Abstract
Yeast and vertebrate nuclear pores display significant morphological similarity by electron microscopy, but sequence similarity between the respective proteins has been more difficult to observe. Herein we have identified a vertebrate nucleoporin, Nup93, in both human and Xenopus that has proved to be an evolutionarily related homologue of the yeast nucleoporin Nic96p. Polyclonal antiserum to human Nup93 detects corresponding proteins in human, rat, and Xenopus cells. Immunofluorescence and immunoelectron microscopy localize vertebrate Nup93 at the nuclear basket and at or near the nuclear entry to the gated channel of the pore. Immunoprecipitation from both mammalian and Xenopus cell extracts indicates that a small fraction of Nup93 physically interacts with the nucleoporin p62, just as yeast Nic96p interacts with the yeast p62 homologue. However, a large fraction of vertebrate Nup93 is extracted from pores and is also present in Xenopus egg extracts in complex with a newly discovered 205-kDa protein. Mass spectrometric sequencing of the human 205-kDa protein reveals that this protein is encoded by an open reading frame, KIAAO225, present in the human database. The putative human nucleoporin of 205 kDa has related sequence homologues in Caenorhabditis elegans and Saccharomyces cerevisiae. To analyze the role of the Nup93 complex in the pore, nuclei were assembled that lack the Nup93 complex after immunodepletion of a Xenopus nuclear reconstitution extract. The Nup93-complex–depleted nuclei are clearly defective for correct nuclear pore assembly. From these experiments, we conclude that the vertebrate and yeast pore have significant homology in their functionally important cores and that, with the identification of Nup93 and the 205-kDa protein, we have extended the knowledge of the nearest-neighbor interactions of this core in both yeast and vertebrates.
INTRODUCTION
The nuclear pore complex (NPC) is a large macromolecular structure that fuses and perforates the two nuclear membranes. The 120-million-dalton nuclear pore provides the major route for the active transport of molecules between the nucleus and cytoplasm (for recent review, see Davis, 1995; Goldberg and Allen, 1995; Panté and Aebi, 1995; Simos and Hurt, 1995). From electron microscopy, the pore complex is seen to have a modular organization, consisting of an octasymmetrical framework of eight spokes sandwiched between cytoplasmic and nuclear rings. The spokes embrace a central channel or “transporter” that is thought to carry out the gated aspects of nucleocytoplasmic transport (Unwin and Milligan, 1982; Akey and Radermacher, 1993). Other microscopically visible features include eight cytoplasmic filaments, which extend from the cytoplasmic ring of the pore, and a nuclear basket, which extends from the nuclear ring of the pore into the nucleoplasm. Electron microscopy reveals that the eightfold symmetry and modular aspects of the pore complex have been largely conserved in evolution from yeast to higher eukaryotes (Rout and Blobel, 1993), although the yeast pore is somewhat smaller with some reduction in domain number. The strong overall conservation in pore morphology, however, suggests that the complex structure is in some way essential to bidirectional nucleocytoplasmic transport.
The cytoplasmic factors required for nuclear transport have also been substantially conserved through evolution. The nuclear localization sequence (NLS) receptor proteins, importin α and β, as well as the accessory cytoplasmic factors Ran and NTF2, are each highly conserved between yeast and vertebrates (Moore and Blobel, 1993; Görlich et al., 1994, 1996; Yano et al., 1994; Becker et al., 1995; Chi et al., 1995; Enenkel et al., 1995; Ren et al., 1995; Clarkson et al., 1996; Corbett et al., 1996). Thus, it has come as a puzzle why much less similarity has been observed between the actual pore proteins themselves in yeast and vertebrates. One possibility is that the disparity is a function of not having identified a significant enough fraction of pore proteins in each species to achieve many matches. As more pore proteins are identified and as the entire yeast genome has become known, this is less likely. A second possibility is that the molecular structure of the pore is less conserved than the morphological structure.
For a number of years, researchers have wished to combine the power of genetics in the yeast system with the reconstitution and immunocytochemical observations possible with vertebrate systems. If such combinations were possible, then theoretically one could arrive at a fuller picture of the pore and the mechanism of nuclear import. In consequence, we have sought proteins that are similar between the evolutionarily distant vertebrate and yeast systems in the expectation that these will allow us to use the genetic deductions derived from yeast and apply them directly to an understanding of the vertebrate pore, with all the ramifications that may have for oncogenesis, signal transduction, viral infection, and other processes influenced by nuclear transport. In addition, conserved proteins would be thought to be those most integral to the machinery of the pore.
To date, only four pairs of unique nucleoporins show homology over their entire amino acid sequence between yeast and vertebrates, rat p62/S.c. Nsp1p, rat Nup107/S.c. Nup84p, rat Nup155/S.c. Nup170p and rat/Xenopus Nup98/S. c. Nup116p (where S.c. is Saccharomyces cerevisiae; Carmo-Fonseca et al., 1991; Radu et al., 1994; Aitchison et al., 1995a; Powers et al., 1995; Radu et al., 1995; Kenna et al., 1996; Siniossoglou et al., 1996). The first pore protein ever identified in vertebrates was p62 and was shown to be vital to the process of nuclear import (Davis and Blobel, 1986; Finlay et al., 1991). Nucleoporin p62 is homologous in sequence to one of the first pore proteins identified in yeast, Nsp1 (Nehrbass et al., 1990; Carmo-Fonseca et al., 1991). Both proteins contain FXFG repeats in their amino-terminal domains, but more importantly, both contain unique carboxyl termini that are identical in length and are 32% identical (Nehrbass et al., 1990; Carmo-Fonseca et al., 1991). Electron microscopy and transport data place p62 and Nsp1p in the functional core of the pore: when Xenopus nuclei were reconstituted without the p62 complex, the altered NPC no longer imported nuclear proteins (Finlay et al., 1991). Mutations in the components of the yeast Nsp1p complex similarly impair nuclear protein import (Grandi et al., 1995b). In vertebrates, the functionally essential epitopes of p62 have been found at both the cytoplasmic and nuclear sides of the central “transporter” (Grote et al., 1995; Guan et al., 1995). It is for this reason that we tried to extend our knowledge of the pore from these centrally located proteins outward.
Early on it was reasoned that identification of the neighbors of known pore proteins would eventually lead to a three-dimensional structure of the pore, which contains up to 100 different proteins, and in addition, would lead to the discovery of novel pore proteins. Lacking genetics this has been the major mode of attack in the vertebrate pore and has also proved highly useful in the study of the yeast pore. Protein p62 was found to be extractable from rat liver pores in a major complex of four nucleoporins, p62, p58, p54, and p45 (Dabauvalle et al., 1990; Finlay et al., 1991; Kita et al., 1993; Buss and Stewart, 1995; Guan et al., 1995; Hu et al., 1996). Xenopus p62 was also seen in a less abundant second complex with the putative oncogenic nucleoporin Nup214/CAN, presumably hinting toward another nearest neighbor in the three-dimensional structure of the pore (Macaulay et al., 1995). In yeast, Nsp1p can be extracted from pores in a tight complex with three proteins, Nup49p, Nup57p, and a large protein, Nic96p (Grandi et al., 1993, 1995a,b), which led to the discovery of these proteins as pore proteins physically associated with Nsp1p. Cloned genes exist for vertebrate p58, p54, and p45 and for yeast Nup49p and Nup57p (Grandi et al., 1993; Hu et al., 1996). Our attention focused then on Nic96p, the protein present in the yeast complex because it is extracted from pores but missing in the extracted vertebrate p62 complex. In yeast, mutants in Nic96p are defective in import, and from genetic and biochemical data, Nic96p is proposed to provide a binding site for the heterotrimeric Nsp1p complex to the large NPC scaffold (Grandi et al., 1993; Nehrbass et al., 1993; Grandi et al., 1995b; Loeb et al., 1995; Shulga et al., 1996). We asked whether we could find any evidence of a homologue for Nic96p in vertebrates.
In this study, we report the finding of a 93-kDa nucleoporin, termed Nup93, in both Xenopus and humans. Nup93 appears to be the vertebrate homologue of yeast Nic96p with homology throughout its length. By immunoelectron microscopy on vertebrate pores, Nup93 localizes both to the basket of the pore and to the nuclear entry of the central gated channel of the pore. Immunoprecipitation of nucleoporin Nup93 shows that it forms a tight complex with a previously undiscovered 205-kDa protein. Partial sequencing of this latter protein by mass spectrometry has led to identification of the human gene encoding the 205-kDa protein, as well as to related homologues in S. cerevisiae and Caenorhabditis elegans. A fraction of Nup93 also interacts with p62. Using a Xenopus nuclear reconstitution system to prepare vertebrate nuclei lacking Nup93, we show that the vertebrate Nup93 complex is required for correct nuclear pore biogenesis.
MATERIALS AND METHODS
Production and Purification of Anti-Nup93 Antibodies
The cDNA sequence corresponding to the NUP93 gene was kindly provided by Dr. Nobuo Nomura (Kazusa DNA Research Institute, Kisarazu Chiba, Japan). This cDNA clone, which was already present in the GenBank sequence data library, has the accession number D42085. The sequence homology of this putative open reading frame (ORF) to yeast Nic96p was reported by Dr. Nomura. The first 654 nucleotides (corresponding to the protein sequence from Asp-2 to Lys-218) of the Nup93 cDNA cloned into pBluescript were amplified by polymerase chain reaction (PCR) using the specific primers XhoI AAAACTCGAGGGATACTGAGGGGTTTGG and MluI+stop AAAAACGCGTTCACTTATCATCCAGCTCTGCGACGG, which also introduced a XhoI site at the 5′ end and a MluI site plus stop codon at the 3′ end of the PCR clone. This PCR fragment was cut with XhoI and MluI restriction enzymes and inserted into a pET8c vector (Studier et al., 1990), giving in-frame fusion with the DNA encoding the (His)6 tag. This plasmid was transformed into Escherichia coli BL21 cells and ampicillin-resistant clones were grown in LB-Amp medium (containing 25 μg/l ampicillin) at 37°C until OD260 = 0.5. The expression of the fusion protein (His)6-Nup93 was induced for 3 h at 23°C by addition of 1 mM isopropyl β-d-thiogalactoside. Bacterial cell lysis, solubilization of inclusion bodies with 8 M urea, and purification of the fusion protein on a Ni-agarose column was done as described previously (Grandi et al., 1995b). Seven hundred OD260 units of cells yielded approximately 2 mg of a 24-kDa (His)6-Nup93 fusion protein. To obtain polyclonal antibodies, rabbits were injected two times with 300 μg of the fusion protein, followed by 200 μg/injection, every 14 d. Bleeds were collected 1 wk after each injection, the clot was discarded, and the crude serum was incubated with the (His)6-Nup93 fusion protein immobilized on an Affi-Gel 15 (Bio-Rad, Richmond, CA) column. After extensive PBS washes, the specific anti-Nup93 antibodies were eluted with 0.2 M glycine hydrochloride, pH 2.8, and 0.5 M NaCl, dialyzed against PBS, and concentrated with Centricon 30 (Amicon, Beverly, MA) to 1 mg/ml total protein (Bio-Rad).
Immunoprecipitation
Rat liver nuclei were prepared according to Blobel and Potter (1966). Rat liver nuclear envelopes (RLNEs) were obtained after salt extraction and DNase I digestion. From 300 g of rat liver, 300 mg of RLNEs were obtained. To optimize the solubilization of Nup93, 0.5 mg of nuclear envelopes (NEs) were resuspended in 250 μl of 20 mM Tris(hydroxymethyl)aminomethane hydrochloride, pH 7.4, 150 mM KCl, 0.2 mM MgCl2, and 1 mM dithiothreitol (DTT) to which Triton X-100 (final concentrations, 0, 0.5, 1, and 2%) and NaCl (final concentrations, 0, 0.5, 1, and 2 M) were added. The samples were incubated for 45 min on ice and then separated into a soluble fraction and a pellet by ultracentrifugation at 140,000 × g for 1 h (60,000 rpm, TL100 rotor).
To prepare HeLa and NRK whole cell extracts, 106 cells grown to 80–90% confluence were scraped from culture plates and washed with PBS, and the cell pellet was frozen at −20°C. After thawing, the cells were lysed by vigorous resuspension in 1.5 ml of 2% Triton lysis buffer [20 mM Tris(hydroxymethyl)aminomethane hydrochloride, pH 7.4, 150 mM KCl, 0.2 mM MgCl2, 1 mM DTT, 2% Triton X-100], kept on ice for 15 min, and cleared by centrifugation at 12,000 rpm in an Eppendorf minifuge at 4°C for 30 min.
Xenopus low-speed egg extracts (Finlay and Forbes, 1990) were also solubilized in 2% Triton lysis buffer, kept on ice for 15 min, and cleared by centrifugation at 12,000 rpm in an Eppendorf minifuge at 4°C for 30 min.
For immunoprecipitation, the supernatants of RLNE, HeLa cells, NRK cells, and Xenopus egg extracts were each preincubated with 200 μg of protein A-Sepharose CL4B (Pharmacia, Uppsala, Sweden) per 1 ml of extract for 45 min at 4°C to minimize nonspecific binding to the Sepharose resin. The cleared supernatant was then incubated for 2 h at 4°C with 5 μg of affinity-purified anti-Nup93 antibodies or 25 μl of rabbit anti-p62 antibodies (crude serum; Finlay et al., 1991) already bound to protein A-Sepharose. The immunopellet was then separated from the immune supernatant by a 2-min centrifugation at 3000 rpm in an Eppendorf minifuge and washed extensively with PBS.
For SDS-PAGE, 1% of the homogenate and immune supernatant and 5% of the immune pellet were loaded on the gels. After blotting, nitrocellulose membranes were incubated with 1:200 diluted anti-Nup93 affinity-purified antibodies and 1:1000 diluted anti-rat p62 rabbit immune serum (Finlay et al., 1991) for 2 h at room temperature. Goat anti-rabbit antibodies conjugated to horseradish peroxidase, 1:1500 diluted, were used as secondary reagents.
Determination of Protein Sequences with Mass Spectroscopy
The 205-kDa protein associated with Nup93 was separated by SDS-PAGE were in-gel digested with trypsin and purified as described (Shevchenko et al., 1996a; Wilm et al., 1996). Specifically, the recovered unseparated peptide mixture was electrosprayed with a nanoelectrospray ion source (Wilm and Mann, 1996) and sequenced by triple quadrupole mass spectrometry, as explained in (Mann and Wilm, 1995). Three peptide sequences were obtained, as described in RESULTS, and these were compared with existing sequences in the GenBank sequence database using Blast analysis. When GenBank was searched for homology, the sequences LLPEQLIK, LTAPEDVFSK, and MLALALLDR were found to be present in a single human protein, the clone for which was present in the nr data base as KIAA0225 (accession number D86978; cloned and sequenced by Nobuo Nomura, Takahiro Nagase, and colleagues [Kazusa DNA Research Institute, Kisarazu Chiba, Japan]). When the fact that mass spectrometry cannot distinguish between the isobaric amino acids isoleucine and leucine is taken into account, the peptide sequences were completely identical to the ones in the database, proving the mass spectrometric sequencing to be highly accurate despite the low amounts of protein on the gel and the presence of an excess of antibody in the band. A C. elegans homologue, CEK12D12 (accession number Z49069), was picked up by comparing human KIAA0225 to all sequences in the nr database with a tblastn search. A S. cerevisiae homologue, ORF YJL039c (accession number Z49314), was picked up by searching the S. cerevisiae protein database with a blastp search. The three sequences KIAA0225, CEK 12D12, and ORF YJL039c were aligned and compared using a ClustalW multiple sequence alignment program and a Boxshade program. The gene encoding human p205 was further analyzed for known domains using the Prosearch program.
Indirect Immunofluorescence
For indirect immunofluorescence, HeLa or NRK cells grown on round coverslips to 50% confluence were washed carefully with PBS, incubated with 0.5% Triton X-100 or 0.3% digitonin in PBS for 3 min on ice, and then fixed with 3% paraformaldehyde for 10 min at room temperature. The excess of paraformaldehyde was quenched by incubation with 0.1 M glycine, and subsequently the cells were incubated in 10% goat serum/PBS for 30 min at room temperature. The anti-Nup93 affinity-purified antibody was then diluted 1:50 or the monoclonal antibody (mAb) 414 was diluted 1:1000 in 10% goat serum/PBS and incubated for 1 h. After three 10-min washes with PBS, the cells were incubated with the secondary antibodies (goat anti-rabbit-Texas red or goat anti-mouse-fluorescein isothiocyanate), 1:100 diluted in 10% goat serum/PBS, and incubated for 1 h at room temperature. DNA staining was done with Hoechst 33285. Cells were visualized with a Zeiss confocal microscope.
Immunoelectron Microscopy
For immunogold labeling, 8-nm-diameter colloidal gold particles were prepared and the affinity-purified anti-Nup93 antibody was conjugated to colloidal gold particles as described by Pantéet al. (1994). Localization of Nup93 was performed in isolated RLNE by preembedding labeling and in rat liver by postembedding labeling. For preembedding labeling, RLNEs (isolated as described by Gerace et al. [1984]) at a concentration of 50 OD260 units/ml were incubated with the gold-conjugated anti-Nup93 antibody at a final concentration of 5 μg/ml for 1 h or 20 μg/ml for 2 h at room temperature. As control, immunogold labeling was also performed with the mAbs QE5 (Pantéet al., 1994) and RL31 (Guan et al., 1995). Samples were centrifuged in an Eppendorf centrifuge at 5000 × g, and the pellets were washed very gentle (without further centrifugation) for three 10-min periods with PBS containing 1% bovine serum albumin (BSA). Next, samples were fixed with 2% glutaraldehyde in PBS at 4°C for 30 min, washed three times with ice-cold PBS, and postfixed for 1 h with 1% OsO4 in PBS at 4°C. Fixed samples were then dehydrated and embedded in Epon 812 resin as described by Jarnik and Aebi (1991). Thin sections were cut on a microtome (Ultracut, Reichert-Jung Optische Werke, Vienna, Austria) using a diamond knife. The sections were collected on carbon/colloidon-coated electron microscope grids, stained with 6% uranyl acetate for 45 min, and poststained with 2% lead citrate for 1 min (Milloning, 1961).
For postembedding labeling, small pieces of fresh rat liver were prefixed at 4°C for 1 h with 4% paraformaldehyde/0.1% glutaraldehyde in PBS containing 0.18 M sucrose, washed three times with ice-cold PBS, and postfixed for 1 h with 1% OsO4 in PBS at 4°C. Fixed samples were then dehydrated and embedded in LR White resin (Polyscience, Northampton, United Kingdom). The resin was polymerized for 24 h at 60°C. Thin sections were obtained and collected on carbon/colloidon-coated electron microscope grids. For immunolabeling, the grids were incubated face down in a moist chamber on drops of 1) 2% BSA in PBS for two 5-min periods and 2) 1 μg/ml gold-conjugated anti-Nup93 antibody for 2 h. The grids were then washed with a constant jet of PBS, fixed for 5 min with 1% glutaraldehyde in PBS, and jet washed in water. The sections were stained as indicated above. Electron micrographs were recorded with a Hitachi H-8000 transmission electron microscope (Hitachi, Tokyo, Japan) operated at an acceleration voltage of 75 kV.
Immunodepletion and Nuclear Import Assays
Methods are as described in (Powers et al., 1995) with the following differences. Xenopus egg cytosol was depleted of Xenopus Nup93 with 1.6 μg of affinity-purified anti-Nup93 antibodies/μl of cytosol. A control extract was treated the same way with nonspecific affinity-purified rabbit IgG (catalogue number 55480, Cappel, Durham, NC). First, 160 μg of antibodies were bound to 25 μl of protein A-Sepharose overnight. The bound antibody was washed with egg lysis buffer: 10 mM HEDES, pH 7.4, 50 mM KCl. 2.5 mM HgCl2, 250 mM sucrose, and 1 mM DTT (ELB), preequilibrated with 2 volumes of 20 mg/ml BSA in ELB for 1 h, and washed again with ELB. At this time, 100 μl of freshly prepared undepleted egg cytosol was added to 12.5 μl of antibody beads and incubated at 4°C tumbling for 1 h. The partially depleted extract was incubated with 12.5 μl of the remaining antibody beads for an additional hour under the above conditions.
Immunodepletion by anti-Nup93 antiserum was of necessity performed on undiluted egg cytosol (40 mg/ml protein), since dilution of the cytosol interferes with subsequent nuclear reconstitution. Immunodepletion removed Nup93, as stated in the RESULTS (see Figure 7, lane 8). The pellet of this immunodepletion could not be effectively analyzed by silver staining for coimmunoprecipitating proteins, because nonspecific silver-staining background from the high concentration of protein obscures any specific protein band. To determine specific partners that were being depleted with Nup93, a separate immunoprecipation was performed (see Figure 5B), revealing the 205-kDa protein. If other nucleoporins are immunodepleted with Nup93, it was not evident under the conditions of this immunoprecipitation; probing such possibilities on the immunodepleted cytosol itself will have to await the development of antibodies against each nucleoporin, followed by immunoblots on Nup93-depleted concentrated cytosol.
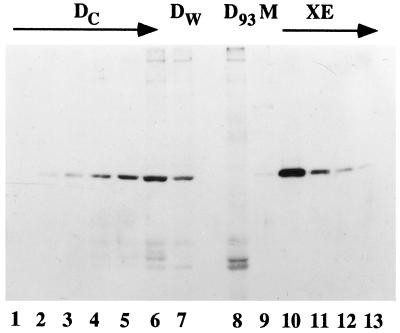
Figure 7.
Nup93 is depleted from Xenopus egg cytosol by immunodepletion with anti-Nup93 antibodies. To determine the extent of depletion of Nup93 from the Nup93-depleted extract and membrane, different amounts of Dc cytosol were electrophoresed on an 8% polyacrylamide gel in lanes 1–6 (0.01, 0.02, 0.04, 0.1, 0.2, and 0.5 μl, respectively). D93 extract was electrophoresed in lane 8 (1 μl). Examination indicates that the D93 cytosol was depleted of more than 99% of Nup93 when compared with Dc in lane 2 (0.02 μl). Washed membranes (1 μl) contained a small percentage of Nup93, as shown in lane 9, estimated to be 0.2% of the total Nup93 normally present in the egg extract.
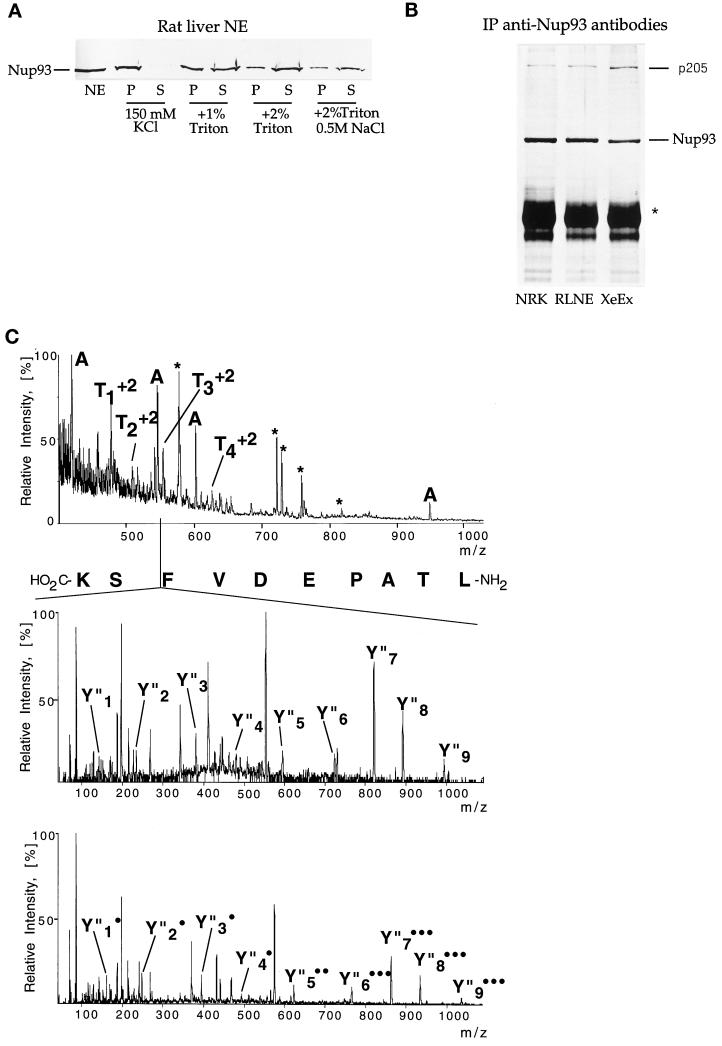
Figure 5.
Nup93, an evolutionarily conserved nucleoporin, is physically associated with a 205-kDa protein. (A) Western blot decorated with anti-Nup93 antibodies. The blot contains purified NEs and derived soluble (S) and insoluble (P) fractions that were obtained by extracting NEs with the indicated salt and detergent concentrations. Note that 2% Triton X-100 solubilizes the majority of Nup93 (70–80%). (B) Silver staining of immunoprecipitates (IP) obtained with anti-Nup93 antibodies from NRK cells (lane NRK), RLNEs (lane RLNE), and Xenopus egg extracts (lane XeEx). The bands corresponding to Nup93 and p205 are indicated. The star indicates the position of the heavy chain of IgG. (C, top) Mass spectrum of the peptide mixture extracted after in-gel tryptic digest of the band with apparent mass 205 kDa. Peaks that are different from known trypsin autolysis products (marked by an asterisk) were fragmented in turn. Peaks designated by A originate from immunoglobulins, as was shown by sequencing and database searching using peptide sequence tags (Mann and Wilm, 1994). (C, middle and bottom) Sequencing of one of the peptide ions of the 205-kDa protein. The doubly charged ion with m/z 553.8 was isolated and fragmented in the mass spectrometer and tandem mass spectra acquired. Collision fragmentation of tryptic peptides resulted in continuous series of fragments containing the carboxyl terminus of the peptide (Y′′ ions). The methylated form of the same peptide (produced by esterification of the whole peptide mixture) was also fragmented (C, bottom). Esterification with methanol identified the carboxyl-terminal ions by their characteristic 14-Da mass shift for every fragment containing the carboxyl terminus and an additional 14-Da shift for each internal Asp and Glu residue present in the fragment. The number of incorporated methyl groups is designated by • (C, bottom). The actual sequence (LTAPEDVFSK) is retrieved by software-assisted comparison of the two sets of sequence data (derivatized and underivatized) by matching precise mass differences between adjacent peaks in the series of Y′′ ions. Two more peptides, Ta (LLPEQLLK) and Tb (MLALALLDR), were sequenced in the same way. Note that L stands both for leucine and isoleucine because mass spectrometry cannot distinguish between these two isobaric amino acids. Only partial sequence information was obtained for the peptide Td but the spectrum was unambiguously matched to the sequence DLPSADSVQYR in the sequence of the 205-kDa protein using a peptide sequence tag, assembled from the tandem mass spectrometric data. (D) Sequence alignment of evolutionarily conserved 205-kDa proteins: the aligned p205 sequences from human KIAA0225 (hum), C. elegans CEK12D12 (Cel), and S. cerevisiae ORF YJL039c (Sc) are shown to document that the proteins align along their complete length. Amino acids common to two or three of the the genes are shaded in black and structurally related amino acids are shaded in gray.
Nuclear reconstitution of Nup93-depleted nuclei was carried out as follows: a 40-μl nuclear assembly reaction typically contained 30 μl of control, Nup93-depleted, or wheat germ agglutinin (WGA)-depleted cytosol, 3.5 μl of washed membranes, an ATP regeneration system, demembranated sperm chromatin, and glycogen. Reconstitution and functional assays were done as described in Powers et al. (1995).
Anti-Nup93 antibodies were added to the following nuclear reconstitution mixture: 10 μl of low-speed Xenopus egg extract and 5 μl of XB buffer [10 mM HEPES, pH 7.4, 250 mM sucrose, 50 mM K(CH3COOH)2, 2.5 mM Mg(CH3COOH)2, 1 mM DTT, 1× protease inhibitors] plus ATP-regenerating system (1×) and 104 demembranated Xenopus sperms. To measure DNA replication, 2.5 mCi of [α-32P]ATP was added, and the samples were incubated at 23°C for a maximum of 2 h. Proteinase K digestions were carried out by adding 1 mg of proteinase K and 1% SDS to each sample and incubating for 2 h at 37°C. The samples were then directly loaded in a 1.2% agarose gel and electrophoresed for 4 h at 100 V. After drying the gel was exposed overnight at −20°C on a Kodak X-Omat film. Nuclear size and NPC biogenesis were measured by immunofluorescence: nuclear reconstitution samples were incubated at 23°C for 1 h, fixed with 3% formaldehyde, and processed for immunofluorescence with mAb414 as described above. The DNA was stained by Hoechst.
Anchored Nuclei and Immunofluorescence Microscopy
Reconstitution of anchored nuclei and immunofluorescence on these nuclei were done as described in Macaulay and Forbes (1996) with the following exceptions. Nuclei were allowed to form at room temperature for 1.5 h. The egg cytosol and membrane mixture was removed. The anchored nuclei were washed five times with ELBK (Macaulay and Forbes, 1996), fixed with 5% formaldehyde in ELBK for 20 min at room temperature, and rinsed with PBS before performing indirect immunofluorescence with mAb414 (1:1000 dilution; BabCo, Richmond, CA).
DNA Replication Assay
The method for performing DNA replication was as described in Powers et al. (1995). Demembranated sperm chromatin was used in the assay. Two microcuries of [α-32P]dCTP were added to a 40-μl reaction. After the samples were run on a 0.8% agarose gel, the gel was dried and exposed to film or to a phosphor screen to quantitate the DNA replication level. Quantitation was done with a Phosphorimager model 445SI (Molecular Dynamics, Sunnyvale, CA) using ImageQuaNT software.
Expression of Human Nup93 in Yeast
To express the human Nup93 cDNA in yeast, the complete ORF (2681 bp) was fused in-frame to the 3′ end of the triple myc tag sequence (triple myc-Nup93) or to the 3′ end of two IgG-binding sequences of protein A (ProtA-Nup93) (Grandi et al., 1993). These fusion genes were put under the control of the yeast NOP1 promoter, inserted into the 2 μ plasmid pRS425, and transformed into yeast cells. The fusion proteins (Triple myc-Nup93 and ProtA-Nup93) were correctly expressed as seen by probing with anti-myc and anti-protein A antibodies on a Western blot of whole yeast cell extracts. To test for complementation, both Nup93 constructs were transformed into the following nic96 mutant strains: nic96 null, nic96–1, and nic96–2 (Zabel et al., 1996). As a control, the fusion gene ProtA-NIC96 also driven by the NOP1 promoter was used (Grandi et al., 1995b).
RESULTS
Identification of Human and Xenopus Homologues of Yeast Nucleoporin Nic96p
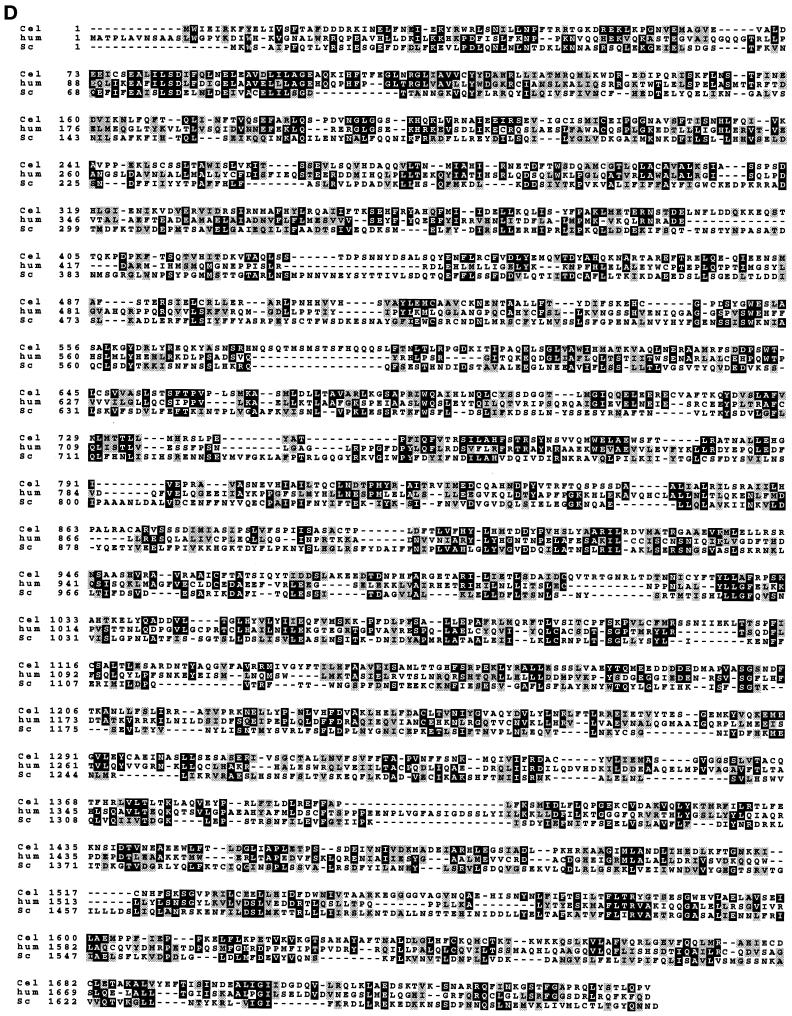
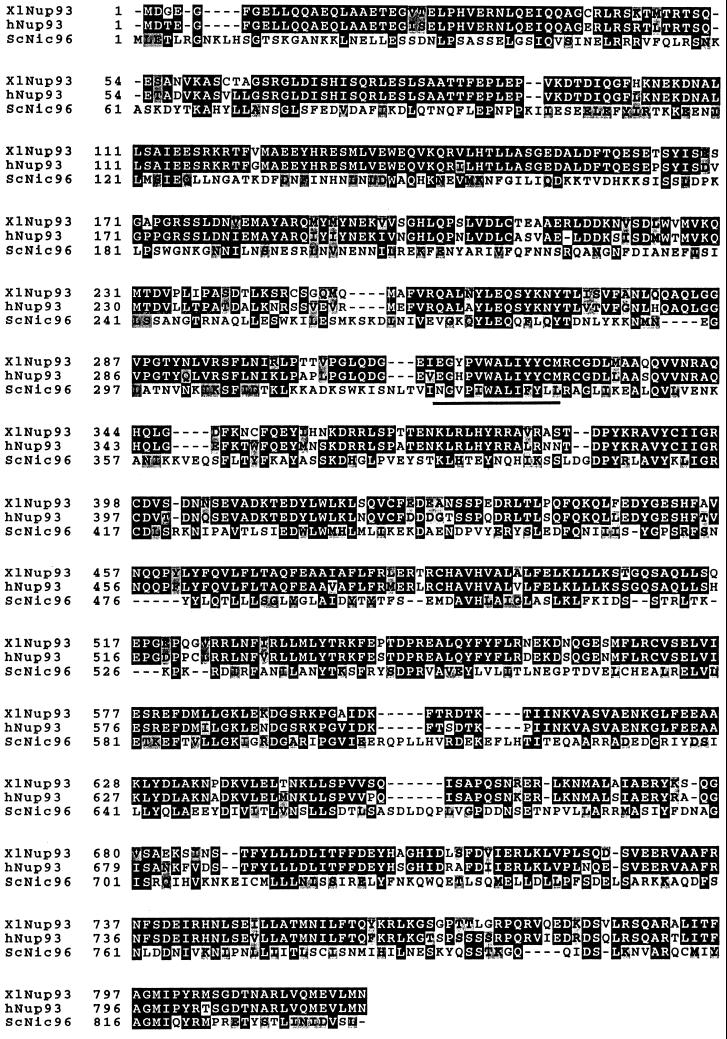
To identify potential vertebrate homologues of essential partners of the yeast Nsp1p complex, a search of vertebrate protein sequence data libraries was done. The search revealed two putative higher eukaryotic homologues of the yeast nucleoporin Nic96p; i.e., two cDNA clones whose deduced amino acid sequences showed significant homology to Nic96p (Figure 1). One cDNA clone is derived from the mRNA of a human immature myeloid cell line and the other is derived from Xenopus laevis mRNA. The putative human Nic96 homologue has 819 amino acids and a calculated molecular weight of 93.4 kDa; the corresponding Xenopus Nic96 homologue has 820 amino acids and a molecular weight of 93.3 kDa. These novel proteins were named Nup93 and are 84% identical. Human Nup93 is 27% identical and 50% similar to yeast Nic96p over nearly its entire sequence (Figure 1). The highest conservation between vertebrates and yeast is found in the central and carboxyl terminal domains of these three proteins (Figure 1). A short sequence (amino acids 328–341), which in Nic96p is essential for cell growth and in which single point mutations yield a thermosensitive phenotype (Grandi et al., 1995b; Zabel et al., 1996), is highly conserved in human and Xenopus Nup93 (Figure 1). During the course of this work, a zebrafish homologue called dead eye, which was found to be essential for embryonic development, was also described (Allende et al., 1996).
Figure 1.
Human and Xenopus Nup93 exhibit significant sequence homology to yeast Nic96p. The deduced amino acid sequence of human and Xenopus Nup93 and yeast Nic96p were aligned using the programs pileup/prettybox. Amino acids common to two or three of the the genes are shaded in black; structurally related amino acids are shaded in gray. A highly conserved short peptide sequence within yeast Nic96p that is essential for cell growth is also indicated. The accession number of the human Nup93 gene is D42085 and of the Xenopus gene is U63919.
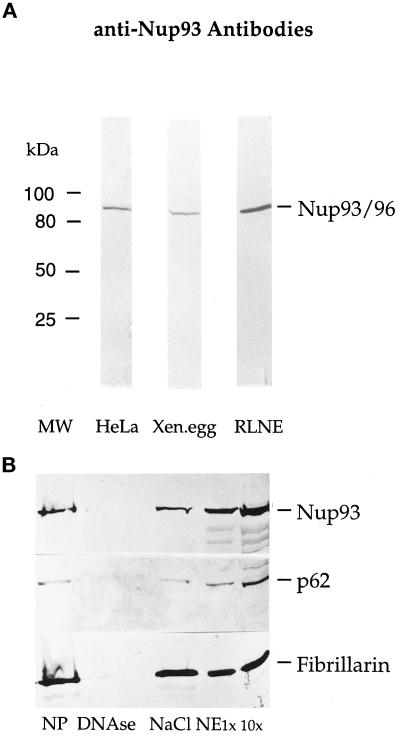
To determine whether human Nup93 is indeed a nucleoporin, polyclonal antibodies were raised in rabbits to the first 200 amino-terminal amino acids of Nup93 (see MATERIAL AND METHODS). Anti-Nup93 antibodies were affinity-purified and tested by SDS-PAGE, followed by Western blotting of whole cell lysates derived from HeLa cells, rat liver nuclei, and Xenopus egg extracts (Figure 2A). In all cases, a single band of ≈90 kDa was stained by the affinity-purified antibodies (Figure 2A). When an immunoblot of isolated NEs was probed with anti-Nup93 affinity-purified antibodies, the Nup93 band was strongly enriched in the NE fraction (Figure 2B, lanes NE), a fraction in which nucleoporin p62 was also enriched. The nucleolar marker protein fibrillarin, however, was significantly extracted from the NEs by salt treatment (Figure 2B, lane NaCl). Thus, Nup93 coenriches with NEs during biochemical fractionation of rat liver nuclei.
Figure 2.
Nup93 is present in cells from human to Xenopus and is enriched in RLNE preparations. (A) Western blot analysis of HeLa whole cell homogenate (lane HeLa), Xenopus egg extract (lane Xen.egg), and RLNEs (lane RLNE) labeled with anti-Nup93 antibodies. The bands visible correspond to human and rat liver Nup93 and Xenopus Nup93. (B) Western blot analysis of the different fractions obtained during RLNE purification were labeled with affinity-purified anti-Nup93 (top), rabbit polyclonal anti-p62 (middle), and rabbit polyclonal anti-fibrillarin antibodies (bottom). The bands corresponding to Nup93, p62, and fibrillarin are indicated. Lane NP, nuclear pellet; lane DNAse, DNase washes; lane NaCl, 0.5 M salt wash; lanes NE 1× and 10×, NEs loaded in 1- or 10-fold volume equivalents.
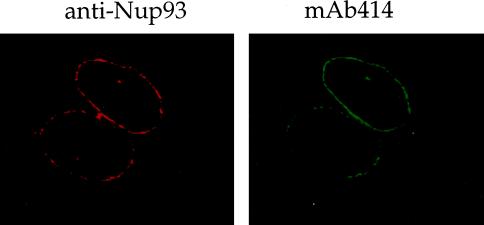
Human Nup93 Is Localized at the Nuclear Pores as Revealed by Its Punctate NE Staining in Indirect Immunofluorescence
To localize Nup93 in cells, affinity-purified anti-Nup93 antibodies were used in indirect immunofluorescence to label HeLa (Figure 3) and NRK (our unpublished results) tissue culture cells. As seen in the confocal microscope, anti-Nup93 antibodies predominantly stained the nuclear periphery in a punctate pattern, although internal nuclear staining was also observed. The NE labeling largely overlapped with the staining produced by mAb414 that recognizes nuclear pores (Wente et al., 1992; Figure 3). Thus, human Nup93 not only is homologous to the yeast nucleoporin Nic96p but also is localized in NPCs.
Figure 3.
Nup93 is a novel nucleoporin. Indirect immunofluorescence staining of HeLa cells using anti-Nup93 antibodies. The punctuate staining of the NE of a confocal section is overlapping with the staining of NPCs obtained with mAb414.
To obtain information as to which side of the nuclear membrane Nup93 is localized, indirect immunofluorescence was performed after permeabilization of HeLa cells with digitonin, which permeabilizes the plasma membrane but leaves the nuclear membrane intact. Under these conditions, little of the punctuate nuclear rim staining normally seen with anti-Nup93 antibodies on Triton X-100-permeabilized cells was observed on the digitonin-permeabilized cells (our unpublished results). In contrast, mAb414, which recognizes nucleoporins on both the cytoplasmic and nuclear sides of the NPCs, still stained the nuclear periphery in a punctate manner in digitonin-permeabilized cells (our unpublished results). This suggested that Nup93 might not be present or not be accessible to the antibodies on the cytoplasmic side of the NPC.
Given that immunofluorescence showed vertebrate Nup93 to be a nucleoporin and to have extended similarity to its yeast counterpart, Nic96p (23% identity, 50% similarity), we asked whether hNup93 could complement yeast Nic96 mutants. We found, however, that hNup93, even when overexpressed from a high-copy-number plasmid (see MATERIAL AND METHODS), failed to complement either a nic96::HIS3 null mutant or two temperature-sensitive Nic96 mutants (Grandi et al., 1993, 1995b; our unpublished results). Human Nup93 also failed to target to the yeast pore, as determined by transformation of a Myc-tagged Nup93 protein and immunofluorescence using anti-Myc antibodies (our unpublished results). Presumably, enough divergence has occurred between yeast and humans to alter certain amino acids crucial both to Nic96/Nup93 function and to pore localization, such that complementation and localization of the human nucleoporin to the pore in yeast does not occur. We also observed that an antibody directed to the first 10 amino acids of yeast Nic96 did not cross-react with human Nup93. Similarly, the antiserum to the first 200 amino acids of human Nup93 used throughout this manuscript did not cross-react with yeast Nic96 (our unpublished results). The lower identity of the two proteins in the amino-terminal region was presumed to explain this lack of antibody cross-reactivity.
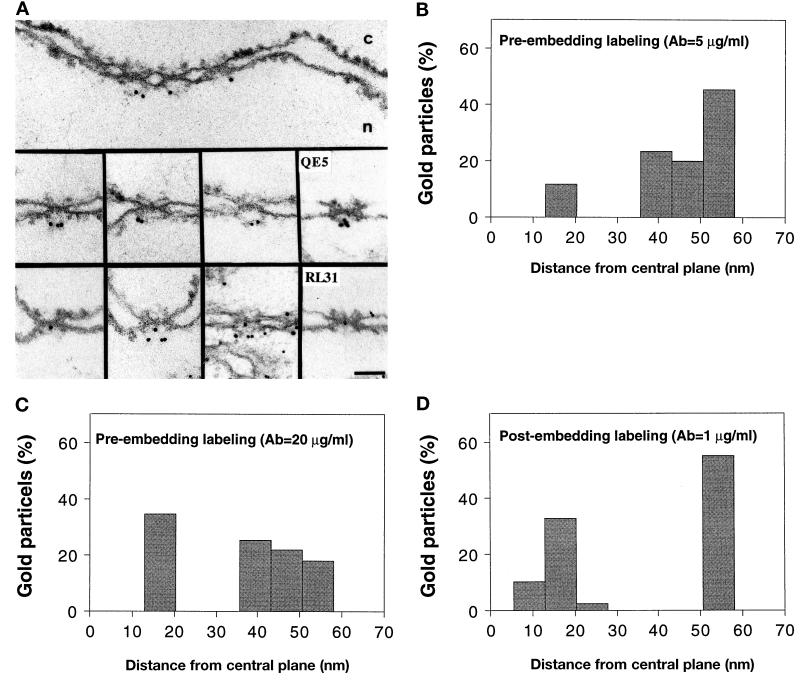
Nup93 Shows a Preferential Location on the Nuclear Basket and at or near the Nuclear Entry to the Central Channel of the NPC
To localize Nup93 more precisely, preembedding immunogold electron microscopy was performed on isolated RLNEs with gold-conjugated anti-Nup93 antibody. As shown in Figure 4A, anti-Nup93 antibody labeled the nuclear side of RLNEs exclusively. The labeling was concentrated at the nuclear periphery of the NPCs at vertical distances within 35 and 55 nm from the central plane of the NPC (Figure 4A, top and middle). This position corresponds to the nuclear baskets, the distal filamentous structures associated with the nuclear side of the pore known (Ris, 1989a,b; Jarnik and Aebi, 1991; Goldberg and Allen, 1992). These structures are also labeled by an anti-nucleoporin mAb, QE5 (Pantéet al., 1994; see also Figure 4A, middle, rightmost example). Labeling at this concentration of antibody was also seen, albeit less frequently, in more central NPC regions (Figure 4A, bottom), similar to the nuclear labeling yield by the mAb RL31 (anti-p62; Guan et al., 1995; see also Figure 4A, bottom, rightmost example).
Figure 4.
Localization of Nup93 in RLNES. (A) Cross-section of a single NE and selected examples of NPCs labeled with the polyclonal anti-Nup93 antibody conjugated to 8-nm colloidal gold by preembedding labeling. The rightmost examples (middle and bottom) are NPCs labeled with the mAbs QE5 and RL31 (anti-p62) conjugated to 8-nm colloidal gold, respectively. Cytoplasmic (c) and nuclear (n) sides of the NE are indicated. Bar, 100 nm. (B–D) Quantitative analysis of gold particles associated with the nuclear side of NPCs after preembedding labeling with 5 μg/ml anti-Nup93 antibody. (B) Sixty particles were scored and 20 μg/ml of anti-Nup93 antibody. (C) Fifty-five particles were scored after postembedding labeling with 1 μg/ml anti-Nup93 antibody. (D) Forty particles were scored.
Quantitative analysis of the distribution of gold particles revealed that 89% of the gold particles were clustered within 35 and 55 nm of the central plane (Figure 4B). The second more-sparse labeling observed at lower vertical distances might be an indication that there is Nup93 epitope at this position but that it is less accessible to the antibody than Nup93 located at the nuclear baskets. Consistent with this explanation, when preembedding labeling was performed with 20 μg/ml (instead of 5 μg/ml) anti-Nup93 antibody, the extent of labeling at lower vertical distances closer to the central gate of the pore increased to 34% (Figure 4C). Similarly, when postembedding labeling of rat liver was performed 45% of the gold particles were found within 10–20 nm from the central plane of the NPC (Figure 4D), although by this method the antibody gave an unspecific labeling at the cytoplasm. Thus, our immunogold electron microscopy studies indicate that Nup93 is located both on the nuclear basket and at or near the nuclear entry to the central channel of the pore.
Human Nup93 and Xenopus Nup93 Are Physically Associated with an Evolutionarily Conserved 205-kDa Protein
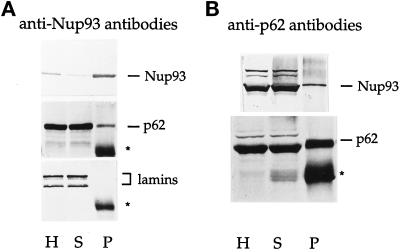
To gain insight into the structural and functional neighbors of Nup93 within the pore, we sought to determine by immunoprecipitation whether Nup93 is physically associated with any known or novel nuclear pore protein(s). RLNEs were extracted with buffers containing different salt and detergent concentrations to determine the optimal conditions to solubilize the pores. As shown in a pilot experiment, the nonionic detergent Triton X-100 at a concentration of 2% was necessary to solubilize the majority of Nup93 from the NEs (Figure 5A, lanes +2% Triton). RLNEs, as well as NRK cell and Xenopus egg extracts, were then treated with 2% Triton X-100 and 150 mM KCl before addition of anti-Nup93 antibodies for immunoprecipitation (see MATERIALS AND METHODS). A silver stain of the immunoprecipitates derived from RLNEs, NRK cells, and Xenopus egg extracts after SDS-PAGE is shown in Figure 5B. In all three cases, a single prominent band of ∼205 kDa was observed to coimmunoprecipitate with Nup93 (Figure 5B). A very slightly faster migrating band, ∼180 kDa rather than 205 kDa, was observed upon immunoprecipitation of HeLa extracts (our unpublished results); this size difference may derive from a slight proteolysis seen with the HeLa extract (see below). Control immunoprecipitation performed with nonrelated rabbit IgGs on the rat liver, NRK, and Xenopus extracts and competition studies using the Nup93 amino-terminal fusion protein indicated that the immunoprecipitation of Nup93 and its coprecipitating p205 component was specific (our unpublished results). p205 is not recognized by the anti-Nup93 antibodies on Western blots (our unpublished results). We conclude not only that is Nup93 highly conserved between mammals and Xenopus at the sequence level (see Figure 1) but also that a physical interaction of Nup93 with a 205-kDa protein is also highly conserved between these species.
To determine the identity of this large protein, the protein was coimmunoprecipitated with Nup93 from HeLa cells, electrophoresed by SDS-PAGE, and cut out of the silver-stained gel. It was then digested in the gel with trypsin and analyzed for peptide sequence by mass spectroscopy (Wilm et al., 1996). A comparison of native and derivatized peptides in tandem mass spectrometry allowed three peptide sequences to be determined unambiguously: LLPEQLLK, LTAPEDVFSK, and MLALALLDR. At the time of the sequence analysis these peptides showed no significant homology to any protein sequence in the database. When GenBank was searched for homology, at a later date, the sequences LLPEQLIK, LTAPEDVFSK, and MLALALLDR were found to be present in a single human protein, the clone for which was present in the database as KIAA0225 (accession number D86978). When the fact that mass spectrometry cannot distinguish between the isobaric amino acids isoleucine and leucine is taken into account, the peptide sequences were completely identical to the ones in the database. The human KIAA0225 gene encodes a protein of 2012 amino acids with a predicted molecular weight of 228 kDa. This relates well to the apparent molecular weight of the p205 band observed from rat and Xenopus extracts in SDS-polyacrylamide gels and led to our conclusion of slightly increased proteolysis in the HeLa extract. One caveat noted by the sequence group that obtained the KIAA0225 sequence is that the actual amino terminus may not be included in the sequence. However, the predicted size of 228 kDa is greater than the observed size of 205 kDa and both the yeast and C. elegans homologues (see below) have amino termini ending within 15–20 amino acids of the presumed human KIAA0225 amino terminus. The gene encoding human p205 is entirely unique in sequence. When searched with a Prosearch program, p205 has numerous potential protein kinase C and CK2 phosphorylation sites. Interestingly, p205 also contains a leucine zipper at amino acids 810–832, which may form a potential interaction domain either for p205 dimerization or for p205 interaction with another nucleoporin. A similar search of Nup93 revealed no obvious leucine zipper.
When Blast analysis was done with human KIAA0225, a C. elegans homologue, CEK12D12, of 1696 amino acids with a predicted molecular weight of 191.2 kDa was observed. Moreover, a S. cerevisiae homologue of 1683 amino acids with a predicted molecular weight of 191.5 kDa, ORF YJL039c, was also observed. The yeast gene is located in the intragenic region between the yeast nucleoporin gene NSP1 and the karyogamy gene KAR2. When the human, C. elegans, and yeast 205 homologues were aligned using the ClustalW sequence alignment protein, the proteins aligned along their complete length (Figure 5D), although the larger human protein appears to have a 230-amino acid additional sequence at its carboxyl-terminal end.
Evidence for Interaction of Nup93 with p62
Because the p62 complex found in rat and Xenopus shares some common characteristics with the yeast Nsp1p complex, we asked whether vertebrate Nup93 could also be found in physical interaction with p62. Although coisolation of a 90-kDa protein with the p62 complex had not previously been reported, we looked for a substoichiometric association of Nup93 with p62. Immunoprecipitation with anti-Nup93 antibodies was performed on HeLa cell extracts and the corresponding Western blot was probed with anti-p62 and anti-Nup93 antibodies, respectively. A significant although small portion of p62 was always found in the immune pellet of Nup93, which was efficiently immunoprecipitated under these conditions (Figure 6A, lane P). The presence of either p58 or p54, members of the p62 complex, in the immune pellet was difficult to assess because they migrate very close to the heavy chain of the immunoglobulins (our unpublished results). Significantly, lamins were never found in the Nup93 immunoprecipitate (Figure 6A), arguing against a nonspecific association of p62 with Nup93. Similarly, we were unable to detect the Nup214/CAN nucleoporin, which forms a separate complex with p62 in Xenopus egg extracts (Macaulay et al., 1995), in the Nup93 immunoprecipitate by immunoblotting the same fractions with mAb414 and QE5 mAbs that recognize Nup214/CAN (Pantéet al., 1994; our unpublished results). Lastly, in a complementary experiment, where anti-p62 antibodies were used for immunoprecipitation, 5–10% of the total Nup93 could be coprecipitated with p62 (Figure 6B, lane P). We conclude that Nup93 binds to a minor fraction of p62 or, alternately, Nup93 binds to a greater fraction of p62 weakly but dissociates during the immunoprecipitation of Nup93, which is done in the presence of 2% Triton X-100. In either case, the interaction of Nup93 and p62 within the pore appears specific.
Figure 6.
Interaction of Nup93 with p62. Western blots of immunoprecipitations using HeLa cell extracts as a source of Nup93. (A) Immunoprecipitation with anti-Nup93 antibodies. Western blots were decorated with anti-Nup93 antibodies (top), anti-p62 antibodies (middle), and anti-lamin antibodies (bottom). (B) Immunoprecipitation with anti-p62 antibodies. Western blots were decorated with anti-Nup93 antibodies (top) and anti-p62 antibodies (bottom). The bands corresponding to Nup93, p62, and lamins are indicated. Stars mark the position of the heavy chain of the IgG. Lane H, homogenate; lane S, immune supernatant; lane P, immune pellet.
Vertebrate Nup93 Is Required for Correct Nuclear Pore Biogenesis
To study the function of the evolutionarily conserved Nup93 protein in more detail, we took advantage of a Xenopus nuclear reconstitution system to form nuclei lacking the Xenopus Nup93 complex. For this, the soluble cytosol of a Xenopus egg extract was immunodepleted of ≥99% of the Nup93 with anti-Nup93 antibodies coupled to protein A-Sepharose (Figure 7, lane 8). This immunodepleted cytosol was then mixed with sperm chromatin and membrane vesicles to assemble nuclei (Powers et al., 1995). An almost quantitative removal of Nup93 from the membrane vesicles was achieved by washing with 0.25 M KCl before use (Figure 7, lane 9). The final Nup93-complex–depleted reaction contained approximately 0.2% of the normal amount of Nup93 in a control reaction. Nuclei were allowed to form for 2 h at room temperature, and then the size of the nuclei formed and the extent of nuclear import of a fluorescent transport substrate were measured. These measurements were also performed in parallel on nuclei assembled with mock-depleted and with WGA-depleted extracts. WGA binds to several nucleoporins in Xenopus extracts including p62, Nup98, and Nup214/CAN and depletes them from the extracts (Finlay et al., 1987; Powers et al., 1995). Interestingly, we found that a fraction of the Nup93 found in egg extracts also binds to WGA-Sepharose, although it does not itself appear to be a glycosylated protein on WGA-horseradish peroxidase blots. Nup93 depletion, however, did not significantly deplete p62, Nup98, or Nup214/CAN, and Nup93 does not immunoprecipitate with those proteins under even mild conditions (our unpublished results).
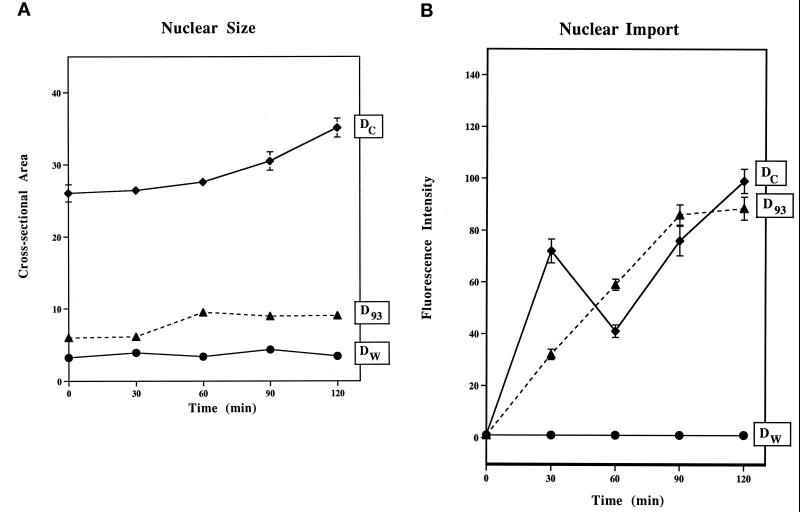
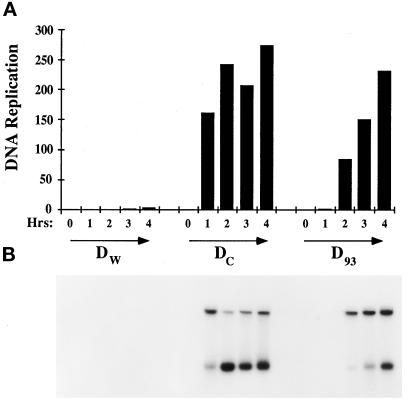
When nuclei were reconstituted in the Nup93-complex–depleted extract, the average cross-sectional area of the nuclei (D93) was found to be fourfold smaller when compared with mock-depleted control nuclei (Dc; Figure 8A). To test for nuclear function, we measured DNA replication in the Nup93-complex–depleted nuclei by assessing the extent of incorporation of [32P]ATP into newly synthesized genomic DNA. In the depleted nuclei, Nup93 depletion caused a 1-h delay in DNA replication, measured by agarose-gel electrophoresis followed by quantitation of the labeled genomic DNA, as compared with mock-depleted nuclei (Figure 9). We found that nuclei lacking the Nup93 complex, although smaller in size, when tested for nuclear import, could import a nuclear transport substrate containing the simian virus 40 large tumor antigen NLS fairly normally (Figure 8B). Import of this substrate was unaffected in the Nup93-complex–depleted nuclei.
Figure 8.
Nup93-complex–depleted nuclei are impaired in nuclear growth but are competent for nuclear import. (A) Nuclear size. Reconstituted nuclei were allowed to form at room temperature for 2 h, and then the karyophilic transport substrate (ss-HSA) was added. At this time, which we called t = 0, nuclei were quantitated for import level and measured for size. Depleted WGA binding protein nuclei (Dw) were done as a negative control for comparison. In A, the cross-sectional area of depleted Nup93 nuclei (D93) is reduced by a factor of 4 compare to mock-depleted nuclei (Dc) over a time period of 4 h. (B) Nuclear import. Nuclear import of the transport substrate appears normal in nuclei lacking Nup93. Transport level was normalized to 16× integration for all time points.
Figure 9.
DNA replication in depleted Nup93 nuclei (D93) is delayed. Replication was performed with demembranated sperm chromatin at 1000 sperm/μl in Dw, Dc, and D93 egg lysate. Replication is seen at 2 h in D93 nuclei, whereas it could be seen in 1 h in the Dc nuclei. To quantitate, a rectangular box was drawn for each lane to include the two bands for measurement. The units on the Y-axis are arbitary units given by the Phosphoimager model 445SI.
Because thermosensitive mutations in the central domain of Nic96p in yeast have been shown to inhibit nuclear pore biogenesis at the restrictive temperature (Zabel et al., 1996), we were most interested to ask whether the presence and distribution of nuclear pores was altered in Nup93-complex–depleted nuclei. For this, anchored nuclei were reconstituted on a glass surface (Macaulay and Forbes, 1996) using Nup93-complex–depleted and control-depleted Xenopus egg cytosol and membrane fractions. Specifically, sperm chromatin was decondensed and attached to a polylysine-coated coverslip, and then egg cytosol and membranes were added. Under control cytosol conditions, a double nuclear membrane replete with numerous nuclear pores is formed (Macaulay and Forbes, 1996).
Once the anchored nuclei were formed, immunofluorescence was performed with the anti-nucleoporin mAb mAb414. mAb414 recognizes four nucleoporins, p62, p214/CAN, Nup153, and RanBP2, and normally gives a bright nuclear rim stain. The intensity of the immunofluorescent signal of the nuclear rim obtained by labeling the nuclear pores was compared for Nup93-complex–depleted and mock-depleted nuclei. The nuclear rims of control and mock-depleted nuclei were extremely bright, so bright that the punctate nature was obscured. We have seen this pattern in the past and have confirmed it to be due to a high number of assembled nuclear pores present. In Nup93-complex–depleted nuclei, however, the signal was greatly reduced and the pattern was visibly punctate (Figure 10). The immunofluorescent nuclear stains typical of control (Dc) nuclei and of Nup93-complex–depleted nuclei (D93) nucleus are shown at equal intensification (32-fold) in Figure 10, a and b. A direct comparison shows the large decrease in mAb414 staining caused by Nup93-complex depletion. When the mAb414 staining of a Nup93-complex–depleted nucleus was intensified 128-fold, focusing on the nuclear rim (Figure 10C) and on the nuclear surface (Figure 10D), individual punctate foci, of the same apparent size as individual pores (or small clusters of pores) as normally observed in tissue culture cells, were seen. A large reduction in apparent pore number is thus seen in the Nup93-complex–depleted nuclei; individual pores are now visible. As stated above, normally the large number of pores per area makes individual pores difficult to visualize in control Dc-reconstituted nuclei (Figure 10A). The apparent reduced number of NPCs in Nup93-complex–depleted nuclei, however, seem to be uniformly distributed at the level of the immunofluorescence resolution (Figure 10D). We also tested the D93 nuclei with a second antiserum to confirm the staining observed: an identical staining pattern as in Figure 10D was observed when anti-Nup214/CAN antiserum, instead of mAb414, was used to analyze the Nup93-complex–depleted nuclei (our unpublished results). Thus, we conclude that the Nup93 complex, which includes Nup93 and the Nup93-interacting protein p205, is required for the complete biogenesis of a normal nuclear pores.
Figure 10.
Anchored nuclei formed in Nup93- complex–depleted Xenopus egg cytosol show greatly decreased staining with mAb414. Egg cytosol was immunodepleted with either anti-Nup93 antibodies (D93) or nonspecific rabbit IgG (Dc). The rim stain of Dc nucleus (a) and the rim stain of a D93 nucleus (b) at 1500× magnification are shown. The mAb414 staining of the D93 nucleus is dimmer and patchy compared with the control. (c) Another D93 nucleus at a higher-intensity integration, which is 4× the integration of a and b. (d) Same nucleus as in c is shown but image is focused on the surface of the nucleus.
The role of the Nup93 complex was also tested in a different way. As it was done to study of the role of nuclear lamins in DNA replication, anti-Nup93 antibodies were added to a normal nuclear reconstitution extract at t = 0. Nuclear growth in the presence of antibody was greatly reduced, DNA replication was inhibited, and there was a strongly reduced mAb414 staining of the nuclei (Figure 11). Thus, the addition of the antibody had a similar effect to the removal of the Nup93 complex from nuclei, reinforcing the conclusion that the Nup93 complex is required for correct nuclear pore biogenesis.
Figure 11.
Antibodies against Nup98 reveal decreased staining of NPCs with mAb414 and inhibit nuclear growth and DNA replication. (A) mAb414 staining and nuclear growth: addition of 3 μl (4.5 μg) of anti-Nup93 antibodies to a Xenopus nuclear reconstitution extract causes a strong decrease in the immunofluorescence staining of nuclear pore proteins recognized by mAb414 and a reduced sperm nuclei size as seen by Hoechst staining. Control experiments were performed by adding 3 μl (4.5 μg) of unspecific rabbit IgG (unspecific antibodies). Two sperms of each sample are shown. Note that the pictures of the samples incubated with anti-Nup93 antibodies were exposed twice as long as the samples treated with unspecific antibodies. (B) DNA replication: the addition of 1 μl (1.5 μg) and 3 μl (4.5 μg) of anti-Nup93 antibodies (lanes 2 and 3, respectively) but not 3 μl of buffer (lanes 1) or 3 μl (4.5 μg) of unspecific rabbit IgGs (lanes 4) to the nuclear reconstitution extract causes a strong inhibition of newly synthesized (32P-labeled) high molecular weight DNA normally produced upon DNA replication. Equal volumes were taken at time points 0 (t = 0 h), after 1 h (t = 1 h), and after 2 h (t = 2 h) of incubation, digested with proteinase K, and loaded onto a agarose gel.
DISCUSSION
Despite the fact that the three-dimensional architecture of the basic framework of the NPC has been solved at 10-nm resolution, we are still far from knowing the identity of many of the nucleoporins, their arrangement within the pore, or indeed, the roles of most nucleoporins in nuclear transport. By maximally exploiting the benefits of several organisms, we predict that it should be possible to overcome the respective restrictions of each and gain insight into these unknown areas. In this article, we have identified and characterized a novel vertebrate nucleoporin, Nup93, finding human and X. laevis Nup93 genes. These counterparts appear to be well conserved sequence homologues of the essential yeast nucleoporin Nic96p. Immunofluorescence and immunoelectron microscopy place Nup93 on the nuclear pore basket and at or near the nuclear entry of the central gated channel of the pore. When RLNEs are treated with Triton X-100, Nup93 is extracted from the nuclear pore in a complex with a novel protein that has an apparent molecular weight of 200 kDa. Its identification in a complex with Nup93 protein that has been extracted directly from nuclear pores likely identifies the 205-kDa protein as a nucleoporin, in much the same manner as Nup58, Nup54, and Nup45 were identified as nucleoporins by virtue of being extracted from nuclear pores in a complex with the known nucleoporin p62. However, definite designation of the 205-kDa protein as a nucleoporin will require the production of an antibody and immunofluorescence microscopy. By peptide sequence analysis, we find that the 205-kDa protein is encoded by the human gene KIAA0225 (actual molecular weight, 228 kDa). We have identified counterparts of this new putative nucleoporin gene in C. elegans and S. cerevisiae. To probe the role of the Nup93 complex in vertebrate nuclear pores, nuclei were reconstituted with pores that lack the Nup93 complex. Absence of the Nup93 complex results in nuclei that are functional for transport, but small in size and greatly reduced in the number of complete nuclear pores. These results indicate that the vertebrate Nup93 complex is required for complete nuclear pore biogenesis.
The landscape of the vertebrate pore, which is expected to contain 50–100 proteins, is at present sparsely populated. Interestingly, the peripheral structures of the pore, the cytoplasmic filaments and the nuclear basket, contain the majority of identified pore proteins, although these structures make up only a fraction of the mass of the pore. Four proteins are known constituents of the cytoplasmic filaments, Nup214/CAN, Nup88, and RanBP2/Nup358, and a fourth protein that associates reversibly with the cytoplasmic filaments of the pore, Crm1 (Kraemer et al., 1994; Wu et al., 1995; Yokoyama et al., 1995; Fornerod et al., 1997; Mahajan et al., 1997; Zimowska et al., 1997). The supposition is that these proteins are involved in the early steps of nuclear import. Two proteins have been localized to the nuclear side of the pore on or near the nuclear basket, Nup153 and Nup98. These also extend intranuclearly (Cordes et al., 1993; Sukegawa and Blobel, 1993; Powers et al., 1995; Radu et al., 1995). A third protein, Tpr, is on intranuclear fibers distinct from the pore basket but attached to it (Byrd et al., 1994; Cordes et al., 1997; Zimowska et al., 1997). Of these, Nup214/CAN, RanBP2/Nup358, Nup153, and Tpr have no known yeast homologues, despite the fact that the entire yeast genome is known. Interestingly, the genes encoding Nup214/CAN, Nup98, and Tpr have been found to be altered in chromosomal translocations causing human leukemias (Byrd et al., 1994; Kraemer et al., 1994; Nakamura et al., 1996). Thus, far from being anonymous members of a large but polyglot cellular structure, each pore protein becomes of interest as a potential altered member of a essential signal cascade that must function normally for correct cell regulation.
Less populated with identified proteins is the bulk of the nuclear pore. This portion consists of the large eight-spoke complex, which in turn is bracketed by the nuclear and cytoplasmic rings and contains at its hub the central transporter. Two integral membrane proteins, POM121 and gp210, have been identified in this area and are presumably key to the initial assembly of the pore and to anchoring the completed pore within the membranes (Greber et al., 1990; Wozniak et al., 1992; Hallberg et al., 1993). Two other vertebrate pore proteins, Nup107 and Nup155, may be localized at this site. The localization of Nup155 has been described as on both the cytoplasmic and nuclear faces of the pore (Radu et al., 1993); Nup107 has been mapped in a general manner to the pore (Radu et al., 1994). These proteins are unique in sequence and stand out from the majority of the vertebrate proteins above in that they have identifiable yeast homologues. Only Nup155 complements its yeast homologue (Aitchison et al., 1995; Kenna et al., 1996). The roles of these two proteins in vertebrates and the identity of their vertebrate neighbors are unknown.
The only other known vertebrate pore proteins, and the most important proteins of these central structures are p62 and its set of complexed proteins p58/p54/p45 (Finlay et al., 1991; Guan et al., 1995; Hu et al., 1996; Schlaich et al., 1996). The p62 complex maps the most centrally in the vertebrate pore to date, and vertebrate p62 shows significant homology to yeast Nsp1p. The p62/Nsp1p proteins are particularly interesting in both yeast and vertebrate because their absence results in greatly diminished nuclear import. Identification of the partners of this pair of nucleoporins, p58, p54, and p45 in vertebrates (Finlay et al., 1991; Guan et al., 1995; Hu et al., 1996) and Nup57p, Nup49p, and Nic96p in yeast (Grandi et al., 1993, 1995b), by coimmunoprecipitation and subsequent cloning has revealed some homologies between p54/Nup57p and p58/Nup49p (Doye and Hurt, 1997). We therefore looked for the existence of a vertebrate Nic96p equivalent. We reasoned that the pore may break apart slightly differently in the two different species and a vertebrate Nic96 equivalent may well exist but become separated from its nearest neighbors, the p62–p58–p54 (p45) complex, in the extraction process. Upon investigation, Nup93, a homologue of yeast Nic96, was indeed discovered to be a component of the vertebrate pore.
Nup93 resembles its yeast counterpart in that it consists entirely of a unique sequence, lacks repeat domains, and is predicted to fold into a coiled-coil domain at the amino terminus. Further examination reveals that Nup93 is extracted from the pores of vertebrates in a complex with a hitherto unknown protein with an apparent molecular weight of 205 kDa. This Nup93–p205 complex is a fairly stable one. Moreover, it appears to represent a subcomplex from which pores are assembled at mitosis, because the Nup93–p205 complex is found to be present in a mitotically disassembled state in Xenopus egg cytosol. The stoichiometry of the Nup93–p205 complex as assessed by silver staining appears not to be 1:1, although this may not be the case because silver staining is often not proportional to protein concentration. For example, the silver staining of individual proteins of the p62–p58–p54–p45 complex fails to accurately reflect their stoichiometry (Miller and Forbes, unpublished results). There is also a formal possibility that other proteins exist in the p93–p205 complex that are not stained with silver; this will be a question addressed in future experiments to identify potential new nucleoporins and nearest neighbors.
As would be predicted from the yeast Nic96p–Nsp1p–Nup57p–Nup49p complex, a small fraction of Nup93 was also extractable from HeLa cells in association with nucleoporin p62. Because of the low amounts of protein, we do not know whether the Nup93–p62 complex also contains p205 or other nucleoporins but believe that this alternate complex most likely arises from the nuclear pore fracturing along a slightly different plane during the extraction procedure. Another explanation as to why this Nup93–p62 complex is not as abundant as the yeast Nsp1p–Nic96p complex is that harsher conditions are required to extract vertebrate pore proteins from the dense vertebrate nuclear lamina; less harsh conditions are required for extraction of yeast NEs, potentially leaving larger complexes intact.
When the different complexes extracted from yeast and vertebrate pores are considered, it is interesting to note that the amino-terminal domain of yeast Nic96p is predicted to fold into a coiled-coil structure, whereas the coiled-coil prediction is less strong for the corresponding amino-terminal domain of Nup93 (our unpublished results). The coiled-coil domain of yeast Nic96p was shown to be required for the direct binding to the Nsp1p–Nup57p–Nup49p complex (Grandi et al., 1995b). Thus, vertebrate Nup93 may associate with the p62–p58–p54–p45 complex less strongly than its yeast counterpart.
Several of the various yeast nucleoporin mutants isolated to date show perturbation of the distribution of NPCs or NE morphology (for example, see Wente and Blobel, 1993; Gorsch et al., 1995; Heath et al., 1995; Li et al., 1995; Schlaich and Hurt, 1995; for reviews see Davis, 1995; Doye and Hurt, 1995). Reconstitution of vertebrate nuclei without the p62–p58–p54 complex shows about normal numbers of nuclear pores. Certain thermosensitive mutants of Nic96 in yeast, however, do inhibit pore biogenesis at the restrictive temperature (Zabel et al., 1996). Nup93 and Nic96p exhibit the highest homology in their central and carboxyl-terminal domains (31–32% identity; Figure 1) and mutations in the central domain of yeast Nic96p inhibit NPC biogenesis. In yeast, both the central and the carboxyl-terminal domains of Nic96p have been functionally linked to two nucleoporins, Nup188p and the transmembrane protein Pom152p, suggested to be part of the yeast NPC core structure (Nehrbass et al., 1996; Zabel et al., 1996); thus the role of Nup93 in pore biogenesis was of particular interest.
When this was assessed, we found that reconstituted Xenopus nuclei depleted of the Nup93 complex were altered primarily in that they showed a dramatic decrease in the assembly of complete nuclear pores. The immunofluorescent stain predictive of complete nuclear pores decreased greatly on the NEs, indicating that the Xenopus Nup93 complex is required for the correct biogenesis of NPCs. This decrease in complete pore assembly was observed with a mAb that recognizes proteins centrally located in the nuclear pore and on both sides of the pore (p62, Nup153, Nup214/CAN, and RanBP2). The decrease was also seen with a polyclonal antibody that recognizes only Nup214/CAN. Two possibilities for the decrease in apparent pore number in Nup93-complex–depleted nuclei exist. The first possibility is that there are indeed fewer total pores. An alternative possibility is that there may be a normal number of pores but that Nup93 is used either for the assembly of p62 and the peripheral pore proteins onto these pores or for the stability and maintenance of these proteins, once assembled, on the pore. In the future, it will be of interest to determine which of these possibilities exists with an extensive electron microscopic analysis, which is beyond the scope of the present study. In either case, however, the conclusion remains that the Nup93 complex is important to the complete biogenesis of nuclear pores in vertebrates.
Why is the pore number not zero in Nup93-complex–depleted nuclei? This may be due to a redundancy of function with another, as yet unknown, protein of the vertebrate pore. In yeast, examples of redundancy have been observed, albeit in a different context; the RNA binding domains of all three of a set of interrelated proteins, Nup100p, Nup116p, and Nup145p, have to be removed before an effect on viability is observed (Fabre et al., 1994). However, if Nup93 is required for stabilizing the pore once it is assembled, an alternate hypothesis to redundancy is that a fraction of pores may manage to remain stable in the absence of the Nup93 complex. A different twist on this stability model would be that pores are continually forming at a normal rate in the Nup93-complex–depleted extract but that there half-life is much shorter and the pore number observed at any one time with immunofluorescence is reduced.
Nup93-complex–depleted nuclei do still contain a fraction of nuclear pores, and moreover, these pores are also functionally active: nuclear import appears fairly normal in Nup98-depleted nuclei, at least for a classical NLS-containing karyophilic substrate. In other experiments, the import of transport substrate into the nuclei of digitonin-permeabilized cells was not inhibited by the addition of anti-Nup93 antiserum (Grandi and Weis, unpublished observations). This would be predicted because the anti-Nup93 antiserum presumably does not reach the nuclear side of the pore where the Nup93 epitope resides, as we have found herein. Additionally, the export of different classes of microinjected labeled RNAs in Xenopus oocytes was unaffected by nuclear injection of anti-Nup93 antibodies (Grandi and Izaurralde, unpublished observations). This experiment has the caveat that we may not have used antibodies of a high enough concentration or that the antibodies may bind to Nup93 in pores but may not inactivate Nup93 function. A fraction of the antibodies raised to any given protein prove capable of functional inactivation. Future experiments, involving a more extensive panel of antisera to different regions of Nup93, may be required to comment further on an involvement of Nup93 in nuclear transport in vertebrates. Finally, we cannot exclude that Nup93 is involved in nuclear protein import of a subset of nuclear proteins required for efficient DNA replication and NPC biogenesis. It is therefore important to test whether the import of other karyophilic substrates (not containing a canonical NLS) that use other nuclear import routes is inhibited in Nup93 depleted nuclei.
Another defect seen in Nup93-depleted nuclei is an initial inhibition of DNA replication. Previously, depletion of another nucleoporin, Nup98/p97, from nuclei resulted in nuclei that could import but that had a profound and seemingly permanent defect in DNA replication (Powers et al., 1995), unlike the delay of 1 h in replication by Nup93-complex–depleted intranuclei. Nup98 is a protein of the nuclear pore basket and the intranuclear interior (Powers et al., 1995; Radu et al., 1995). Antibodies to Nup98, when injected into oocyte nuclei, block the export of multiple classes of RNA (Powers et al., 1995). Antibodies to Nup93 to date, however, do not block RNA export (Grandi and Izaurralde, unpublished results). We think it likely that in the case of the Nup93-depleted nuclei, the lesser number of complete nuclear pores has led to a secondary effect on DNA replication. Interestingly, however, when anti-Nup93 antiserum was included in a nuclear reconstitution reaction from t = 0, replication was severely blocked (Figure 11). In this experiment, Nup93 with attached antibody might assemble into pores and block further pore assembly and function, so that import, export, and replication are all blocked. Further experiments will have to be done to assess the cause of the defects seen in the presence of antibody.
In yeast, immunolocalization is frequently not possible. Thus, it was of interest to identify the localization of Nup93, as the sequence homologue of yeast Nic96p and as a protein required for complete pore biogenesis. We found by immunocytochemical methods that vertebrate Nup93 was localized to distinct substructures of the nuclear pore, i.e., the nuclear baskets and at or near the nuclear entry to the central gated channel of the NPC. One can imagine that a protein located in these central and basket regions, when absent, would affect the ability of proteins such as p62 and Nup153 to assemble correctly. Their absence could in turn affect the assembly of Nup214/CAN, to which p62 can be found complexed, leading to incorrect or incomplete nuclear pore biogenesis.
Of interest to gaining knowledge of the central ring/scaffold portion of the nuclear pore is our discovery of a new potential vertebrate pore protein, p205. This protein, identified by high-sensitivity mass spectrometry, immunoprecipates in a complex with Nup93 both from extracts of vertebrate pores and from the cytosol of Xenopus eggs that contains unassembled subcomplexes of the pore. Powerful mass spectrometric methods have previously been developed which, in combination with the completely sequenced genome of S. cerevisiae, allow rapid identification of low amounts of gel-separated yeast proteins (Shevchenko et al., 1996b). As we found herein in the analysis of the novel 205-kDa protein, mass spectrometric sequencing, while more difficult in vertebrates, can also readily be used to determine protein sequence in the vertebrate system. The human peptide sequences obtained by such means allowed identification of a single human gene in the database that encoded a protein containing identical peptide sequences, KIAA0225. Homologues in size and in their presence in a Nup93 complex are present in humans, rats, and Xenopus. A search of sequence databases reveals related homologues in C. elegans and S. cerevisiae of 192 kDa. We are now raising antisera to determine whether the vertebrate complex between Nup93 and p205 can also be observed in extracts of yeast nuclear pores. Analysis of the predicted homologue in yeast, ORF YJL039c, will allow us to complete the circle that was begun by using a yeast protein, Nic96p, to search for a vertebrate homologue, analyze its vertebrate localization, use nuclear reconstitution systems to assess vertebrate function, identify a putative pore protein, p205, and return to the yeast system for genetic analysis of the yeast homologue. Thus, we can use each system for distinct purposes and synergistically extend the knowledge of nuclear pore structure and function.
Using knowledge from an evolutionarily distant organism, we have made inroads on identifying proteins central to the vertebrate nuclear pore, Nup93, and a partner protein, p205, and have elucidated a fundamental molecular similarity between yeast and vertebrate nuclear pores.
ACKNOWLEDGMENTS
We thank Nobuo Nomura (Kazusa DNA Research Institute, Kisarazu Chiba, Japan) for the NUP93 cDNA clone, Athina Pyrpasopulou (European Molecular Biology Laboratory [EMBL], Heidelberg Germany) for help in the preparation of RLNE, Augustin Alconada (EMBL, Germany) for help with tissue culture, Isabel Palacios (EMBL, Germany) for help in confocal microscopy, and Elisa Izaurralde (EMBL, Heidelberg Germany) for performing the RNA export experiments. We are grateful to Drs. B. Burke (University of Calgary, Canada), L. Gerace (Scripps Research Institute, La Jolla, CA) for the kind gift of the mAbs QE5 and RL31, and Brian Miller and Stuart Tugendreich (University of California at San Diego, La Jolla) for help in the computer analysis. This work was supported by the Kanton Basel-Stadt, the M.E. Müller Foundation of Switzerland, and research grants from the Swiss National Science Foundation (31–39691.93) and the Human Frontier Science Program Organization.
REFERENCES
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–3155. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran-TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- Blobel G, Potter VR. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 1966;154:1662–1665. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- Buss F, Stewart M. Macromolecular interactions in the nucleoporin p62 complex of rat nuclear pores: binding of nucleoporin p54 to the rod domain of p62. J Cell Biol. 1995;128:251–261. doi: 10.1083/jcb.128.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd DA, Sweet DJ, Panté N, Konstantinov KN, Guan T, Saphire ACS, Mitchell PJ, Cooper CS, Aebi U, Gerace L. Tpr, a large coiled coil protein whose amino terminus is involved in activation of oncogenic kinases, is localized to the cytoplasmic surface of the nuclear pore complex. J Cell Biol. 1994;127:1515–1526. doi: 10.1083/jcb.127.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Kern H, Hurt EC. Human nucleoporin p62 and the essential yeast nuclear pore protein NSP1 show sequence homology and a similar domain organization. Eur J Cell Biol. 1991;55:17–30. [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson WD, Kent HM, Stewart M. Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J Mol Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Köhler A, Stuurman N, van Driel R, Franke WW. Intranuclear filaments containing a nuclear pore complex protein. J Cell Biol. 1993;123:1333–1344. doi: 10.1083/jcb.123.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabauvalle M-C, Loos K, Scheer U. Identification of a soluble precursor complex essential for nuclear pore assembly. Chromosoma. 1990;100:56–66. doi: 10.1007/BF00337603. [DOI] [PubMed] [Google Scholar]
- Davis LI. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt EC. Genetic approaches to nuclear pore structure and function. Trends Genet. 1995;11:193–199. doi: 10.1016/s0168-9525(00)89057-5. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt EC. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EC. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Newmeyer DD, Price TM, Forbes DJ. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace, L., Comeau, C., and Benson, M. (1984). Organization and modulation of nuclear lamina structure. J. Cell Sci. 1(suppl), 137–160. [DOI] [PubMed]
- Goldberg MW, Allen TD. High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J Cell Biol. 1992;119:1429–1440. doi: 10.1083/jcb.119.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Allen TD. Structural and functional organization of the nuclear envelope. Curr Opin Cell Biol. 1995;7:301–309. doi: 10.1016/0955-0674(95)80083-2. [DOI] [PubMed] [Google Scholar]
- Görlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey R A, Mattaj IW, Izaurralde E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with ’GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt EC. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J Cell Biol. 1995a;130:1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995b;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber UF, Senior A, Gerace L. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 1990;9:1495–1502. doi: 10.1002/j.1460-2075.1990.tb08267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote M, Kubitscheck U, Reichelt R, Peters R. Mapping of nucleoporins to the center of the nuclear pore complex by post-embedding immunogold electron microscopy. J Cell Sci. 1995;108:2963–2972. doi: 10.1242/jcs.108.9.2963. [DOI] [PubMed] [Google Scholar]
- Guan T, Müller S, Klier G, Panté N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg E, Wozniak RW, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol. 1993;122:513–521. doi: 10.1083/jcb.122.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TH, Guan TL, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarnik M, Aebi U. Toward a more complete 3-D structure of the nuclear pore complex. J Struct Biol. 1991;107:291–308. doi: 10.1016/1047-8477(91)90054-z. [DOI] [PubMed] [Google Scholar]
- Kenna MA, Petranka JG, Reilly JL, Davis LI. Yeast Nle3p-Nup170p is required for normal stoichiometry of FG nucleoporins within the nuclear pore complex. Mol Cell Biol. 1996;16:2025–2036. doi: 10.1128/mcb.16.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K, Omata S, Horigome T. Purification and characterization of a nuclear pore glycoprotein complex containing p62. J Biochem (Tokyo) 1993;113:377–382. doi: 10.1093/oxfordjournals.jbchem.a124054. [DOI] [PubMed] [Google Scholar]
- Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li O, Heath CV, Amberg DC, Dockendorff TC, Copeland CS, Snyder M, Cole CN. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol Biol Cell. 1995;6:401–417. doi: 10.1091/mbc.6.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JDJ, Schlenstedt G, Pellman D, Kornitzer D, Silver PA, Fink GR. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C, Forbes DJ. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTPgammaS, and BAPTA. J Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mann M, Wilm M. Electrospray mass spectrometry for protein characterisation. Trends Biochem Sci. 1995;20:219–223. doi: 10.1016/s0968-0004(00)89019-2. [DOI] [PubMed] [Google Scholar]
- Mann M, Wilm MS. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- Milloning G. A modified procedure for lead staining of thin sections. J Biophys Biochem Cytol. 1961;11:736–739. doi: 10.1083/jcb.11.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Nakamura T, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Fabre E, Dihlmann S, Herth W, Hurt EC. Analysis of nucleo-cytoplasmic transport in a thermosensitive mutant of the nuclear pore protein NSP1. Eur J Cell Biol. 1993;62:1–12. [PubMed] [Google Scholar]
- Nehrbass U, Kern H, Mutvei A, Horstmann H, Marshallsay B, Hurt EC. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell. 1990;61:979–990. doi: 10.1016/0092-8674(90)90063-k. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panté, N., and Aebi, U. (1995). Exploring nuclear pore complex structure and function in molecular detail. J. Cell Sci. 108(suppl 19), 1–11. [DOI] [PubMed]
- Panté N, Bastos R, McMorrow I, Burke B, Aebi U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J Cell Biol. 1994;126:603–617. doi: 10.1083/jcb.126.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Blobel G, Wozniak RW. Nup107 is a novel nuclear pore complex protein that contains a leucine zipper. J Biol Chem. 1994;269:17600–17605. [PubMed] [Google Scholar]
- Radu A, Blobel G, Wozniak R. Nup155 is a novel nuclear pore complex protein that contains neither repetitive sequence motifs nor reacts with WGA. J Cell Biol. 1993;121:1–9. doi: 10.1083/jcb.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Ren M, Villamarin A, Shih A, Coutavas E, Moore MS, Locurcio M, Clarke V, Oppenheim JD, D’Eustachio P, Rush MG. Separate domains of the ran GTPase interact with different factors to regulate nuclear protein import and RNA processing. Mol Cell Biol. 1995;15:2117–2124. doi: 10.1128/mcb.15.4.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ris H. Three-dimensional structure of the nuclear pore complex. J Cell Biol. 1989a;109:134a. doi: 10.1083/jcb.109.3.955. (Abstract). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ris, H. (1989b). Three-dimensional imaging of cell ultrastructure with high resolution low-voltage SEM. Inst. Phys. Conf. Ser. No. 98: Chapter 16, (EMAG-MICRO 89, Lond.), 657–662.
- Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaich NL, Häner M, Lustig A, Aebi U, Hurt EC. In vitro reconstitution of a heterotrimeric nucleoporin complex consisting of recombinant Nsp1p, Nup49p and Nup57p. Mol Biol Cell. 1997;8:33–46. doi: 10.1091/mbc.8.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaich NL, Hurt EC. Analysis of nucleocytoplasmic transport and nuclear envelope structure in yeast disrupted for the gene encoding the nuclear pore protein Nup1p. Eur J Cell Biol. 1995;67:8–14. [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996b;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained gels. Anal Chem. 1996a;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for Hsp70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Hurt EC. Nucleocytoplasmic transport: factors and mechanisms. FEBS Lett. 1995;369:107–112. doi: 10.1016/0014-5793(95)00674-x. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosemberg HA, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:62–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sukegawa JC, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Unwin PN, Milligan RA. A large particle associated with the perimeter of the nuclear pore complex. J Cell Biol. 1982;93:63–75. doi: 10.1083/jcb.93.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm M, Mann M. Analytical properties of the nano electrospray ion source. Anal Chem. 1996;66:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gel by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Wozniak RW, Blobel G. The single transmembrane segment of gp210 is sufficient for sorting to the pore membrane domain of the nuclear envelope. J Cell Biol. 1992;119:1441–1449. doi: 10.1083/jcb.119.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yano R, Oakes ML, Tabb MM, Nomura M. Yeast Srp1p has homology to armadillo-plakoglobin-beta-catenin and participates in apparently multiple nuclear functions including the maintenance of the nucleolar structure. Proc Natl Acad Sci USA. 1994;91:6880–6884. doi: 10.1073/pnas.91.15.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Panté N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, Fukui M, Nishimoto T. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska G, Aris JP, Paddy MR. BX34: a large, filamentous protein localized to the extrachromosomal channel network and nuclear pore complexes in Drosophila. J Cell Sci. 1997;110:927–944. doi: 10.1242/jcs.110.8.927. [DOI] [PubMed] [Google Scholar]