Abstract
Spinal muscular atrophy (SMA) is a recessive neuromuscular disease caused by mutations in the human survival motor neuron 1 (SMN1) gene. The human SMN protein is part of a large macromolecular complex involved in the biogenesis of small ribonucleoproteins. Previously, we showed that SMN is a sarcomeric protein in flies and mice. In this report, we show that the entire mouse Smn complex localizes to the sarcomeric Z-disc. Smn colocalizes with α-actinin, a Z-disc marker protein, in both skeletal and cardiac myofibrils. Furthermore, this localization is both calcium- and calpain-dependent. Calpains are known to release proteins from various regions of the sarcomere as a part of the normal functioning of the muscle; however, this removal can be either direct or indirect. Using mammalian cell lysates, purified native SMN complexes, as well as recombinant SMN protein, we show that SMN is a direct target of calpain cleavage. Finally, myofibers from a mouse model of severe SMA, but not controls, display morphological defects that are consistent with a Z-disc deficiency. These results support the view that the SMN complex performs a muscle-specific function at the Z-discs.
INTRODUCTION
Loss-of-function mutations in the human survival motor neuron 1 (SMN1) gene result in spinal muscular atrophy (SMA), a devastating neuromuscular disorder. SMN1 is currently the only gene known to cause SMA, however, roughly 4% of the cases are unlinked to this locus (1). SMN protein is predominantly found as a part of a large macromolecular complex, consisting of nine different proteins: SMN, Gemins 2–8 and UNRIP/STRAP (2,3). SMN is involved in essential housekeeping functions as well as tissue-specific ones (4–7). These include biogenesis of small nuclear ribonucleoproteins (snRNPs), transcription, pre-mRNA splicing, axonal mRNA transport, neurite outgrowth, neuromuscular junction (NMJ) formation, myoblast fusion and myofibrillogenesis (8–23). A common theme among many of these processes is the actin cytoskeleton, however, the molecular details by which loss of SMN function results in SMA are not known.
Although a majority of studies have focused on roles for SMN in motoneurons, SMA patients display a pattern of muscle weakness that is more reminiscent of a myopathic disorder than a neurogenic one (4). SMN-depleted myoblasts from humans and mice show marked defects in fusion and proliferation (12,15). Moreover, co-cultures of SMA patient-derived muscle cells with wild-type motoneurons revealed that SMN expression in muscle is required for maintaining stable innervation (24,25). However, transgenic experiments in flies (17) and mice (26) have shown that SMN expression in muscle does not ameliorate the phenotype in the absence of neuronal co-expression. Overexpression of SMN in neurons alone also does not rescue the SMA phenotype (17,26). Thus SMA might not be a cell-autonomous disease. The observed motor defects could be caused by primary abnormalities in both muscles and motoneurons, or perhaps due to a failure of communication between these two tissues. Indeed, SMA has recently been described as an NMJ synaptopathy (27,28). Clearly, a better understanding of SMN’s role(s) in muscle cell function will help to distinguish among these possibilities.
Muscle fibers (myofibers) are composed of bundles of contractile filaments, termed myofibrils. At the ultrastructural level, each myofibril consists of hundreds of individual contractile units, called sarcomeres. Sarcomeres work in tandem to produce the mechanical force of muscle contraction. Within each sarcomere is an actin-rich I-band that is bisected by the Z-disc (or Z-line) and a myosin-rich A-band that is bisected by the M-line (29). The boundaries of each sarcomere are defined by the Z-discs, which function to interlock adjacent sarcomeres and to anchor the actin-rich thin filaments. Myofibrillar dynamics at the Z-discs and overall muscle maintenance are regulated by two distinct proteolytic pathways, the calcium-dependent calpain system and the ubiquitin proteasome system (30,31). Disruptions within both of these proteolytic pathways have been linked to a number of muscular disorders (32–35).
The ubiquitin proteasome is the principal pathway involved in muscle growth, remodeling and atrophy (36,37). Although the proteasome is the final degradative mechanism utilized by muscle during wasting, it is clear that additional proteolytic systems operate upstream to disassemble the sarcomere, as proteasomes are not able to degrade intact myofibrils (38). Calpain-mediated proteolysis is important in this regard, and this pathway also contributes to muscle protein breakdown and remodeling under catabolic conditions (37,39,40). Thus calpains are calcium-dependent proteases that act upstream of the proteasome to release proteins from the myofibrillar Z-disc (40–42).
We recently reported that reduced expression of dSMN within the adult Drosophila thorax results in severe neuromuscular dysfunction, with prominent myofibrillogenesis defects (9). Surprisingly, we found that SMN is a sarcomeric protein that forms a complex with α-actinin and colocalizes at the Z-discs of both Drosophila and mouse myofibrils. Here, we show that the entire Smn complex localizes to the Z-discs of wild-type mouse myofibrils. Furthermore, Smn is present at the Z-disc in both skeletal and cardiac muscles and, interestingly, this localization is both calcium- and calpain-dependent. In accordance with the view that calpains regulate Z-disc protein turnover (31), we also demonstrate that Smn is a direct target of calpain cleavage in vitro and in vivo. Importantly, myofibers from a mouse model of SMA, but not controls, display morphological defects that are consistent with a Z-disc deficiency. Taken together with the fact that calpains are regulatory proteases with functions in diverse cell types, the results have direct implications not only for muscle-specific functions of SMN but also for neuronal ones as well.
RESULTS
Smn localization in striated muscle
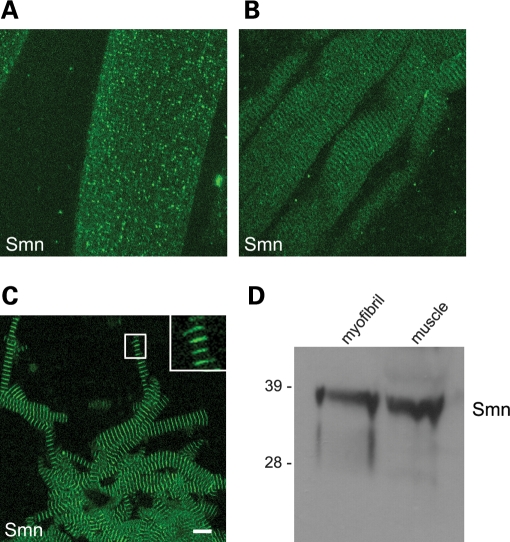
Previously, we showed that Smn localizes to the sarcomeres of native Drosophila and purified mouse striated muscle myofibrils (9). Although there was some variability in the anti-dSMN staining patterns of the native Drosophila myofibrils (i.e. those that are mechanically teased apart), the more highly purified mouse myofibrils (see Materials and Methods) showed prominent Z-disc anti-Smn staining (9). We therefore investigated the distribution of Smn in native myofibers and purified myofibrils from wild-type mice (Fig. 1). Similar to the situation in Drosophila, we observed two different patterns of Smn staining in the native mouse fibers: granular and striated (Fig. 1A and B). In the majority of muscle fibers, the granular staining pattern was predominant (Fig. 1A), whereas in a subset of fibers, a more striated pattern was revealed (Fig. 1B). A significant amount of diffuse Smn signal was overlaid upon both of the patterns (Fig. 1A and B). We hypothesized that the diffuse and granular staining was primarily due to sarcoplasmic signal and that removal of this material would allow better visualization of the sarcomeric Smn protein. Consistent with this interpretation, Smn was found to localize in a distinctly striated pattern throughout the length of the myofibrils following the purification procedure (Fig. 1C). Importantly, western blot analysis (Fig. 1D) demonstrated the presence of Smn in purified myofibrils and confirmed the specificity of the Smn signal observed by immunostaining. Thus, Smn localizes in a sarcomeric pattern on mouse skeletal muscle myofibrils.
Figure 1.
Smn localizes to skeletal myofibrils. Mouse muscle fibers and purified myofibrils were subjected to immunofluorescence imaging (average projection of a Z-stack of 13 focal planes, imaged at 0.5 µm per section) and western blotting. Antibodies against Smn (mAb Clone 8) were used to stain muscle fibers (A, B) and purified myofibrils (C); scale bar represents 10 µm. The presence of Smn was verified by western blot of whole muscle lysate and purified myofibrils (D).
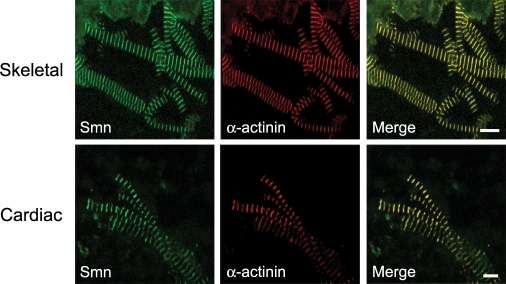
The Z-disc, which marks the boundary of each sarcomere, is composed of many different components, several of which localize to multiple subsarcomeric compartments (43). Thus we wanted to determine whether Smn colocalized with other known Z-disc proteins. α-Actinin plays an important role in the sarcomere as an actin filament cross-linker and is often used as a Z-disc marker. Because we previously observed that Smn colocalizes with α-actinin in mouse skeletal muscle myofibrils (9), we tested whether this colocalization also occurs in cardiac myofibrils, especially given the fact that both acute and chronic SMA patients are reported to have cardiac problems (see Discussion). As shown in Figure 2, Smn colocalizes with α-actinin in purified myofibrils from both skeletal and cardiac muscles. We conclude that Smn localization to the Z-disc of striated muscles is a conserved feature among metazoans.
Figure 2.
Smn localizes to the Z-discs of striated muscle. Purified myofibrils from skeletal and cardiac muscle were co-stained with antibodies against Smn and the Z-disc protein α-actinin; the scale bar for skeletal myofibrils is 10 and 5 µm for cardiac myofibrils.
Smn complex is a component of the Z-disc
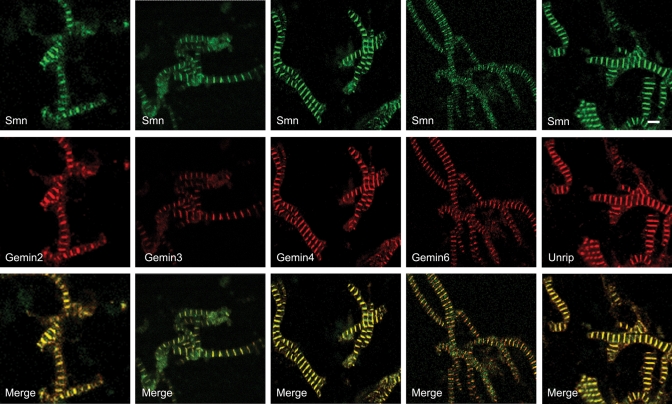
Smn forms a large, oligomeric complex with at least eight other binding partners, collectively known as Gemins (2,3). In order to determine whether Smn was present in the sarcomere as part of a larger complex, we investigated the sarcomeric localization of Gemins 2–6 and 8, as well as that of Unrip, another integral member of the Smn complex (44–52). Each of the Gemins we tested, with the possible exception of Gemin 5, colocalized precisely with Smn in skeletal muscle myofibrils (Fig. 3 and data not shown). Although it was certainly detectable, the Gemin 5 staining was weaker than that of the other Gemins (data not shown). Furthermore, consistent with the cytoplasmic origin of the myofibril, we found that Unrip, a protein that copurifies with the Smn complex preferentially in the cytoplasm of other cell types (45,46) also colocalized with Smn at the Z-disc (Fig. 3). Although colocalization does not necessarily mean these factors are complexed together at the Z-disc, the most parsimonious interpretation of the data leads us to conclude that the entire cytoplasmic Smn complex is a part of the sarcomeric Z-disc ensemble.
Figure 3.
Smn complex is present at Z-Discs. Purified mouse hindlimb skeletal myofibrils were co-stained with antibodies against Smn and Gemins 2–4, 6 and Unrip. Because the anti-Gemin 3 antibody (mAb 12H12) does not cross-react with mouse, but does cross-react with the hamster protein (http://www.abcam.com/index.html?datasheet=10305), purified myofibrils from hamster hindlimb were used for the Gemin 3 panel. Scale bar represents 5 µm.
U snRNPs do not localize to sarcomeres
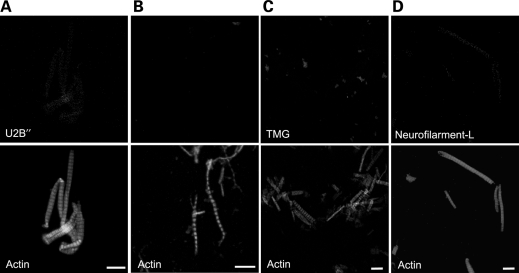
The function of the Smn complex in cytoplasmic assembly and nuclear import of Sm-class snRNPs is well documented (14,22,23,53,54). However, Sm-class snRNPs are not thought to be a part of the muscle myofibril. To confirm the absence of splicing snRNPs from the myofibril preparations, we used three markers: U2B′′, a protein specific to the U2 snRNP, SmB, a member of the spliceosomal Sm core, and the trimethylguanosine (TMG) cap, a structure common to all mature Sm-class snRNPs. Antibodies targeting a non-muscle protein, neurofilament-L, were used as a negative control; actin (visualized by phalloidin) was used as a positve control. When purified skeletal muscle myofibrils were probed using antibodies targeting these markers, no specific signals were detected (Fig. 4). These findings are therefore most consistent with a function for sarcomeric Smn that is muscle-specific and not related to its role in spliceosomal snRNP biogenesis.
Figure 4.
U snRNPs do not localize to myofibrils. Purified skeletal myofibrils were co-stained with an antibody against the U2-specific protein, U2B′′, an antibody specific to the U snRNP Sm core (mAb Y12), an Sm-core protein, SmB (mAb 12F5) and an antibody specific to the U snRNP 5′-TMG cap, TMG (mAb K121) (A–C). The myofibrils were counter stained with phaloidin conjugated with FITC. A negative control antibody (neurofilament-L) was also used on purified myofibrils (D); scale bars represent 10 µm.
Smn is removed from myofibrils by calcium or calpain treatment
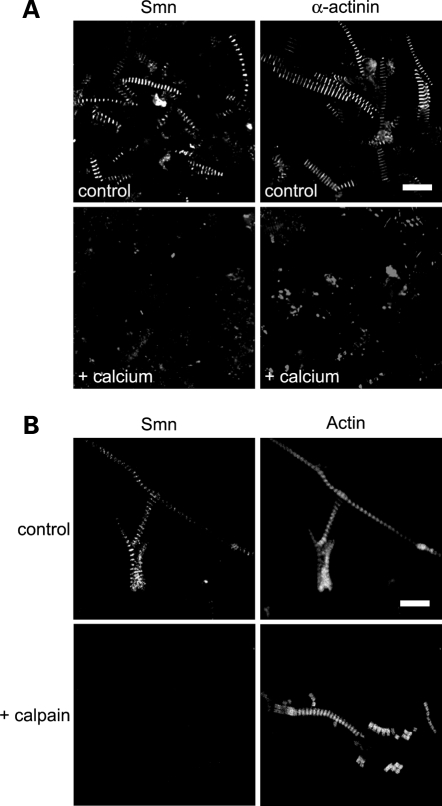
Calpains are thought to be regulatory proteases (as opposed to degradative ones) and are essential for numerous cellular processes virtually in every tissue in the body, including muscle (55–57). The ubiquitous calpains 1 and 2, as well as the muscle-specific isoform, calpain 3, are involved in aspects of muscle maintenance, including myofibril degeneration and sarcomeric remodeling (31,42,58). Increasing the endogenous calpain activity with calcium is known to trigger disassembly of the sarcomere, including removal of Z-disc proteins such as α-actinin (59–62). To test whether Smn could be removed by these procedures, muscle tissue was incubated overnight in Ringer’s buffer that contained either 10 mm Ca++ or 1 mm ethylene glycol tetraacetate (EGTA) (control). Myofibrils were then purified by standard procedures and analyzed by fluorescence microscopy. As shown in Figure 5A (upper panels), the control myofibrils remained intact, as visualized by Z-disc staining with Smn or α-actinin. The calcium-treated preparations showed loss of the signals for both Smn and α-actinin (Fig. 5A, lower panels).
Figure 5.
Calpain activity removes Smn from myofibrils. Increased endogenous calpain activity was indirectly assessed by incubating skeletal muscle tissue either in a buffer with 10 mm calcium, which activates calpain, or in a buffer lacking calcium; purified myofibrils were either stained with antibodies against Smn or α-actinin as a positive control (A). Exogenous calpain 1 was incubated with purified skeletal myofibrils (controls were incubated without added calpain) and co-stained with antibodies against Smn and conjugated phalloidin (B). Scale bars 10 µm.
Although suggestive of calpain activity, the previous results demonstrate that endogenous proteases are responsible for the removal of Smn and α-actinin from the Z-disc. In order to test whether calpain activity is required, we purified myofibrils under standard conditions and then treated them with or without exogenous calpain for 20 min at room temperature (RT). Figure 5B clearly shows that calpain treatment removed Smn from the Z-discs, leaving the myofibrils intact (as evidenced by the actin counterstain).
Smn is a direct target of calpain-mediated proteolytic cleavage
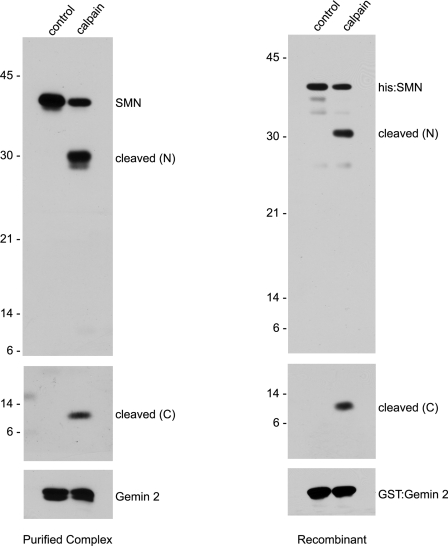
Calpains can remove Z-disc proteins from myofibrils either by direct cleavage of the target protein or indirectly by cleavage of an interacting partner. For example, calpain treatment of myofibrils is known to remove α-actinin from the Z-disc, but calpains do not directly target skeletal muscle α-actinin for cleavage (62). We therefore assayed whether SMN was a direct target of calpain. We incubated purified native human SMN complexes or recombinant SMN/Gemin 2 heterodimers in the presence or absence of calpain and analyzed the cleavage products by western blotting with N- and C-terminal-specific anti-SMN monoclonal antibodies. As shown in Figure 6, a ∼28 kDa N-terminal SMN cleavage product and a ∼10 kDa C-terminal product were detected only upon addition of calpain. The N-terminal fragments were visualized using a commercial anti-SMN monoclonal antibody (Clone 8, BD Biosciences) that recognizes residues mapping within human exon 2B, whereas the C-terminal fragments were detected using a monoclonal antibody (mAb 9F2) that recognizes residues near the polyproline motif in exon 5 (Supplementary Material, Fig. S1 and L. Pellizzoni, unpublished data). Notably, glutathione-s-transferase (GST)-Gemin 2, which was co-expressed in the bacteria expressing His-SMN, was unaffected by the calpain treatment (Fig. 6). Similarly, native Gemin 2 was also uncleaved when purified SMN complexes were treated with calpain (Fig. 6). We conclude that calpain is necessary and sufficient for cleavage of SMN in vitro.
Figure 6.
Calpain directly cleaves SMN in vitro. To verify that calpain cleaves SMN directly, purified SMN complexes or recombinant His-SMN/GST-Gemin 2 heterodimers were incubated with 0.06 and 0.15 U of calpain, respectively. Antibodies recognizing the N- (Clone 8) and C-terminal (mAb 9F2) domains of SMN were used to visualize the two fragments of SMN from the purified SMN complexes. His-probe reagent was used to visualize recombinant His-SMN.
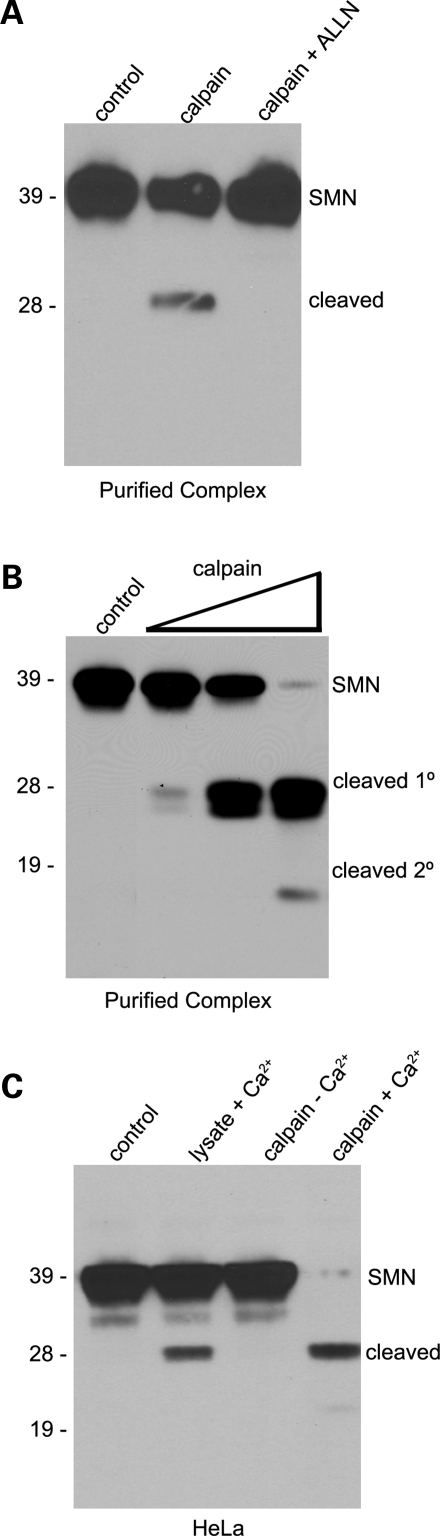
Cleavage of SMN was inhibited by addition of N-acetyl-leucyl-leucyl-norleucinal (ALLN), a synthetic inhibitor of calpain and other neutral cysteine proteases. Purified native SMN complexes (Fig. 7A) or cellular lysates (Supplementary Material, Fig. S2) were treated with calpain in the presence or absence of the ALLN peptide. In the presence of ALLN, no cleavage product was detected. Titration of the reaction showed that the appearance of the 28 kDa cleavage product and disappearance of the full-length protein was dependent upon the amount of added calpain (Fig. 7B). At very high concentrations of calpain, the full-length SMN protein was fully cleaved and shorter, secondary SMN cleavage products were also detected (Fig. 7B). Thus calpain targets SMN in the context of the native complex.
Figure 7.
Calpain inhibition, titration and endogenous activation. Purified native SMN complexes were treated with calpain (0.05 U) or with calpain and ALLN inhibitor. The cleavage of SMN was compared with an undigested control lane (A). Increasing amounts of calpain (0.003 to 3.0 U) were incubated with purified SMN complexes and the cleavage profile of SMN was analyzed (B). Western blotting was performed on HeLa lysates using anti-SMN (Clone 8) antibodies. In lanes 2 and 4, 1 mm calcium was added to the lysates. In lanes 3 and 4, 1.5 U of calpain were added. Lysates were incubated for 15 min. at 30°C and SMN cleavage profiles were compared with the control (lane 1) (C).
To examine whether SMN cleavage could be detected in vivo, we incubated total HeLa cell lysates with or without added calcium and/or calpain. Importantly, we found that addition of 1 mm Ca++ to the lysate was sufficient to induce SMN cleavage, presumably by endogenous calpains (Fig. 7C, lane 2). Incubation of control lysates with exogenous calpain but without added Ca++ also showed no cleavage (Fig. 7C, lane 3). When lysates were incubated in 1 mm Ca++ along with exogenous calpain, SMN cleavage was nearly complete (Fig. 7C, lane 4). Note that ∼10-fold higher concentrations of exogenous calpain were used in the reactions performed on total cellular lysates than on those using purified SMN proteins (recombinant or native). The fact that the same-sized cleavage products were detected in the presence or absence of added calpain (Fig. 7C) suggests that SMN is a target of calpain cleavage in vivo.
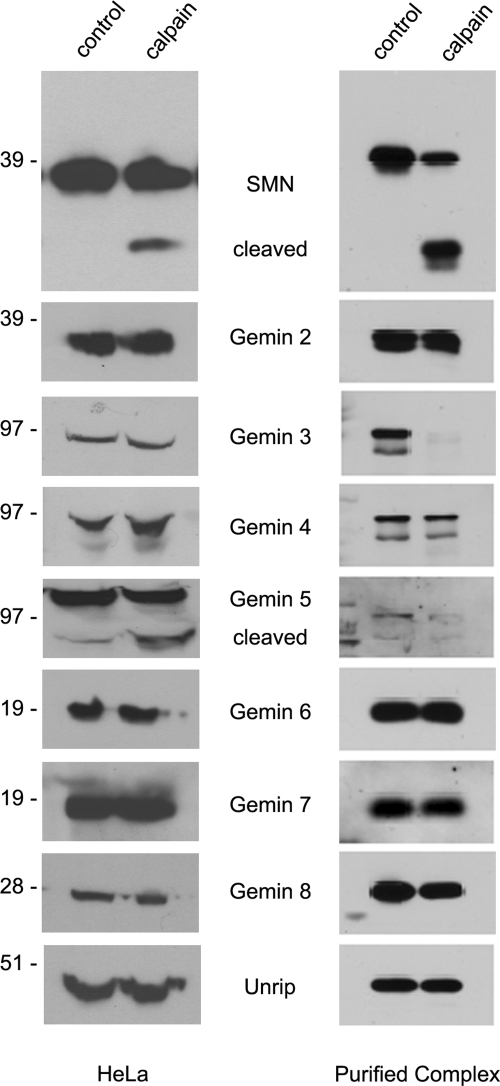
We also assayed for cleavage of other members of the SMN complex by calpain. Western blotting for SMN, Gemins 2–8 and Unrip following treatment of either HeLa lysate or purified native SMN complexes with calpain showed that Gemin 5 was cleaved in both HeLa lysate and the purified SMN complex. In contrast, Gemin 3 cleavage was only detected when the purified complex was used as the substrate (Fig. 8). Other members of the SMN complex were relatively unaffected. Unfortunately, we were unable to detect the ∼10 kDa C-terminal cleavage products in total HeLa lysate using mAb 9F2 (data not shown). Although we have not measured the stability of the SMN cleavage products in HeLa lysate, it is likely that these products are ultimately degraded. We were similarly unable to detect endogenous SMN cleavage products in wild-type mouse muscle tissues (data not shown). Thus, the steady-state level of these proteolytic isoforms of SMN is likely to be quite low.
Figure 8.
Calpain cleaves a subset of the Gemins within the SMN complex in vitro and in vivo. HeLa lysate and purified SMN complexes were treated with exogenous calpain, 1.4 and 0.06 U, respectively. Western blotting was performed with antibodies to each member of the SMN complex. Note control and calpain lanes in the Gemin 5 and Gemin 3 panels.
Myofibrillar defects in a mouse model of severe SMA
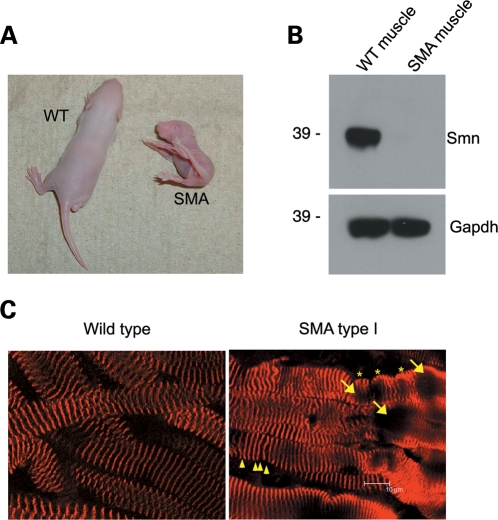
To determine whether reduced Smn expression leads to sarcomeric defects, we investigated the integrity of skeletal muscle myofibrils in the severe hypomorphic background of SMA type I mice (Smn−/−;SMN2+/+). These animals recapitulate much of the human SMA pathology and are characterized by reduced size, proximal muscle paralysis and a general failure to thrive (63,64). As shown in Figure 9A, the mutant pups were significantly smaller than their wild-type littermates at postnatal day 5 (P5) and the amount of Smn protein in skeletal muscle lysates of the SMA mice was dramatically reduced (Fig. 9B). Myofiber preparations from wild-type and SMA mutant mice were stained with antibodies targeting the Z-disc marker protein, α-actinin, and imaged in 3D by laser-scanning confocal microscopy. Two-dimensional (2D) views of 3D data sets (average intensity projections) are shown in Figure 9C. The images reveal that wild-type myofibrils contain the expected ordered sarcomeric staining pattern throughout their lengths, whereas the SMA mutant muscles exhibit numerous morphological defects, including vacuoles (Fig. 9C, arrows), wave-like lobulations (Fig. 9C, stars) and altered Z-disc spacing (Fig. 9C, arrowheads). The ‘wavy’ myofibers and associated α-actinin negative vacuolar areas in the SMA type I muscles are reminiscent of the phenotype seen in the Drosophila model of SMA (9). The vacuolar areas lacking α-actinin may represent focal dissolution of the myofibrils, one of the key features found in myofibrillar myopathies (65–67). Although the vacuoles are invariably associated with convolutions of the myofiber, the Z-disc malformations include occasional streaming and variations in spacing that indicate an overall loss of sarcomeric uniformity (Fig. 9C). Consequently, many sarcomeres are out of alignment with their neighbors and appear to be either overstretched or reduced in length. Whether this phenotype is primarily myopathic or if it is a secondary consequence of denervation is not known and remains to be determined. However, these results establish that myofibrils from SMA type I mice display defects that are consistent with those observed in other myopathies (65,67–73).
Figure 9.
SMA type 1 mice have aberrant myofibrils. Muscle fibers from SMA and wild-type littermates (A, B) were fixed and then stained with an antibody against α-actinin (average projection of a Z-stack of 18 focal planes at 0.5 µm per section) and compared (C). See text for details.
DISCUSSION
Role of the Smn complex in the sarcomere
We have shown that the entire cytoplasmic Smn complex localizes to the sarcomeric Z-disc in both cardiac and skeletal myofibrils of the mouse. Numerous case reports have suggested congenital heart defects in SMA patients (74–80). Our discovery that SMN is a sarcomeric protein provides a plausible explanation for the observed cardiac involvement in SMA. We speculate that SMN performs a tissue-specific function in striated muscles because spliceosomal snRNPs and related protein factors do not localize to purified myofibrils. Thus it is unlikely that the sarcomeric SMN complex participates in snRNP biogenesis. Instead, we suggest three non-mutually exclusive possibilities for SMN function in the sarcomere: maintenance of Z-disc integrity, signaling to nucleus, and mRNP transport.
The alignment of sarcomeres between adjacent myofibrils provides a means to coordinate contractions between individual myofibrils. This precise alignment is accomplished by a complex network of protein–protein interactions within and physically linked to the Z-disc (29). The Z-discs also link peripheral myofibrils to the nuclear membrane, to the sarcolemmal costameres and to the mitochondria. These lateral associations are mediated by intermediate filaments and associated cytoskeletal proteins such as desmin, vinculin, plectin and alpha-B crystallin (29). It is interesting to note that SMN has been shown to form complexes with α-actinin (9) and alpha-B crystallin (81). Given the sarcomeric malformations observed in the muscles of SMA type I mice (Fig. 9), it is possible that the sarcomeric SMN complex is involved in Z-disc homeostasis.
In addition to its canonical roles in anchoring actin filaments and transmitting the contractile force, the Z-disc has recently emerged as a platform for intracellular signaling (82,83). The SMN complex localizes to both the nucleus and cytoplasm of many cell types, including muscles. SMN is known to interact with components of the epidermal growth factor receptor signaling pathway (84–86) and is, therefore, a candidate Z-disc signaling factor.
Cell types with vast cytoplasmic extensions, such as muscles and neurons, face tremendous challenges in maintaining the asymmetric protein distributions required for proper functioning of these highly specialized cells. Localized translation is thought to be important for setting up these cellular asymmetries. Previous work has shown that the Smn complex colocalizes with β-actin mRNP granules in axons and growth cones of motoneurons and that reduced Smn expression correlates with a reduction in β-actin protein and mRNA staining at these distal sites (11,19). Thus SMN is thought to play a role in the assembly, transport and/or localization of β-actin mRNP complexes. We hypothesize that Smn plays a similar role in the transport and localized translation of mRNPs in muscle cells. In support of this model, Morris and Fulton (87) showed that three different Z-disc associated proteins (desmin, vinculin and vimentin) colocalize with their respective mRNAs in a striated pattern in primary cultures of embryonic chicken skeletal muscle. Similar to β-actin (88), vimentin mRNA is also thought to contain an mRNA ‘zipcode’ that targets the mRNP to a specific subcellular locale (89,90). Interestingly, a protein that binds to the vimentin mRNA zipcode (EF-1γ) is known to copurify with Z-disc proteins MuRF1 and MuRF2 (91). Thus it is plausible that localized translation takes place within the sarcomeric Z-disc. In light of the putative role for the SMN complex in mRNP transport (11) and its demonstrated localization to the Z-disc (9), SMN’s sarcomeric function may be regulated by calcium and/or calpain.
Calpain cleavage of SMN: functional implications
We have shown that SMN is a direct target of calpain cleavage in vitro and in vivo. As mentioned earlier, calpain is a regulatory protease, whose action typically activates or inactivates a given protein target; calpain activity is compartmentalized and works in microenvironments where the Ca++ flux can be controlled. Calpains 1 and 2 are ubiquitously expressed and share a common subunit, calpain 4. The genes encoding calpains 2 and 4, Capn2 and Capn4, are essential (92–94). In contrast, calpain 1 knockout mice are viable and fertile (95). Calpain 3 is a muscle-specific isoform, but is not essential (34). However, mutations in human calpain 3 (CAPN3), are known to cause limb girdle muscular dystrophy, type 2A (58). Notably, patients with mutations in CAPN3 have been misdiagnosed with SMA type III (96), illustrating a degree of phenotypic overlap between the two diseases. Although calpain 3 is a sarcomeric protein, it is not thought to localize to the Z-disc (97,98). Thus, while we do not anticipate that SMN is a target of calpain 3, additional experiments will be required to address the question of which calpains cleave SMN in vivo.
The structural cues for cleavage by calpain are incompletely understood, as the protease does not have a clear target site preference. Instead, 3D features within the substrate (99,100) as well as the sequence context (101,102) appear to be more important than the actual scissile peptide linkage. Although we have not mapped the precise cleavage site on SMN, the epitope for the monoclonal antibody used to identify the ∼10 kDa C-terminal cleavage product lies within the region encoded by exon 5 of the human gene (L. Pellizzoni, unpublished data). We therefore place the cleavage site of the human protein somewhere between amino acids 190 and 230, separating the N-terminal Tudor domain of SMN from its C-terminal Y-G box motif.
Importantly, the C-terminal domain of SMN (residues 235–294) has been implicated in regulating the G- to F-actin ratio, and expression of this C-terminal fragment can rescue neurite outgrowth defects in PC-12 cells that have been depleted of endogenous Smn (8). Moreover, SMN was shown to modulate the inhibitory effect of profilin IIa on spontaneous actin polymerization in vitro (13). SMN was also shown to associate with the actin cytoskeleton in fibroblasts (103) and to colocalize with F-actin in neuronal growth cones (21). Finally, regulation of the actin cytoskeleton was recently shown to be a key factor in SMA type III pathology, as overexpression of an F-actin stabilizing protein rescued the disease phenotype in humans (104). These results firmly establish the importance of actin cytoskeletal dynamics in SMN function and SMA pathology. Calpains are ubiquitously expressed and are important for the function of many cell types, including neurons. Thus the identification of SMN as a target of calpain cleavage is of general interest. In the future, it will be interesting to test whether proteolytic isoforms of SMN play a role in actin cytoskeletal dynamics.
MATERIALS AND METHODS
Mouse lines and crosses
Wild-type strains of FVB/NJ, C57BL/6J and 129 genetic backgrounds were used throughout the study. FVB.Cg-Tg(SMN2)89Ahmb Smntm1Msd/J mice carrying the SMA allele were obtained from the Jackson Laboratory. All strains were maintained on a standard diet of 50/10 food pellets and sterile water. These mice were housed in micro-isolation chambers. Breeding pairs consisted of mice that were homozygous for the transgene and heterozygous for the knockout allele, which resulted in pups that display the SMA phenotype and control littermates. All mice were humanely euthanized according to protocols set forth by Institutional Animal Care and Use Committee (IACUC) standards.
Antibodies
The antibodies used are as follows: anti-α-actinin (Abcam), anti-Gemin 2 (2E17, Abcam), anti-Gemin 3 (12H12), anti-Gemin 4 (30-H3), anti-Gemin 6 (20H8-E5), anti-Gemin 8 (1F8-B6), anti-neurofilament-L (DA2, Cell Signaling), anti-SmB (125F), N-terminal anti-SMN (Clone 8, BD bioscience), C-terminal anti-SMN (9F2) and anti-Unrip (3G6).
Western blotting
Muscle, myofibril and cell lystates were prepared, electrophoresed, and blotted using standard protocols. Antibodies directed against Smn (anti-mouse, 1:2500; BD biosciences) and GST (anti-mouse, 1:3000) were used. Goat anti-mouse secondary conjugated to horseradish peroxidase (HRP) was used for detection at a dilution of 1:10 000. Detection of His-SMN was carried out by use of Ni conjugated to HRP (Peirce) at a dilution of 1:5000.
Immunofluorescence
Muscle fibers were prepared primarily from excision of the gastrocnemius of P5 pups. Muscle fibers were fixed in 4% paraformaldehyde/1X Triton X-100. Myofibrils were prepared from striated muscle taken from adult mice, which were subsequently fixed in 4% formaldehyde. Immunostaining was performed following established protocols. Certain preparations were also stained for filamentous actin by adding 1 µM FITC-conjugated phalloidin (Sigma-Aldrich) 20 min before the secondary antibody incubation was complete. Images were taken using either a TCS SP2 laser scanning confocal microscope or a DM6000 microscope (both Leica), and assembled using Photoshop (Adobe). The Leica Confocal Scanner is interfaced with Leica Confocal Software, and the DM6000 microscope is interfaced with Volocity software.
Myofibril preparations and staining
Mouse skeletal and cardiac myofibrils were prepared by following the protocols of Knight and Trinick (105). Striated muscle was depleted of calcium by incubating overnight in EGTA-Ringer’s solution (100 mm NaCl, 2 mm KCl, 2 mm MgCl2, 6 mm KH2PO4, 1 mm EGTA, 0.1% glucose, pH 7.0 at 0°C) at 4°C. The samples were then transferred to rigor buffer (0.1 M KCl, 2 mm MgCl2, 1 mm EDTA, 0.5 mm DTT added fresh each time, 10 mm KH2PO4, pH 7.0 at 0°C) and homogenized using a glass Dounce tissue grinder. The homogenate was cleared of cell membrane and other debris when spun at 200g for 5 min. The resulting supernatant was then spun at 2000g for 5 min and washed by repeated cycles until a pure preparation of myofibrils (as monitored by phase-contrast microscopy) was obtained. Purified myofibrils were used for western blotting or were fixed to gelatin coated slides (0.05%) and subjected to immunofluorescence analysis using standard protocols. Note that for dual staining of Smn and α-actinin or Gemins, mouse monoclonal antibodies were incubated with purified myofibrils, followed by incubation with secondary antibodies conjugated to Alexa Fluor 594. Following extensive washing, the preparations were incubated with FITC-conjugated monoclonal anti-SMN antibodies.
Calpain treatments
Cell lysates prepared in a gentle binding buffer (50 mm Tris pH 7.5, 200 mm NaCl, 0.2 mm EDTA, 0.05% NP40) were incubated with either calpain 1 or calpain 2 (Calbiochem) at 1.4 units for 10 min at 30°C. Note for each reaction 1 mm CaCl2 is used to activate the calpain protease. The resulting lysates were then electrophoresed and analyzed by standard western blot procedures. Purified SMN complexes and His-SMN/GST-Gemin 2 heterodimers were treated with calpain as described earlier with the following exception: 0.003–0.3 units of calpain 1 were used. Myofibril preps were incubated in 1 ml of calcium activating buffer (100 mm KCl, 20 mm Tis-acetate pH 7.0, 10 mm CaCl2, 1 mm NaN3) with calpain 1 (<40 units), for 30 min at RT on a nutator. After incubation the myofibrils were spun down and washed 3× in rigor buffer at 2000g at 4°C and fixed to slides. When activation of endogenous calpains was analyzed fresh skeletal muscle was incubated in calcium activating buffer overnight at RT on a nutator. The muscle tissue was then prepared for myofibril extraction as described earlier. Standard immunostaining protocols were used for analysis.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Muscular Dystrophy Association (RG-4070) and the National Institutes of Health (R01-NS41617, R01-GM53034). During early phases of the project, M.P.W. was supported in part by National Institutes of Health predoctoral traineeship (T32-GM08613).
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Lorson, G. Morris and G. Dreyfuss for generously providing antibodies.
Conflict of Interest statement. The authors declare no conflicting interests.
REFERENCES
- 1.Wirth B., Brichta L., Hahnen E. Spinal muscular atrophy: from gene to therapy. Semin. Pediatr. Neurol. 2006;13:121–131. doi: 10.1016/j.spen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolb S.J., Battle D.J., Dreyfuss G. Molecular functions of the SMN complex. J. Child Neurol. 2007;22:990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 4.Sumner C.J. Molecular mechanisms of spinal muscular atrophy. J. Child Neurol. 2007;22:979–989. doi: 10.1177/0883073807305787. [DOI] [PubMed] [Google Scholar]
- 5.Eggert C., Chari A., Laggerbauer B., Fischer U. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 2006;12:113–121. doi: 10.1016/j.molmed.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Monani U.R. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Briese M., Esmaeili B., Sattelle D.B. Is spinal muscular atrophy the result of defects in motor neuron processes? Bioessays. 2005;27:946–957. doi: 10.1002/bies.20283. [DOI] [PubMed] [Google Scholar]
- 8.van Bergeijk J., Rydel-Konecke K., Grothe C., Claus P. The spinal muscular atrophy gene product regulates neurite outgrowth: importance of the C terminus. FASEB J. 2007;21:1492–1502. doi: 10.1096/fj.06-7136com. [DOI] [PubMed] [Google Scholar]
- 9.Rajendra T.K., Gonsalvez G.B., Walker M.P., Shpargel K.B., Salz H.K., Matera A.G. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J. Cell Biol. 2007;176:831–841. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., Xing L., Rossoll W., Wichterle H., Singer R.H., Bassell G.J. Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J. Neurosci. 2006;26:8622–8632. doi: 10.1523/JNEUROSCI.3967-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shafey D., Cote P.D., Kothary R. Hypomorphic Smn knockdown C2C12 myoblasts reveal intrinsic defects in myoblast fusion and myotube morphology. Exp. Cell Res. 2005;311:49–61. doi: 10.1016/j.yexcr.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Sharma A., Lambrechts A., Hao le T., Le T.T., Sewry C.A., Ampe C., Burghes A.H., Morris G.E. A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp. Cell Res. 2005;309:185–197. doi: 10.1016/j.yexcr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Shpargel K.B., Matera A.G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl Acad. Sci. USA. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnold A.S., Gueye M., Guettier-Sigrist S., Courdier-Fruh I., Coupin G., Poindron P., Gies J.P. Reduced expression of nicotinic AChRs in myotubes from spinal muscular atrophy I patients. Lab. Invest. 2004;84:1271–1278. doi: 10.1038/labinvest.3700163. [DOI] [PubMed] [Google Scholar]
- 16.Yong J., Golembe T.J., Battle D.J., Pellizzoni L., Dreyfuss G. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell. Biol. 2004;24:2747–2756. doi: 10.1128/MCB.24.7.2747-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan Y.B., Miguel-Aliaga I., Franks C., Thomas N., Trulzsch B., Sattelle D.B., Davies K.E., van den Heuvel M. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum. Mol. Genet. 2003;12:1367–1376. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- 18.McWhorter M.L., Monani U.R., Burghes A.H., Beattie C.E. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossoll W., Jablonka S., Andreassi C., Kroning A.K., Karle K., Monani U.R., Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillai R.S., Grimmler M., Meister G., Will C.L., Luhrmann R., Fischer U., Schumperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan L., Simard L.R. Survival motor neuron (SMN) protein: role in neurite outgrowth and neuromuscular maturation during neuronal differentiation and development. Hum. Mol. Genet. 2002;11:1605–1614. doi: 10.1093/hmg/11.14.1605. [DOI] [PubMed] [Google Scholar]
- 22.Pellizzoni L., Yong J., Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 23.Meister G., Buhler D., Pillai R., Lottspeich F., Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 24.Braun S., Croizat B., Lagrange M.C., Warter J.M., Poindron P. Constitutive muscular abnormalities in culture in spinal muscular atrophy. Lancet. 1995;345:694–695. doi: 10.1016/s0140-6736(95)90869-2. [DOI] [PubMed] [Google Scholar]
- 25.Guettier-Sigrist S., Hugel B., Coupin G., Freyssinet J.M., Poindron P., Warter J.M. Possible pathogenic role of muscle cell dysfunction in motor neuron death in spinal muscular atrophy. Muscle Nerve. 2002;25:700–708. doi: 10.1002/mus.10081. [DOI] [PubMed] [Google Scholar]
- 26.Gavrilina T.O., McGovern V.L., Workman E., Crawford T.O., Gogliotti R.G., DiDonato C.J., Monani U.R., Morris G.E., Burghes A.H. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum. Mol. Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 29.Clark K.A., McElhinny A.S., Beckerle M.C., Gregorio C.C. Striated muscle cytoarchitecture: an intricate web of form and function. Annu. Rev. Cell Dev. Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 30.Nury D., Doucet C., Coux O. Roles and potential therapeutic targets of the ubiquitin proteasome system in muscle wasting. BMC Biochem. 2007;8(Suppl. 1):S7. doi: 10.1186/1471-2091-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartoli M., Richard I. Calpains in muscle wasting. Int. J. Biochem. Cell Biol. 2005;37:2115–2133. doi: 10.1016/j.biocel.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Wang J., Maldonado M.A. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell. Mol. Immunol. 2006;3:255–261. [PubMed] [Google Scholar]
- 33.Abu-Baker A., Messaed C., Laganiere J., Gaspar C., Brais B., Rouleau G.A. Involvement of the ubiquitin-proteasome pathway and molecular chaperones in oculopharyngeal muscular dystrophy. Hum. Mol. Genet. 2003;12:2609–2623. doi: 10.1093/hmg/ddg293. [DOI] [PubMed] [Google Scholar]
- 34.Tagawa K., Taya C., Hayashi Y., Nakagawa M., Ono Y., Fukuda R., Karasuyama H., Toyama-Sorimachi N., Katsui Y., Hata S., et al. Myopathy phenotype of transgenic mice expressing active site-mutated inactive p94 skeletal muscle-specific calpain, the gene product responsible for limb girdle muscular dystrophy type 2A. Hum. Mol. Genet. 2000;9:1393–1402. doi: 10.1093/hmg/9.9.1393. [DOI] [PubMed] [Google Scholar]
- 35.Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 36.Jagoe R.T., Goldberg A.L. What do we really know about the ubiquitin-proteasome pathway in muscle atrophy? Curr. Opin. Clin. Nutr. Metab. Care. 2001;4:183–190. doi: 10.1097/00075197-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 37.Taillandier D., Aurousseau E., Meynial-Denis D., Bechet D., Ferrara M., Cottin P., Ducastaing A., Bigard X., Guezennec C.Y., Schmid H.P., et al. Coordinate activation of lysosomal, Ca2+-activated and ATP-ubiquitin-dependent proteinases in the unweighted rat soleus muscle. Biochem. J. 1996;316:65–72. doi: 10.1042/bj3160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solomon V., Goldberg A.L. Importance of the ATP-ubiquitin-proteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J. Biol. Chem. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 39.Gissel H. The role of Ca2+ in muscle cell damage. Ann. NY Acad. Sci. 2005;1066:166–180. doi: 10.1196/annals.1363.013. [DOI] [PubMed] [Google Scholar]
- 40.Hasselgren P.O., Fischer J.E. Muscle cachexia: current concepts of intracellular mechanisms and molecular regulation. Ann. Surg. 2001;233:9–17. doi: 10.1097/00000658-200101000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramerova I., Kudryashova E., Venkatraman G., Spencer M.J. Calpain 3 participates in sarcomere remodeling by acting upstream of the ubiquitin-proteasome pathway. Hum. Mol. Genet. 2005;14:2125–2134. doi: 10.1093/hmg/ddi217. [DOI] [PubMed] [Google Scholar]
- 42.Goll D.E., Neti G., Mares S.W., Thompson V.F. Myofibrillar protein turnover: the proteasome and the calpains. J. Anim. Sci. 2008;86:E19–E35. doi: 10.2527/jas.2007-0395. [DOI] [PubMed] [Google Scholar]
- 43.Lange S., Ehler E., Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16:11–18. doi: 10.1016/j.tcb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Carissimi C., Saieva L., Gabanella F., Pellizzoni L. Gemin8 is required for the architecture and function of the survival motor neuron complex. J. Biol. Chem. 2006;281:37009–37016. doi: 10.1074/jbc.M607505200. [DOI] [PubMed] [Google Scholar]
- 45.Grimmler M., Otter S., Peter C., Muller F., Chari A., Fischer U. Unrip, a factor implicated in cap-independent translation, associates with the cytosolic SMN complex and influences its intracellular localization. Hum. Mol. Genet. 2005;14:3099–3111. doi: 10.1093/hmg/ddi343. [DOI] [PubMed] [Google Scholar]
- 46.Carissimi C., Baccon J., Straccia M., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L. Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett. 2005;579:2348–2354. doi: 10.1016/j.febslet.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 47.Baccon J., Pellizzoni L., Rappsilber J., Mann M., Dreyfuss G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 2002;277:31957–31962. doi: 10.1074/jbc.M203478200. [DOI] [PubMed] [Google Scholar]
- 48.Pellizzoni L., Baccon J., Rappsilber J., Mann M., Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 49.Charroux B., Pellizzoni L., Perkinson R.A., Yong J., Shevchenko A., Mann M., Dreyfuss G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charroux B., Pellizzoni L., Perkinson R.A., Shevchenko A., Mann M., Dreyfuss G. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gubitz A.K., Mourelatos Z., Abel L., Rappsilber J., Mann M., Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- 52.Liu Q., Fischer U., Wang F., Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 53.Narayanan U., Achsel T., Luhrmann R., Matera A.G. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell. 2004;16:223–234. doi: 10.1016/j.molcel.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 54.Grimmler M., Bauer L., Nousiainen M., Korner R., Meister G., Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;6:70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H.Y., Lynch D.R. Calpain and synaptic function. Mol. Neurobiol. 2006;33:215–236. doi: 10.1385/MN:33:3:215. [DOI] [PubMed] [Google Scholar]
- 56.Stockholm D., Bartoli M., Sillon G., Bourg N., Davoust J., Richard I. Imaging calpain protease activity by multiphoton FRET in living mice. J. Mol. Biol. 2005;346:215–222. doi: 10.1016/j.jmb.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 57.Goll D.E., Thompson V.F., Li H., Wei W., Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 58.Duguez S., Bartoli M., Richard I. Calpain 3: a key regulator of the sarcomere? FEBS J. 2006;273:3427–3436. doi: 10.1111/j.1742-4658.2006.05351.x. [DOI] [PubMed] [Google Scholar]
- 59.Busch W.A., Stromer M.H., Goll D.E., Suzuki A. Ca2+-specific removal of Z lines from rabbit skeletal muscle. J. Cell Biol. 1972;52:367–381. doi: 10.1083/jcb.52.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dayton W.R., Goll D.E., Zeece M.G., Robson R.M., Reville W.J. A Ca2+-activated protease possibly involved in myofibrillar protein turnover. Purification from porcine muscle. Biochemistry. 1976;15:2150–2158. doi: 10.1021/bi00655a019. [DOI] [PubMed] [Google Scholar]
- 61.Dayton W.R., Reville W.J., Goll D.E., Stromer M.H. A Ca2+-activated protease possibly involved in myofibrillar protein turnover. Partial characterization of the purified enzyme. Biochemistry. 1976;15:2159–2167. doi: 10.1021/bi00655a020. [DOI] [PubMed] [Google Scholar]
- 62.Goll D.E., Dayton W.R., Singh I., Robson R.M. Studies of the alpha-actinin/actin interaction in the Z-disc by using calpain. J. Biol. Chem. 1991;266:8501–8510. [PubMed] [Google Scholar]
- 63.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 64.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 65.Nakano S., Engel A.G., Waclawik A.J., Emslie-Smith A.M., Busis N.A. Myofibrillar myopathy with abnormal foci of desmin positivity. I. Light and electron microscopy analysis of 10 cases. J. Neuropathol. Exp. Neurol. 1996;55:549–562. doi: 10.1097/00005072-199605000-00008. [DOI] [PubMed] [Google Scholar]
- 66.De Bleecker J.L., van den Abeele K.G., De Reuck J.L. Variable involvement of rat skeletal muscles in paraoxon-induced necrotizing myopathy. Res. Commun. Chem. Pathol. Pharmacol. 1992;75:309–322. [PubMed] [Google Scholar]
- 67.Selcen D., Engel A.G. Mutations in myotilin cause myofibrillar myopathy. Neurology. 2004;62:1363–1371. doi: 10.1212/01.wnl.0000123576.74801.75. [DOI] [PubMed] [Google Scholar]
- 68.Fanin M., Nardetto L., Nascimbeni A.C., Tasca E., Spinazzi M., Padoan R., Angelini C. Correlations between clinical severity, genotype and muscle pathology in limb girdle muscular dystrophy type 2A. J. Med. Genet. 2007;44:609–614. doi: 10.1136/jmg.2007.050328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramerova I., Kudryashova E., Tidball J.G., Spencer M.J. Null mutation of calpain 3 (p94) in mice causes abnormal sarcomere formation in vivo and in vitro. Hum. Mol. Genet. 2004;13:1373–1388. doi: 10.1093/hmg/ddh153. [DOI] [PubMed] [Google Scholar]
- 70.Chae J., Minami N., Jin Y., Nakagawa M., Murayama K., Igarashi F., Nonaka I. Calpain 3 gene mutations: genetic and clinico-pathologic findings in limb-girdle muscular dystrophy. Neuromuscul. Disord. 2001;11:547–555. doi: 10.1016/s0960-8966(01)00197-3. [DOI] [PubMed] [Google Scholar]
- 71.Sussman M.A., Baque S., Uhm C.S., Daniels M.P., Price R.L., Simpson D., Terracio L., Kedes L. Altered expression of tropomodulin in cardiomyocytes disrupts the sarcomeric structure of myofibrils. Circ. Res. 1998;82:94–105. doi: 10.1161/01.res.82.1.94. [DOI] [PubMed] [Google Scholar]
- 72.Kvist A.P., Latvanlehto A., Sund M., Eklund L., Vaisanen T., Hagg P., Sormunen R., Komulainen J., Fassler R., Pihlajaniemi T. Lack of cytosolic and transmembrane domains of type XIII collagen results in progressive myopathy. Am. J. Pathol. 2001;159:1581–1592. doi: 10.1016/S0002-9440(10)62542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haldar S.M., Ibrahim O.A., Jain M.K. Kruppel-like Factors (KLFs) in muscle biology. J. Mol. Cell. Cardiol. 2007;43:1–10. doi: 10.1016/j.yjmcc.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moller P., Moe N., Saugstad O.D., Skullerud K., Velken M., Berg K., Nitter-Hauge S., Borresen A.L. Spinal muscular atrophy type I combined with atrial septal defect in three sibs. Clin. Genet. 1990;38:81–83. doi: 10.1111/j.1399-0004.1990.tb03553.x. [DOI] [PubMed] [Google Scholar]
- 75.Mulleners W.M., van Ravenswaay C.M., Gabreels F.J., Hamel B.C., van Oort A., Sengers R.C. Spinal muscular atrophy combined with congenital heart disease: a report of two cases. Neuropediatrics. 1996;27:333–334. doi: 10.1055/s-2007-973805. [DOI] [PubMed] [Google Scholar]
- 76.Jong Y.J., Chang J.G., Wu J.R. Large-scale deletions in a Chinese infant associated with a variant form of Werdnig-Hoffmann disease. Neurology. 1998;51:878–879. doi: 10.1212/wnl.51.3.878. [DOI] [PubMed] [Google Scholar]
- 77.El-Matary W., Kotagiri S., Cameron D., Peart I. Spinal muscle atrophy type 1 (Werdnig-Hoffman disease) with complex cardiac malformation. Eur. J. Pediatr. 2004;163:331–332. doi: 10.1007/s00431-004-1437-6. [DOI] [PubMed] [Google Scholar]
- 78.Cook A.L., Curzon C.L., Milazzo A.S. An infant with hypoplastic left heart syndrome and spinal muscular atrophy. Cardiol. Young. 2006;16:78–80. doi: 10.1017/S1047951105002131. [DOI] [PubMed] [Google Scholar]
- 79.Vaidla E., Talvik I., Kulla A., Sibul H., Maasalu K., Metsvaht T., Piirsoo A., Talvik T. Neonatal spinal muscular atrophy type 1 with bone fractures and heart defect. J. Child Neurol. 2007;22:67–70. doi: 10.1177/0883073807299954. [DOI] [PubMed] [Google Scholar]
- 80.Menke L.A., Poll-The B.T., Clur S.A., Bilardo C.M., van der Wal A.C., Lemmink H.H., Cobben J.M. Congenital heart defects in spinal muscular atrophy type I: a clinical report of two siblings and a review of the literature. Am. J. Med. Genet. A. 2008;146A:740–744. doi: 10.1002/ajmg.a.32233. [DOI] [PubMed] [Google Scholar]
- 81.den Engelsman J.G.D., de Jong W.W., Robbins J., Kato K., Boelens W.C. Nuclear import of {alpha}B-crystallin is phosphorylation-dependent and hampered by hyperphosphorylation of the myopathy-related mutant R120G. J. Biol. Chem. 2005;280:37139–37148. doi: 10.1074/jbc.M504106200. [DOI] [PubMed] [Google Scholar]
- 82.Frank D., Kuhn C., Katus H.A., Frey N. The sarcomeric Z-disc: a nodal point in signalling and disease. J. Mol. Med. 2006;84:446–468. doi: 10.1007/s00109-005-0033-1. [DOI] [PubMed] [Google Scholar]
- 83.Pyle W.G., Solaro R.J. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 2004;94:296–305. doi: 10.1161/01.RES.0000116143.74830.A9. [DOI] [PubMed] [Google Scholar]
- 84.Mishra A.K., Gangwani L., Davis R.J., Lambright D.G. Structural insights into the interaction of the evolutionarily conserved ZPR1 domain tandem with eukaryotic EF1A, receptors, and SMN complexes. Proc. Natl Acad. Sci. USA. 2007;104:13930–13935. doi: 10.1073/pnas.0704915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gangwani L., Flavell R.A., Davis R.J. ZPR1 is essential for survival and is required for localization of the survival motor neurons (SMN) protein to Cajal bodies. Mol. Cell. Biol. 2005;25:2744–2756. doi: 10.1128/MCB.25.7.2744-2756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Doran B., Gherbesi N., Hendricks G., Flavell R.A., Davis R.J., Gangwani L. Deficiency of the zinc finger protein ZPR1 causes neurodegeneration. Proc. Natl Acad. Sci. USA. 2006;103:7471–7475. doi: 10.1073/pnas.0602057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morris E.J., Fulton A.B. Rearrangement of mRNAs for costamere proteins during costamere development in cultured skeletal muscle from chicken. J. Cell Sci. 1994;107:377–386. doi: 10.1242/jcs.107.3.377. [DOI] [PubMed] [Google Scholar]
- 88.Condeelis J., Singer R.H. How and why does beta-actin mRNA target? Biol. Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- 89.Chabanon H., Mickleburgh I., Hesketh J. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief. Funct. Genomic. Proteomic. 2004;3:240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- 90.Al-Maghrebi M., Brule H., Padkina M., Allen C., Holmes W.M., Zehner Z.E. The 3’ untranslated region of human vimentin mRNA interacts with protein complexes containing eEF-1gamma and HAX-1. Nucleic Acids Res. 2002;30:5017–5028. doi: 10.1093/nar/gkf656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Witt C.C., Witt S.H., Lerche S., Labeit D., Back W., Labeit S. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J. 2008;27:350–360. doi: 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dutt P., Croall D.E., Arthur J.S., Veyra T.D., Williams K., Elce J.S., Greer P.A. m-Calpain is required for preimplantation embryonic development in mice. BMC Dev. Biol. 2006;6:3. doi: 10.1186/1471-213X-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zimmerman U.J., Boring L., Pak J.H., Mukerjee N., Wang K.K. The calpain small subunit gene is essential: its inactivation results in embryonic lethality. IUBMB Life. 2000;50:63–68. doi: 10.1080/15216540050176610. [DOI] [PubMed] [Google Scholar]
- 94.Arthur J.S., Elce J.S., Hegadorn C., Williams K., Greer P.A. Disruption of the murine calpain small subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol. Cell. Biol. 2000;20:4474–4481. doi: 10.1128/mcb.20.12.4474-4481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Azam M., Andrabi S.S., Sahr K.E., Kamath L., Kuliopulos A., Chishti A.H. Disruption of the mouse mu-calpain gene reveals an essential role in platelet function. Mol. Cell. Biol. 2001;21:2213–2220. doi: 10.1128/MCB.21.6.2213-2220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Starling A., de Paula F., Silva H., Vainzof M., Zatz M. Calpainopathy: how broad is the spectrum of clinical variability? J. Mol. Neurosci. 2003;21:233–236. doi: 10.1385/jmn:21:3:233. [DOI] [PubMed] [Google Scholar]
- 97.Sorimachi H., Kinbara K., Kimura S., Takahashi M., Ishiura S., Sasagawa N., Sorimachi N., Shimada H., Tagawa K., Maruyama K., et al. Muscle-specific calpain, p94, responsible for limb girdle muscular dystrophy type 2A, associates with connectin through IS2, a p94-specific sequence. J. Biol. Chem. 1995;270:31158–31162. doi: 10.1074/jbc.270.52.31158. [DOI] [PubMed] [Google Scholar]
- 98.Keira Y., Noguchi S., Minami N., Hayashi Y.K., Nishino I. Localization of calpain 3 in human skeletal muscle and its alteration in limb-girdle muscular dystrophy 2A muscle. J. Biochem. 2003;133:659–664. doi: 10.1093/jb/mvg084. [DOI] [PubMed] [Google Scholar]
- 99.Sakai K., Akanuma H., Imahori K., Kawashima S. A unique specificity of a calcium activated neutral protease indicated in histone hydrolysis. J. Biochem. 1987;101:911–918. doi: 10.1093/oxfordjournals.jbchem.a121959. [DOI] [PubMed] [Google Scholar]
- 100.Stabach P.R., Cianci C.D., Glantz S.B., Zhang Z., Morrow J.S. Site-directed mutagenesis of alpha II spectrin at codon 1175 modulates its mu-calpain susceptibility. Biochemistry. 1997;36:57–65. doi: 10.1021/bi962034i. [DOI] [PubMed] [Google Scholar]
- 101.Tompa P., Buzder-Lantos P., Tantos A., Farkas A., Szilagyi A., Banoczi Z., Hudecz F., Friedrich P. On the sequential determinants of calpain cleavage. J. Biol. Chem. 2004;279:20775–20785. doi: 10.1074/jbc.M313873200. [DOI] [PubMed] [Google Scholar]
- 102.Cuerrier D., Moldoveanu T., Davies P.L. Determination of peptide substrate specificity for mu-calpain by a peptide library-based approach: the importance of primed side interactions. J. Biol. Chem. 2005;280:40632–40641. doi: 10.1074/jbc.M506870200. [DOI] [PubMed] [Google Scholar]
- 103.Zhang H.L., Pan F., Hong D., Shenoy S.M., Singer R.H., Bassell G.J. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oprea G.E., Krober S., McWhorter M.L., Rossoll W., Muller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Knight P.J., Trinick J.A. Preparation of myofibrils. Methods Enzymol. 1982;85:9–12. doi: 10.1016/0076-6879(82)85004-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.