to DNA, the capsid proteins were first incubated with the karyopherins. The protein complex of capsid protein and karyopherin(s) was then incubated with linearized plasmid DNA. The nucleoprotein complexes were analyzed by EMSA to determine whether the capsid protein bound by karyopherins maintained the ability to bind DNA. As shown in Fig. 2A, mPy VP1 capsid protein (2.6 μM) associated with DNA migrated slower than DNA alone (Fig. 2A, compare lanes 1 and 2). VP1 prebound with karyopherin α2, β1, or α2β1 did not significantly inhibit DNA binding by VP1 as measured by the migration of the nucleoprotein complex in the agarose gel (Fig. 2A, compare lanes 2 through 5). The order of karyopherin addition to VP1 was examined by adding karyopherin α2 to VP1 prior to karyopherin β1 (lane 5), karyopherin β1 to VP1 prior to karyopherin α2 (lane 6), or mixing karyopherins α2 and β1 together followed by the addition of VP1 to the binding reaction (lane 7). The order of karyopherin addition to VP1 did not significantly affect VP1 DNA binding (compare lane 2 with lanes 5 to 7). These data suggest that the binding of karyopherins to VP1 capsomeres does not inhibit VP1 binding to DNA.
FIG. 2.
Karyopherins differentially affect the DNA binding of VP1 and L1 to DNA. Shown are results for recombinant mPy VP1 or VP1NΔ6 (A) or L1 or L1CΔ29 (B) capsomeres incubated with karyopherins α2, β1, and α2β1 and DNA. (C) Recombinant HPV11 L1 capsomeres incubated with karyopherins and DNA (top panel). Associated proteins were detected with antibodies against L1 (panel second from top), karyopherin α2 (panel second from bottom), or karyopherin β1 (bottom panel). DPC designates migration of the DNA-protein complex. (D) GST-L1 or GST bound to glutathione-Sepharose incubated with karyopherin α2, β1, or α2β1. Bound protein complexes were identified by SDS-PAGE followed by Western blotting.
Similar DNA binding reactions were performed using HPV11 L1 capsomeres alone or in the presence of karyopherins. In contrast to VP1 capsomeres, the binding of L1 capsomeres to DNA was inhibited by karyopherin α2β1 as measured by the change in DNA migration through an agarose gel (Fig. 2B, compare lanes 2 and 5). Interestingly, L1 binding to DNA was also inhibited when L1 was preincubated with karyopherin β1 alone (Fig. 2B, compare lanes 1, 2, and 4). When L1 was incubated with karyopherin α2 and DNA, the DNA protein complex was supershifted relative to L1-bound DNA (Fig. 2B, compare lanes 2 and 3). The latter results suggest that capsomeres of L1 bound to karyopherin α2 can also bind DNA. The order of karyopherin addition to L1 was examined by adding karyopherin α2 to L1 prior to karyopherin β1 (lane 5) or karyopherin β1 to L1 prior to karyopherin α2 (lane 6), or mixing karyopherins α2 and β1 together followed by the addition of L1 to the binding reaction mixture (lane 7). The order of karyopherin addition to L1 did not significantly affect the inhibition of DNA binding by L1 (compare lane 2 with lanes 5 to 7).
Proteins can localize to the nucleus in a karyopherin α-independent, β-dependent manner (22, 34, 47). Because karyopherin β1 alone inhibited the association of HPV11 L1 capsomeres with DNA, the interaction between karyopherin β1 and L1 was investigated by immunoblotting of the proteins associated with DNA after EMSA. HPV11 L1 capsomeres alone or bound by karyopherin α2, β1, or α2β1 were subsequently incubated with plasmid DNA, and the resultant complexes were resolved by agarose gel electrophoresis. Consistent with the HPV11 L1 DNA binding experiment shown in Fig. 2B, the addition of karyopherin α2 alone to the L1-DNA complex resulted in a DNA supershift, whereas addition of karyopherin β1 alone or karyopherin α2β1 completely inhibited L1 DNA binding (Fig. 2C, top panel). Western blot analysis of proteins transferred from agarose gels shown in the top panel of Fig. 2C identified L1 and karyopherin α2 in the supershifted nucleoprotein complex (Fig. 2C, anti-L1 and anti-α2 panels). A protein complex comprised of L1 and karyopherin β1 or L1 and karyopherin α2β1 was observed in the lanes where DNA was no longer bound by L1 (Fig. 2C, anti-L1, anti-α2, and anti-β1 panels). In contrast to the formation of an L1-karyopherin α2-DNA complex, these data suggest that when karyopherin β1 or α2β1 binds L1, L1 is subsequently unable to bind DNA. While a significant amount of DNA is released from L1 with the addition of karyopherin β1 or α2β1 to the DNA binding reaction, it is possible that some DNA remains associated with L1, accounting for the L1 migration pattern shown in Fig. 2C. Western blot analysis of HPV11 L1 incubated with or without DNA followed by agarose gel electrophoresis demonstrated that the migration of L1 in native agarose gels was not altered when DNA was present in the binding reaction (not shown).
To further validate the interaction between karyopherin β1 and HPV11 L1, full-length recombinant L1 protein fused to GST was incubated with karyopherin α2, β1, or α2β1. Proteins bound to GST-L1 were detected by SDS-PAGE followed by Western blot analysis using antibodies against HPV11 L1 or karyopherin α2 or β1. As shown in Fig. 2D, GST-L1 bound karyopherin α2 and the α2β1 heterodimer. Interestingly, GST-L1 was also capable of binding karyopherin β1 independent of karyopherin α2.
mPy VP1 and HPV11 L1 capsomeres can bind karyopherin α2 and DNA simultaneously.
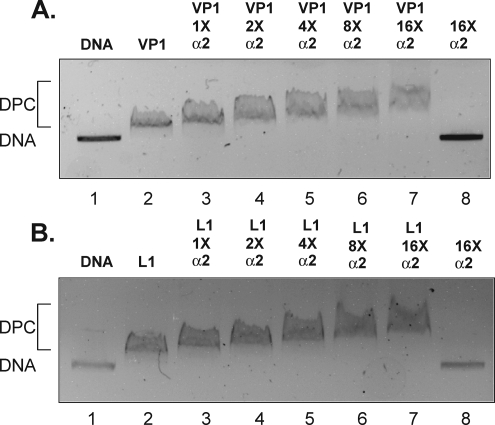
Our initial experiments showed that karyopherin α2 (0.5 μM) did not inhibit DNA (6.9 nM) binding by either mPy VP1 or HPV11 L1 (2.6 μM) capsid proteins. Because the amino acid sequence comprising the NLS of mPy VP1 and HPV11 L1 overlaps their DNA binding domain, we tested whether binding karyopherin α2 would inhibit DNA binding when karyopherin α2 is in molar excess relative to VP1 or L1. Karyopherin α2, ranging from an equal molar concentration up to a 16-fold molar excess relative to the capsid protein monomer, was preincubated with either mPy VP1 or HPV11 L1 protein prior to addition of plasmid DNA (Fig. 3A and B). The resultant DNA-protein complexes were then resolved by agarose gel electrophoresis. A decrease in the mobility of the VP1 or L1 plasmid DNA complex was observed when the capsid protein (2.6 μM) was bound with an equal molar amount of karyopherin α2 (Fig. 3A and B, compare lanes 2 and 3). Increasing the amount of karyopherin α2 added to the binding reaction from a 2-fold to 16-fold molar excess relative to the capsid protein concentration further decreased the mobility of the nucleoprotein complex corresponding to the amount of karyopherin α2 added to the reaction (Fig. 3A and B). These results suggest that karyopherin α2 binding to VP1 or L1 capsomeres was insufficient to disrupt DNA binding and that mPy VP1 or HPV11 L1 capsomeres can bind karyopherin α2 and DNA simultaneously.
FIG. 3.
mPy VP1 and HPV11 L1 capsomeres bind DNA and karyopherin α2 simultaneously. Shown are results for recombinant mPy VP1 (A) or HPV11 L1 (B) capsomeres incubated with DNA and karyopherin α2 at equal molar concentrations relative to capsid protein (1×) or karyopherin α2 in molar excess (2×, 4×, 8×, and 16×). DPC designates migration of the DNA-protein complex.
DNA competes for karyopherin α2 binding to VP1 and L1.
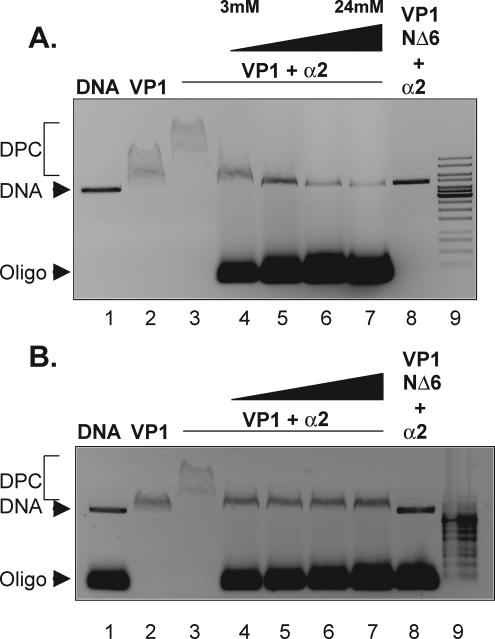
When karyopherin α disassociates from the capsid proteins after nuclear import is currently unkown. One possibility is that the “NLS-like” sequences at the amino terminus of karyopherin α compete the karyopherin from the VP1 or L1 NLS (17, 18). Alternatively, because their DNA binding domain overlaps their NLS, binding DNA to VP1 or L1 may release α2 from the NLS. The results shown in Fig. 3 suggested that VP1 and L1 can form a nucleoprotein complex that includes the capsid protein, karyopherin α2, and DNA, as evidenced by the slower-migrating complex. We tested whether increasing the DNA concentration in the DNA binding reaction mixture containing VP1 and karyopherin α2 could compete karyopherin α2 from the nucleoprotein complex. VP1 (2.6 μM) was first incubated with the karyopherin α2 (0.5 μM) and linearized plasmid DNA (6.9 nM). In an attempt to compete karyopherin α2 from the nucleoprotein complex, 60-bp duplex oligonucleotides were then added to the binding reaction mixture to a final concentration of 3, 6, 12, or 24 μM. The migration of the nucleoprotein complex containing plasmid DNA was assessed by agarose gel electrophoresis. As shown in Fig. 4A, the oligonucleotides competed VP1 and karyopherin α2 binding to plasmid DNA when the concentration of oligonucleotides was 6 μM or greater, as determined by the migration of the plasmid DNA in the agarose gel (Fig. 4A, compare lanes 3 and 5). Interestingly, the nucleoprotein complex from the reaction mixture containing 3 μM oligonucleotide migrated to the same position as the nucleoprotein complex containing VP1 and plasmid DNA (Fig. 4A, compare lanes 2 and 4). These results suggest that the addition of oligonucleotide DNA above 6 μM to the binding reaction mixture competed both VP1 and karyopherin α2 from the plasmid DNA while addition of 3 μM competed only karyopherin α2 from the VP1-DNA complex. Similar results were also observed for HPV11 L1 (data not shown). The competition between DNA and karyopherin α2 binding to VP1 capsomeres was further analyzed by titrating the concentration of oligonucleotides into the binding reaction from 3 μM to 5 μM (Fig. 4B). As shown in Fig. 4B, karyopherin α2 was competed from the nucleoprotein complex with the addition of oligonucleotide DNA ranging from 3 to 5 μM (Fig. 4B, compare lanes 2 and 4 to 7). We interpret these results to suggest that DNA is able to compete karyopherin α2 from the VP1 NLS.
FIG. 4.
DNA competes karyopherin α2 from the NLS of mPy VP1. Shown are EMSA results for VP1-karyopherin α2 binding to DNA in the presence of completing duplexed DNA oligonucleotides ranging from 3 to 24 μM (A) and from 3 to 6 μM (B). DPC designates migration of the DNA-protein complex.
Karyopherin binding inhibits in vitro capsid assembly of VP1 and L1 capsomeres.
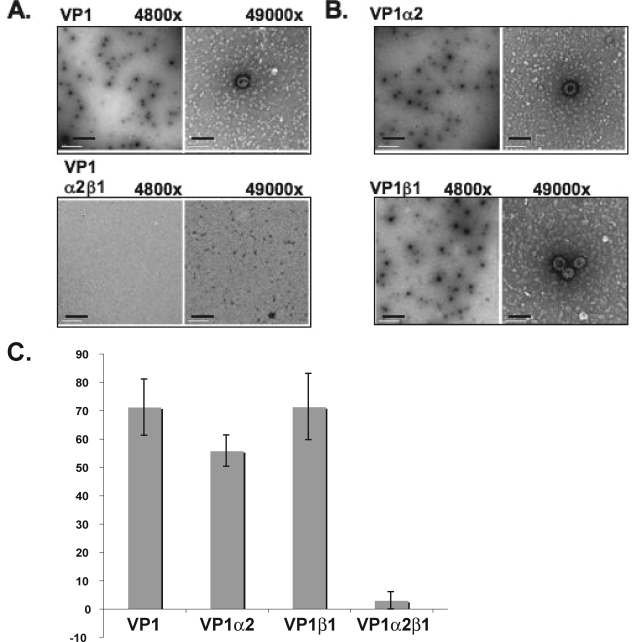
The subcellular location of papovavirus assembly is controlled such that virions are formed in the cell nucleus and not in the cytoplasm (13, 33). The hsp70 family of proteins has been shown to bind polyomavirus VP1 and has been implicated in regulating polyomavirus capsid assembly (7, 9). Karyopherin proteins associate with VP1 capsomeres in the cytoplasm and remain associated with the capsomeres as they localize to the nucleus. Therefore, a potential role for karyopherins in inhibiting mPy VP1 capsid formation has been tested. In vitro calcium-mediated assembly of polyomavirus VP1 capsomeres was used as an assay to test how factors that bind VP1 capsomeres in vivo may affect assembly (7). Assembly reactions with mPy VP1 were performed in the presence or absence of karyopherin α2, β1, or α2β1 by overnight dialysis in assembly buffer containing 0.5 mM calcium chloride. The VP1 concentration did not change during dialysis, as detected by soluble protein concentration. The samples were then analyzed by TEM. Viruslike particles were counted using six images per grid from four independent experiments. VP1 assembled into T=7 particles in the absence of karyopherins, but not in the presence of the karyopherin α2β1 heterodimer (Fig. 5A). VP1 also assembled into T=7 particles in the presence of either α2 or β1 karyopherin alone (Fig. 5B). Quantitation of assembled particles demonstrated that calcium-mediated in vitro assembly of VP1 viruslike particles was reduced by more than 20-fold in the presence of karyopherin α2β1 (Fig. 5C).
FIG. 5.
Karyopherin α2β1 inhibits in vitro assembly of VP1. Assembly reactions of VP1 with purified karyopherin α2, β1, or α2β1 were visualized by negative staining and TEM. (A) Electron micrograph of a VP1 assembly reaction (top row) or VP1 with karyopherin α2β1 (bottom row) Magnifications, ×4,800 and ×49,000. (B) Electron micrograph of a VP1-karyopherin α2 (top row) or VP1-karyopherin β1 (bottom row) assembly reaction. Magnifications, ×4,800 and ×49,000. (C) Quantitation of the average number of assembled particles per image (n = 6) from four independent assembly reactions. The scale bars are 1 μm at ×4,800 and 100 nm at ×49,000.
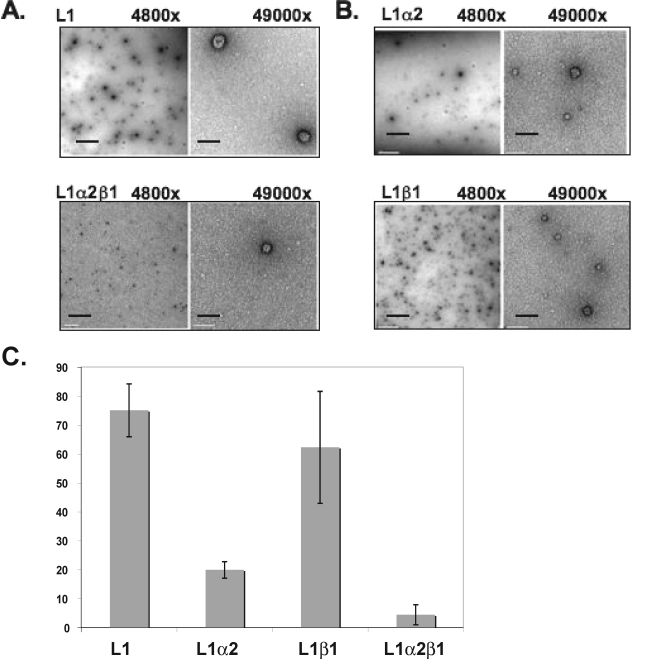
In vitro assembly of papillomavirus L1 capsomeres was used to assess the effects of karyopherins on papillomavirus capsid assembly. Assembly reactions were performed with L1 in the presence or absence of karyopherin α2, β1, or α2β1 followed by an overnight dialysis in assembly buffer that allowed formation of interpentameric disulfide bonds. As observed with VP1 capsomeres, L1 capsomeres assembled into T=7 particles in the absence of karyopherins, but not in the presence of the karyopherin α2β1 heterodimer (Fig. 6A). In contrast to VP1, we also observed an inhibition of L1 particle assembly in the presence of karyopherin α2 (Fig. 6B). Additionally, we observed a shift toward the formation of T=1 and T=3 particles in assembly reaction mixtures containing L1 and β1. Quantitation of assembled particles demonstrated that assembly of L1 capsids was reduced by more than 15-fold in the presence of karyopherin α2β1 (Fig. 6C). Individually, karyopherin α2 inhibited L1 capsid assembly by threefold (Fig. 6C).
FIG. 6.
Karyopherins α2 and α2β1 inhibit in vitro assembly of L1. In vitro assembly reactions of L1 capsomeres in the presence of purified karyopherin α2, β1, or α2β1 were analyzed by electron microscopy. (A) L1 assembly reaction (top row) or L1 assembled in the presence of karyopherin α2β1 (bottom row) Magnifications, ×4,800 and ×49,000. (B) L1-karyopherin α2 (top row) or L1-karyopherin β1 (bottom row) assembly reaction. Magnifications, ×4,800 and ×49,000. (C) Quantitation of the average number of assembled particles per image (n = 5) from three independent assembly reactions. The scale bars are 1 μm at ×4,800 and 100 nm at ×49,000.
DISCUSSION
In order to determine whether karyopherin binding to papovavirus capsid protein affects capsid assembly, the interaction between karyopherins and the HPV11 L1 or mPy VP1 capsid proteins was investigated. As previously observed for the L1 capsid proteins of HPV11 and HPV45 (38), the HPV11 L1 capsid protein bound both karyopherins α2 and α2β1. mPy VP1 also interacted with karyopherins α2 and α2β1. The interaction between karyopherins and L1 or VP1 was dependent on an intact NLS (Fig. 1).
Nuclear import of proteins that contain a canonical NLS, such as mPy VP1 and HPV11 L1, occurs by binding the adaptor protein karyopherin α2 to the NLS that then recruits karyopherin β. An alternative pathway for nuclear import utilizes karyopherin β without the adaptor protein karyopherin α (22, 34, 47). Although HPV11 L1 capsomeres interact with karyopherin α2 or the α2β1 heterodimer in an NLS-dependent manner, L1 capsomeres also interacted with karyopherin β1 in the absence of karyopherin α2 (Fig. 2). Moreover, karyopherin β1 strongly inhibited DNA binding by L1 capsomeres, suggesting that the interaction of L1 with karyopherin β1 competes for L1 binding to DNA. However, it is unclear whether L1 binding by karyopherin β1 alone is biologically significant or restricted to an in vitro association. Our current understanding of L1 import suggests that karyopherin β1 would not be present in the nucleus to inhibit the interaction between L1 capsomeres and DNA. More work is needed to evaluate the potential significance of the interaction between HPV11 L1 and karyopherin β1 in vivo.
Karyopherin-mediated nuclear import may also be important during the initial phase of virus infection when viral nucleoprotein complexes, composed of capsid proteins bound to the viral genome, exits a membrane compartment (the endosome for papillomavirus or the endoplasmic reticulum for polyomavirus) into the cytoplasm (11, 16). In the cytoplasm, karyopherins may bind the exposed NLS(s) of capsomeres or the minor capsid proteins, which are bound to the viral DNA, and then transport the viral protein complex into the nucleus. In order for this transport pathway to function, capsomeres must be able to bind the viral genome and karyopherins simultaneously. We found that VP1 (2.6 μM) complexed with karyopherin α2 (0.5 μM) was able to bind plasmid DNA (6.9 nM). Additionally, increasing karyopherin α2 to a 16-fold molar excess over the capsid protein led to slower migration of the DNA-protein complex by EMSA (Fig. 3). We interpret these results to mean that the five DNA binding domains of a VP1 pentamer are not fully occupied by DNA, because unoccupied NLSs continued to bind karyopherin α2. However, the gel mobility of the complex also indicated that the interaction between karyopherin α2 and the NLSs of mPy VP1 was insufficient to displace DNA from the DNA binding domain. These data suggest that in vitro, a pentamer bound to plasmid DNA does not have all five DNA binding domains of the pentamer in contact with the DNA. However, during infection the polyomavirus genome is configured with nucleosomes and how the DNA binding domain of VP1 or L1 will interact with nucleosome-bound DNA is unclear.
Similar to the results of mPy VP1, we found that HPV11 L1 (2.6 μM) complexed with karyopherin α2 (0.5 μM) was able to bind plasmid DNA (6.9 nM). Additionally, increasing karyopherin α2 to a 16-fold molar excess over the capsid protein led to slower migration of the DNA-protein complex by EMSA (Fig. 3). However, DNA binding by HPV11 L1 capsomers was inhibited by karyopherin α2β1 (Fig. 2B), and karyopherin α2 cannot alone mediate nuclear import in the absence of karyopherin β1 import receptor. These results suggest that it is unlikely that HPV11 L1 mediates import of the viral genome during the initial phase of infection as L1 binding of karyopherin α2β1 in the cytoplasm would release the viral DNA from the capsid protein prior to being transported into the nucleus. Consistent with this reasoning, L1 has not been detected in the nucleus in the initial phase of the papillomavirus infection and, instead, L2 seems to colocalize in the nucleus with the incoming viral genome (10).
The basic amino acids that make up the NLSs for both papillomavirus and polyomavirus also function in DNA binding. The DNA binding constant for recombinant VP1 has been determined by an immunoprecipitation assay to be 1 × 10−11 M to 2 × 10−11 M (35). The DNA binding affinity of L1 has not been quantitated, but L1 has been shown to bind DNA by Southwestern blotting and EMSA gel assay (26, 46). Karyopherin α2β1 competed DNA from HPV11 L1, but not mPy VP1. These data suggest that the DNA binding affinity of L1 may be less than that of VP1. The relatively tight binding affinity of VP1 may serve a functional role during capsid assembly, with the DNA serving as scaffolding around which VP1 capsomeres assemble. The weaker binding affinity of L1 for DNA may reflect a greater dependence on the viral L2 protein to bind the viral genome and serve as an interface for encapsidation.
As the capsomeres enter the nucleus, RanGTP dissociates karyopherin β from the cargo protein complex. However, what mediates the dissociation of karyopherin α from the capsid proteins is unknown. One possibility is that DNA binding to VP1 or L1 releases karyopherin α2 from the NLS. We observed that addition of oligonucleotide DNA ranging from 3 to 6 μM to a nucleoprotein complex composed of capsid protein (2.6 μM), karyopherin α2 (0.5 μM), and plasmid DNA (6.9 nM) was able to compete karyopherin α2 from the NLS of mPy VP1 (Fig. 4B). These data raise the possibility that in the nucleus, the viral DNA may displace karyopherin α2 from the capsid protein, allowing for capsid assembly around the viral genome.
The mPy VP1 and HPV11 L1 capsid proteins have the intrinsic ability to self-assemble in vitro and in vivo. Given these intrinsic self-assembly properties, there must be biological controls in vivo that regulate the quality and subcellular location of viral capsid assembly. The hsp70 family of chaperones previously has been shown to modulate assembly and disassembly of polyomavirus and papillomavirus capsid protein (6, 7). Our data suggest that another potential mechanism for modulating capsid assembly may involve the karyopherin proteins that bind the NLS of HPV11 L1 and mPy VP1 and that karyopherin proteins may have “chaperone” functions. The karyopherin heterodimer α2β1 inhibited in vitro assembly of both HPV11 L1 and mPy VP1. Karyopherin α2β1 heterodimers (157.5-kDa complex) bind HPV11 L1 at the NLS located at the carboxyl terminus of the protein. Binding to the carboxyl terminus may inhibit assembly through steric inhibition, because the L1 carboxyl terminus is also used for interpentamer bonding (25, 32). Karyopherin α2 also had an inhibitory effect on L1 assembly, although to a lesser extent than karyopherin α2β1. This finding supports the idea that steric hindrance may inhibit assembly and that by recruiting karyopherin β1 to the L1-karyopherin α2 complex assembly is further inhibited. Interestingly, karyopherin β1 appeared to shift assembly in favor of the T=3 and T=1 products. These data suggest that karyopherin β1 may interact with the carboxyl terminus of L1 and alter the interpentamer contacts mediated by the carboxyl-terminal domain. The NLS of VP1 is located at its amino terminus, which for SV40 is involved in interpentamer contacts through the formation of disulfide bonds (43). Binding of karyopherin α2β1 to the VP1 amino terminus may prevent these interpentameric disulfide bonds between VP1 molecules as well as prevent the carboxyl-terminal domains of VP1 from forming interpentameric bonds through an indirect steric effect. Inhibition of in vitro assembly of mPy VP1 and HPV11 L1 capsids by karyopherin α2β1 supports the hypothesis that karyopherins may play a role in chaperoning VP1 and L1 capsid assembly in vivo. Karyopherins are abundant in the cytosol, ranging from 1 to 2 μM of each karyopherin (23). This high level of karyopherins would allow them to function as chaperones in addition to their nuclear import function. Because the nuclear localization of many proteins is mediated by karyopherins, the chaperone function observed with karyopherins in modulating viral protein complex assembly and DNA binding may be applicable to proteins other than the capsid proteins investigated in this study.
Acknowledgments
We thank Laura Chromy for purification of recombinant capsid proteins mPy VP1 and HPV11 L1 and Dorothy Dill for technical assistance with electron microscopy.
This work was supported by National Institutes of Health grant CA094898 (to J.M.), National Institutes of Health grant CA37667 (to R.L.G.), and National Institutes of Health postdoctoral training grant T32CA082086 (to G.B. and R.L.G.).
Footnotes
Published ahead of print on 13 August 2008.
REFERENCES
- 1.Belnap, D. M., N. H. Olson, N. M. Cladel, W. W. Newcomb, J. C. Brown, J. W. Kreider, N. D. Christensen, and T. S. Baker. 1996. Conserved features in papillomavirus and polyomavirus capsids. J. Mol. Biol. 259249-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady, J. N., V. D. Winston, and R. A. Consigli. 1977. Dissociation of polyoma virus by the chelation of calcium ions found associated with purified virions. J. Virol. 23717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, D., J. I. Haynes, Jr., J. N. Brady, and R. A. Consigli. 1993. Identification of amino acid sequences in the polyomavirus capsid proteins that serve as nuclear localization signals. Trans. Kans. Acad. Sci. 9635-39. [PubMed] [Google Scholar]
- 4.Chen, P. L., M. Wang, W. C. Ou, C. K. Lii, L. S. Chen, and D. Chang. 2001. Disulfide bonds stabilize JC virus capsid-like structure by protecting calcium ions from chelation. FEBS Lett. 500109-113. [DOI] [PubMed] [Google Scholar]
- 5.Chi, N. C., E. J. Adam, and S. A. Adam. 1995. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J. Cell Biol. 130265-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chromy, L. R., A. Oltman, P. A. Estes, and R. L. Garcea. 2006. Chaperone-mediated in vitro disassembly of polyoma- and papillomaviruses. J. Virol. 805086-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chromy, L. R., J. M. Pipas, and R. L. Garcea. 2003. Chaperone-mediated in vitro assembly of polyomavirus capsids. Proc. Natl. Acad. Sci. USA 10010477-10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook, A., F. Bono, M. Jinek, and E. Conti. 2007. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76647-671. [DOI] [PubMed] [Google Scholar]
- 9.Cripe, T. P., S. E. Delos, P. A. Estes, and R. L. Garcea. 1995. In vivo and in vitro association of hsc70 with polyomavirus capsid proteins. J. Virol. 697807-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day, P. M., C. C. Baker, D. R. Lowy, and J. T. Schiller. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. USA 10114252-14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day, P. M., D. R. Lowy, and J. T. Schiller. 2003. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology 3071-11. [DOI] [PubMed] [Google Scholar]
- 12.Florin, L., F. Schafer, K. Sotlar, R. E. Streeck, and M. Sapp. 2002. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology 29597-107. [DOI] [PubMed] [Google Scholar]
- 13.Forstova, J., N. Krauzewicz, S. Wallace, A. J. Street, S. M. Dilworth, S. Beard, and B. E. Griffin. 1993. Cooperation of structural proteins during late events in the life cycle of polyomavirus. J. Virol. 671405-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcea, R. L., D. M. Salunke, and D. L. Caspar. 1987. Site-directed mutation affecting polyomavirus capsid self-assembly in vitro. Nature 32986-87. [DOI] [PubMed] [Google Scholar]
- 15.Gorlich, D., F. Vogel, A. D. Mills, E. Hartmann, and R. A. Laskey. 1995. Distinct functions for the two importin subunits in nuclear protein import. Nature 377246-248. [DOI] [PubMed] [Google Scholar]
- 16.Helenius, A. 1984. Semliki Forest virus penetration from endosomes: a morphological study. Biol. Cell 51181-185. [DOI] [PubMed] [Google Scholar]
- 17.Hodel, A. E., M. T. Harreman, K. F. Pulliam, M. E. Harben, J. S. Holmes, M. R. Hodel, K. M. Berland, and A. H. Corbett. 2006. Nuclear localization signal receptor affinity correlates with in vivo localization in Saccharomyces cerevisiae. J. Biol. Chem. 28123545-23556. [DOI] [PubMed] [Google Scholar]
- 18.Hodel, M. R., A. H. Corbett, and A. E. Hodel. 2001. Dissection of a nuclear localization signal. J. Biol. Chem. 2761317-1325. [DOI] [PubMed] [Google Scholar]
- 19.Imamoto, N., T. Shimamoto, S. Kose, T. Takao, T. Tachibana, M. Matsubae, T. Sekimoto, Y. Shimonishi, and Y. Yoneda. 1995. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 368415-419. [DOI] [PubMed] [Google Scholar]
- 20.Ishii, N., A. Nakanishi, M. Yamada, M. H. Macalalad, and H. Kasamatsu. 1994. Functional complementation of nuclear targeting-defective mutants of simian virus 40 structural proteins. J. Virol. 688209-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishizu, K.-I., H. Watanabe, S.-I. Han, S.-N. Kanesashi, M. Hoque, H. Yajima, K. Kataoka, and H. Handa. 2001. Roles of disulfide linkage and calcium ion-mediated interactions in assembly and disassembly of virus-like particles composed of simian virus 40 VP1 capsid protein. J. Virol. 7561-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakel, S., and D. Gorlich. 1998. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 174491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakel, S., J. M. Mingot, P. Schwarzmaier, E. Hartmann, and D. Gorlich. 2002. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutay, U., F. R. Bischoff, S. Kostka, R. Kraft, and D. Gorlich. 1997. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 901061-1071. [DOI] [PubMed] [Google Scholar]
- 25.Li, M., P. Beard, P. A. Estes, M. K. Lyon, and R. L. Garcea. 1998. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J. Virol. 722160-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M., T. P. Cripe, P. A. Estes, M. K. Lyon, R. C. Rose, and R. L. Garcea. 1997. Expression of the human papillomavirus type 11 L1 capsid protein in Escherichia coli: characterization of protein domains involved in DNA binding and capsid assembly. J. Virol. 712988-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, P. P., A. Nakanishi, D. Shum, P. C.-K. Sun, A. M. Salazar, C. F. Fernandez, S.-W. Chan, and H. Kasamatsu. 2001. Simian virus 40 Vp1 DNA-binding domain is functionally separable from the overlapping nuclear localization signal and is required for effective virion formation and full viability. J. Virol. 757321-7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liddington, R. C., Y. Yan, J. Moulai, R. Sahli, T. L. Benjamin, and S. C. Harrison. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354278-284. [DOI] [PubMed] [Google Scholar]
- 29.Matsuura, Y., A. Lange, M. T. Harreman, A. H. Corbett, and M. Stewart. 2003. Structural basis for Nup2p function in cargo release and karyopherin recycling in nuclear import. EMBO J. 225358-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy, M. P., W. I. White, F. Palmer-Hill, S. Koenig, and J. A. Suzich. 1998. Quantitative disassembly and reassembly of human papillomavirus type 11 viruslike particles in vitro. J. Virol. 7232-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merle, E., R. C. Rose, L. LeRoux, and J. Moroianu. 1999. Nuclear import of HPV11 L1 capsid protein is mediated by karyopherin alpha2beta1 heterodimers. J. Cell Biochem. 74628-637. [PubMed] [Google Scholar]
- 32.Modis, Y., B. L. Trus, and S. C. Harrison. 2002. Atomic model of the papillomavirus capsid. EMBO J. 214754-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montross, L., S. Watkins, R. B. Moreland, H. Mamon, D. L. D. Caspar, and R. L. Garcea. 1991. Nuclear assembly of polyomavirus capsids in insect cells expressing the major capsid protein VP1. J. Virol. 654991-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore, J. D., J. Yang, R. Truant, and S. Kornbluth. 1999. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J. Cell Biol. 144213-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreland, R. B., and R. L. Garcea. 1991. Characterization of a nuclear localization sequence in the polyomavirus capsid protein VP1. Virology 185513-518. [DOI] [PubMed] [Google Scholar]
- 36.Moroianu, J., M. Hijikata, G. Blobel, and A. Radu. 1995. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. USA 926532-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakanishi, A., D. Shum, H. Morioka, E. Otsuka, and H. Kasamatsu. 2002. Interaction of the Vp3 nuclear localization signal with the importin α2/β heterodimer directs nuclear entry of infecting simian virus 40. J. Virol. 769368-9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson, L. M., R. C. Rose, L. LeRoux, C. Lane, K. Bruya, and J. Moroianu. 2000. Nuclear import and DNA binding of human papillomavirus type 45 L1 capsid protein. J. Cell Biochem. 79225-238. [PubMed] [Google Scholar]
- 39.Nelson, L. M., R. C. Rose, and J. Moroianu. 2002. Nuclear import strategies of high risk HPV16 L1 major capsid protein. J. Biol. Chem. 27723958-23964. [DOI] [PubMed] [Google Scholar]
- 40.Qu, Q., H. Sawa, T. Suzuki, S. Semba, C. Henmi, Y. Okada, M. Tsuda, S. Tanaka, W. J. Atwood, and K. Nagashima. 2004. Nuclear entry mechanism of the human polyomavirus JC virus-like particle: role of importins and the nuclear pore complex. J. Biol. Chem. 27927735-27742. [DOI] [PubMed] [Google Scholar]
- 41.Rose, R. C., W. Bonnez, R. C. Reichman, and R. L. Garcea. 1993. Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J. Virol. 671936-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sapp, M., C. Volpers, M. Muller, and R. E. Streeck. 1995. Organization of the major and minor capsid proteins in human papillomavirus type 33 virus-like particles. J. Gen. Virol. 762407-2412. [DOI] [PubMed] [Google Scholar]
- 43.Schelhaas, M., J. Malmstrom, L. Pelkmans, J. Haugstetter, L. Ellgaard, K. Grunewald, and A. Helenius. 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131516-529. [DOI] [PubMed] [Google Scholar]
- 44.Stehle, T., S. J. Gamblin, Y. Yan, and S. C. Harrison. 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure 4165-182. [DOI] [PubMed] [Google Scholar]
- 45.Stehle, T., and S. C. Harrison. 1997. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 165139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touze, A., D. Mahe, S. El Mehdaoui, C. Dupuy, A. L. Combita-Rojas, L. Bousarghin, P. Y. Sizaret, and P. Coursaget. 2000. The nine C-terminal amino acids of the major capsid protein of the human papillomavirus type 16 are essential for DNA binding and gene transfer capacity. FEMS Microbiol. Lett. 189121-127. [DOI] [PubMed] [Google Scholar]
- 47.Truant, R., and B. R. Cullen. 1999. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol. Cell. Biol. 191210-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 2681049-1053. [DOI] [PubMed] [Google Scholar]
- 49.Wychowski, C., D. Benichou, and M. Girard. 1986. A domain of SV40 capsid polypeptide VP1 that specifies migration into the cell nucleus. EMBO J. 52569-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]