Abstract
Many mutations occur as a result of DNA synthesis past the site of DNA damage by DNA damage bypass polymerases. The frequency and types of mutations not only depend on the nature of the damage, but also on the sequence context, as revealed from analysis of mutation spectra of DNA exposed to mutagens. Herein we report a new method for the rapid determination of the effect of sequence context on mutagenesis called SAMS for serial analysis of mutation spectra. This technique makes use of the methodology that underlies serial analysis of gene expression (SAGE) to analyze mutations that result from DNA synthesis past a DNA lesion site-specifically embedded in a library of DNA sequences. To illustrate our technique we determined the effect of sequence context on mutations generated by DNA synthesis past a tetrahydrofuran abasic site model by the DNA damage bypass polymerase yeast polymerase η.
INTRODUCTION
In human cells, both normal metabolic activities and environmental factors such as UV light can cause DNA damage (1). Many of these lesions cause mutations during replication of the DNA, thereby altering the coding properties of the gene that the affected DNA encodes. Specialized DNA damage bypass polymerases have evolved to synthesize past DNA damage and invariably produce mutations as a result (2,3). The types and frequencies of mutations, or the mutation spectrum, produced by a single type of lesion differs widely depending on its position within the genome results in mutation hotspots (4,5). The precise origin of these hotspots for a particular type of lesion is not well understood, but sequence context certainly plays an important role. The effect of sequence context on the mutation spectrum of a lesion has been investigated by three general types of approaches. In one approach, the progeny of bypass products of a randomly damaged template are sequenced following transfection into bacteria (6). In a second, related approach, the progeny of the bypass products of homogeneous, site-specifically damaged templates are sequenced (7–9). In a third approach, the selectivity of nucleotide insertion opposite a homogenous substrate is determined by steady-state or pre-steady-state kinetic experiments (10,11). The first approach is limited to the sequence contexts present in the heterogeneous template and involves uncharacterized products that may be produced with varying frequencies at different sites. The second approach has the advantage of using a characterized product, but requires the preparation of separate templates for each sequence context. The third approach also suffers from having to prepare separate templates, and is in addition highly laborious and only directly provides information on specific types of mutations.
Herein, we report a new approach to obtaining mutation spectra as a function of sequence context in which the bypass products of a homogeneous lesion in a library of nearly equally represented sequence contexts is amplified by PCR, restricted, polymerized, cloned and sequenced (Figure 1). The advantages of this approach are (i) that a pure lesion is used, (ii) that each sequence context is equally represented and (iii) each clone gives information on about multiple sequence contexts, thereby greatly increasing throughput. We illustrate how this new technique can be used to rapidly uncover flanking sequence-dependent substitution and deletion mutations produced in the bypass of a tetrahydrofuran abasic site model (F) by a pol Y family polymerase.
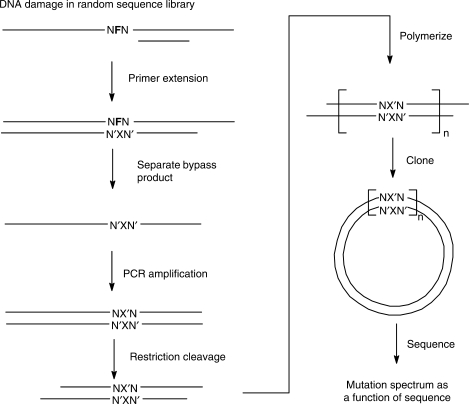
Figure 1.
Serial analysis of mutation spectra (SAMS) scheme. N represents a random nucleotide, F the tetrahydrofuran abasic site model, N′ the nucleotide complementary to N, X, the nucleotide inserted opposite F. Polymerization was carried out with DNA ligase.
MATERIALS AND METHODS
Enzymes and substrate preparation
DNA templates and primers were synthesized on an Applied Biosystems Inc. Expedite 8909 DNA synthesizer using standard solid-phase phosphoramidite synthesis or were obtained from Integrated DNA Technologies, Inc. A randomized nucleotide was introduced site-specifically by using a 1.5 : 1.25 : 1.15 : 1.0 mixture of A, C, G and T phosphoramidites, respectively, during the coupling step (12). All ODNs were purified by 15% or 20% denaturing polyacrylamide gel electrophoresis, followed by extraction with phenol/chloroform and ethanol precipitation. The catalytic core of yeast pol η was expressed and purified as described previously (13).
Primer extension reactions
Primer-templates were annealed in a 1.5 : 1 ratio by heating at 85°C and allowing to slowly cool to 25°C. All reactions (30 µl) were run at 37°C and contained 100 nM template and primer and 100 µM dATP, dGTP, dCTP and dTTP. For DNA pol η, reactions contained 40 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 10 mM dithiothreitol, 7.5 µg of bovine serum albumin, 2.5% glycerol and 5 nM DNA polymerase η. At time intervals from 1 to 30 min, aliquots were removed from the reaction and quenched by the addition of an equal volume of 95% formamide in 25 mM EDTA.
Isolation and cloning of the bypass product
The bypass reaction products were separated from template and excess primer on a 20% denaturing polyacrylamide gel, and recovered by the procedure described below for PCR fragments. PCR was carried out with 1 U of platinum pfx polymerase and 1 μM d(AGCTCGGAATTCGG) and d(AGCTCGAGAATTCAG) primers for 30 cycles of 95°C for 30 s, 50°C for 30 s and 72°C for 30 s in 100 μl reaction mixture containing 160 μM of each dNTP in PCR buffer. After amplification, the reaction mixture was concentrated and loaded onto a 20% denaturing PAGE gel and electrophoresed to separate the PCR products, extracted with phenol/chloroform, followed by ethanol precipitation. The purified PCR products were digested by EcoRI, and labeled by an exchange reaction with 1× reaction buffer (Fermentas Life Science, 50 mM imidazole-HCl, 18 mM MgCl2, 5 mM DTT, 0.1 mM spermidine, 0.1 mM EDTA and 0.1 mM ADP) with PEG6000 solution and 10 U of T4 polynucleotide kinase and 40 pmol [γ-32P]–ATP. The labeled products were loaded onto a 20% denaturing PAGE gel and electrophoresed to separate the 14- and 15-mer products which were then ligated with T4 DNA ligase at 16°C for 2 h. Concatemers were isolated using the QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA), following the manufacturer's instructions. The mixture was then electrophoresed on a 1% agarose gel (TAE) and fractions with 300–500-bp length were excised. Concatemers were inserted into the EcoRI site of pZErO-1 (Invitrogen,, San Diego, CA) with T4 DNA ligase for 4 h at 16°C and then transfected into competent Escherichia coli (Oneshot® Top10, Invitrogen) following the manufacturer's manual. The transfectants were plated on low salt Luria–Bertani (LB) plates containing 50 µg/ml ZeocinTM (Invivogen) and incubated for about 18 h at 37°C. ZeocinTM resistant transformants were picked using pipette tips, incubated in 5 ml SOB containing 50 µl/ml ZeocinTM and grown overnight at 37°C. Plasmid DNA was prepared by the Plasmid mini prep kit (QIAGEN). Plasmids containing sizable inserts were then forwarded to the Washington University Genome Center for sequencing.
Primer-extension opposite homogeneous substrates
Ten micromolar [γ-32P]-ATP labeled d(CAGGTCGATAATTCA) and the indicated template were mixed in a 1.5 : 1 ratio and heated to 85°C and then slowly cooled to 25°C. Reactions were run at 37°C for 30 min with 5 nM DNA pol η, 100 nM substrate and 150 nM primer and 100 μM dATP, dGTP, dCTP and dTTP, in buffer containing 40 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 10 mM dithiothreitol, 7.5 µg of bovine serum albumin, 2.5% glycerol. The reactions were then heated to 100°C in water bath for 5 min, cooled to RT, after which 10 U of EcoRI was added with 1× EcoRI buffer, and after incubating for 1 h at 37°C, was quenched by adding to an equal volume of 95% formamide in 25 mM EDTA and loaded on a PAGE gel.
RESULTS
The SAMS method
The SAMS method is diagramed in Figure 1. The template is designed to have a site-specific damage embedded approximately in the center of the template within any desired type of random sequence library. The damage site is flanked by restriction enzyme sites, and the primer is made to start to the 5′-side of the 3′-end of the template so that the bypass products can be separated from the template prior to the PCR step by size. The primer is extended by a polymerase and the bypass products separated by PAGE. The bypass products are amplified by PCR, restricted, 32P-end labeled and isolated by PAGE. The labeled products are then polymerized by DNA ligase, and DNA fragments of 300–500 bp isolated by agarose gel electrophoresis. The purified concatemers are then cloned into the pZero-1 vector, transfected into E. coli, plated and clones identified from plasmid minipreps as have 20 or so inserts are then sequenced.
Design and synthesis of an abasic site model containing template
As an example, we chose to investigate the sequence dependence of the mutagenicity of a synthetic tetrahydrofuran abasic site model (F) (14) when bypassed by the DNA damage bypass polymerase pol η. This abasic site model lacks the 1′-OH of an actual abasic site, and as a result locks the abasic site into its cyclic structure. This lesion and polymerase were chosen because the mutagencity of its bypass by pol η has been extensively studied, and because the abasic site model can be site-specifically incorporated into a DNA molecule via solid-phase synthesis utilizing a commercially available building block. The 45-mer template shown in Figure 2a was designed to have F roughly in the center of the template, and flanked by randomized nucleotides to the 5′- and 3′-side, for a total of 16 sequence contexts. The template was also designed to have an EcoRI sequence to the 5′- and 3′-sides so as to enable excision of a 15-mer sized fragment for polymerization following PCR amplification. Outside of the randomized flanking sequences, the sequence was designed to be asymmetric so that the template and primer strands could be distinguished following DNA sequencing. The template was synthesized by standard automated phosphoramidite DNA synthesis, and the random flanking sequences were introduced using a mixture of A, C, G & T phosphoramidites during coupling in a ratio reported to result in their incorporation with approximately equal frequency.
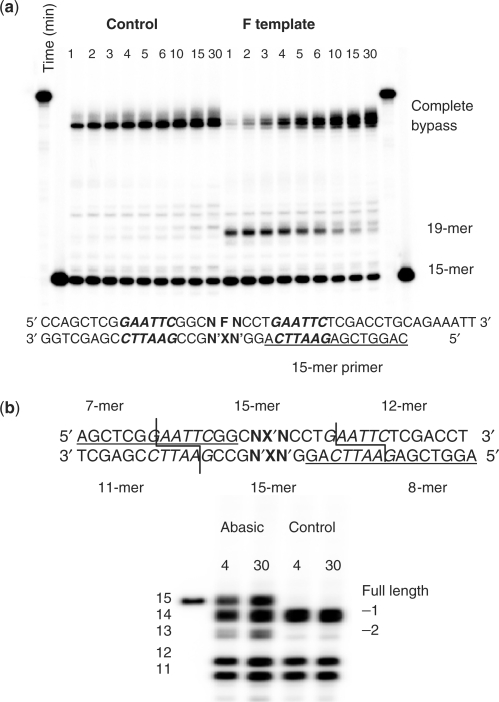
Figure 2.
(a) Primer extension opposite an abasic site model in a random flanking sequence library of 16 possible sequences. The abasic site model F and its flanking random sequence are in bold. The EcoRI restriction sites are italicized, the 15-mer primer is underlined. The control template is identical to the F-containing template, except that F was not incorporated. (b) Formation of the sequence tags for oligomerization. Polyacrylamide gel electrophoresis of the products of PCR amplification of the bypass products of the abasic site and control sequence followed by restriction digestion with EcoRI. PCR primers are underlined.
Primer extension reactions
Primer-extension reactions were carried out with yeast pol η and dNTPs on the 45-mer abasic-template and an otherwise identical 44-mer control template lacking the abasic site model but bearing the random NN library. Under the experimental conditions used, we found that about 50% of the abasic site was bypassed by yeast pol η in 4 min, and that almost all was bypassed in about 30 min (Figure 2a). The bypass products from the control template, and the 4- and 30-min bypass products of the abasic template were isolated by PAGE and then amplified by PCR. The PCR products were digested with EcoRI, and 5′-end labeled with [γ-32P]-ATP and T4 polynucleotide kinase in an exchange reaction and analyzed by PAGE (Figure 2b). It was immediately apparent that more than 50% of the bypass products of the abasic template were −1 deletions (14-mer band), 56% for the 4-min reaction and 54% for the 30-min reaction. There was also a significant amount of −2 deletion detected, whereas the control template showed very little −1 deletion. Because of the similar amounts of −1 deletion product in the 4 and 30-min reactions, we decided to focus on the bypass products of the 30-min reaction, to be sure that the mutation spectra for all sequence contexts would be represented.
Mutation spectra
The 14- and 15-mer PCR bands corresponding to the full length and −1 deletion bypass products were carried on through the polymerization and cloning steps. About 100 plasmids containing about 10 bypass sequences each were sequenced. The bypass sequences were readily identified and extracted from between the EcoRI recognition sequences, GAATTC and further assigned to the template or primer strand and to substitution or −1 mutations from the sequence context. A sequence of the form GAATTCAGGN′XN′GCCGAATTC could be assigned to the primer strand, where the 5′-N′ corresponds to the nucleotide complementary to the random nucleotide flanking the 3′-side of the abasic site (3′-N), and X is the nucleotide inserted opposite the abasic site. On the other hand, a sequence of the form GAATTCGGCNX′NCCTGAATTC corresponds to the template strand, where X′ is the base complementary to the nucleotide inserted opposite the abasic site. Single nucleotide deletions appear as a sequenced shortened by 1 nt, for example, GAATTCAGGN′N′GCCGAATTC, which corresponds to the primer strand. Analysis of the 985 sequenced bypass products revealed that 533 or 54% were −1 deletions, and 452 or 46% were substitutions (Table 1). This ratio is exactly what was found by PAGE analysis of the PCR products (Figure 2b) demonstrating that the cloning statistics accurately represent the frequencies of substitution and −1 deletions that were cloned.
Table 1.
SAMS spectrum of −1 deletion and substitution mutations for 5′-NFN-3′
| Nucleotide X inserted opposite F (% insertion)c |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NN | Total | −1 | (% −1)a | Sub | (% Sub)b | A | (% A) | C | (% C) | G | (% G) | T | (% T) |
| AA | 45 | 7 | (16) | 38 | (84) | 14 | (37) | 2 | (5) | 22 | (58) | 0 | (0) |
| AC | 94 | 68 | (72) | 26 | (28) | 7 | (27) | 0 | (0) | 19 | (73) | 0 | (0) |
| AG | 57 | 18 | (32) | 39 | (68) | 27 | (69) | 0 | (0) | 12 | (31) | 0 | (0) |
| AT | 49 | 25 | (51) | 24 | (49) | 11 | (46) | 0 | (0) | 13 | (54) | 0 | (0) |
| CA | 83 | 64 | (77) | 19 | (23) | 16 | (84) | 0 | (0) | 0 | (0) | 3 | (16) |
| CC | 42 | 28 | (67) | 14 | (33) | 9 | (64) | 2 | (14) | 1 | (7) | 2 | (14) |
| CG | 81 | 53 | (65) | 28 | (35) | 24 | (86) | 0 | (0) | 1 | (4) | 3 | (11) |
| CT | 64 | 52 | (81) | 12 | (19) | 6 | (50) | 0 | (0) | 3 | (25) | 3 | (25) |
| GA | 69 | 6 | (9) | 63 | (91) | 46 | (73) | 4 | (6) | 12 | (19) | 1 | (2) |
| GC | 60 | 44 | (73) | 16 | (27) | 8 | (50) | 5 | (31) | 3 | (19) | 0 | (0) |
| GG | 69 | 25 | (36) | 44 | (64) | 29 | (66) | 9 | (20) | 6 | (14) | 0 | (0) |
| GT | 60 | 33 | (55) | 27 | (45) | 17 | (63) | 5 | (19) | 3 | (11) | 2 | (7) |
| TA | 72 | 21 | (29) | 51 | (71) | 9 | (18) | 0 | (0) | 41 | (80) | 1 | (2) |
| TC | 28 | 20 | (71) | 8 | (29) | 0 | (0) | 0 | (0) | 5 | (63) | 3 | (38) |
| TG | 95 | 57 | (60) | 38 | (40) | 8 | (21) | 1 | (3) | 29 | (76) | 0 | (0) |
| TT | 17 | 12 | (71) | 5 | (29) | 0 | (0) | 2 | (40) | 3 | (60) | 0 | (0) |
| Total | 985 | 533 | (54) | 452 | (46) | 231 | (51) | 30 | (7) | 173 | (38) | 18 | (4) |
a(−1 Deletion)/(−1 deletion + substitutions) %.
b(Substitution)/(−1 deletions + substitutions) %.
c(X inserted opposite F)/(all insertions opposite F) %.
Not all combinations of flanking sequence appeared to be represented equally, however. If one sums the number of substitution and −1 mutations for each sequence context, the observed frequencies differed significantly (χ2 = 117) from an even distribution at the 0.01 significance level (χ20.99 = 30.6). This could either mean that the A, C, G and T were not incorporated equally during synthesis, or that certain sequence contexts are bypassed more slowly than others and are therefore not equally represented as the others in the 30-min reaction products. It is also possible that the under-represented sequences resulted in −2 deletion mutations that were excluded from the SAMS analysis. To test whether the bias was being introduced during DNA synthesis, we also sequenced 177 bypass products of the control sequence which lacked the abasic site, but included the random NN sequence library. In this case, the observed frequencies did not differ significantly (χ2 = 25) from the expected distribution at the 0.05 significance level (χ20.95 = 25), suggesting that bypass efficiency and/or −2 mutations were biasing the results (Supplementary Table 1). As one will see later, −2 mutations were a significant contributing factor to the under-representation of −1 and substitution mutations in some sequence contexts.
Substitution mutations
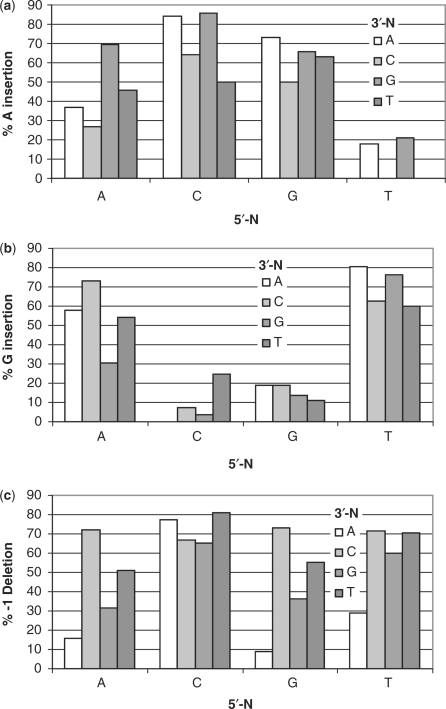
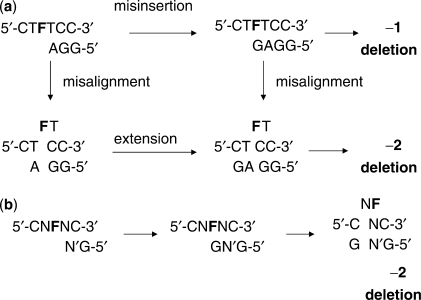
The major substitution mutations observed corresponded to an average insertion of A (51%) or G (38%) opposite the abasic site in a given sequence context whereas insertion of C (7%) and T (4%) was much less frequent (Table 1). The four sites showing the highest amount of A insertion opposite the abasic site were CFG, CFA, GFA and AFG (86, 84, 73 and 69%, respectively), while the four sites showing the highest G insertion were TFA, TFG, AFC and TFC (80, 76, 73 and 63%, respectively) (Figure 3a and b). There did not appear to be any obvious correlation between either flanking nucleotide and the insertion of A or G that might be expected if the flanking base was templating the insertion, which would have led to the formation of NFN → TTN, NTT, CCN, or NCC sequences (Figure 4 mechanisms a and b). On the other hand, A insertion appeared to be favored by a 3′-A or G flanking the abasic site which might be explained by their better ability to base stack with the dATP or dGTP when inserting opposite the abasic site model (Table 2). The low frequency of A incorporation with T flanking the 3′-side of F and the low frequency of G incorporation with a flanking C can be attributed to preferential −1 deletion formation by a misinsertion-mediated misalignment mechanism to be discussed (Figure 4d).
Figure 3.
Insertion and −1 deletion frequencies as a function of sequence context. (a) Percentage of A and (b) percentage of G insertion opposite F of all substitution mutations. (c) Percentage of −1 deletion of all −1 and substitution mutations. The base at the bottom refers to the 5′-N of 5′-NFN-3′, while the color-coded bar refers to the N-3′.
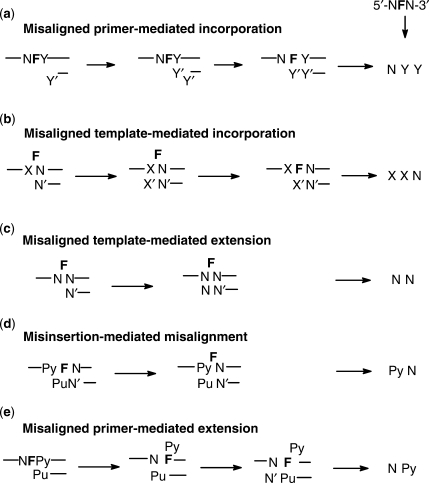
Figure 4.
Possible mechanisms to explain the sequence specificity of substitution and −1 deletion mutations. (a) Nucleotide incorporated opposite F is encoded by the 3′-flanking base through misalignment-mediated insertion followed by realignment; (b) nucleotide incorporated opposite F encoded by 5′-flanking base through misalignment-mediated insertion followed by re-alignment; (c) non-sequence-specific −1 deletion involving an extrahelical or bulged out F in the template, also referred to as a dNTP-stabilized misalignment; (d) sequence-specific −1 deletion resulting preferential insertion of A or G opposite F and misalignment to pair with a 5′-T or C flanking F; (e) sequence-specific −1 deletion results from preferential misalignment of a primer terminating in A or G opposite a 3′-T or C flanking F to align opposite the abasic site after which extension takes place.
Table 2.
Correlation of −1 deletion and major substitution mutations with flanking sequence in 5′-NFN-3′
| Avg. % −1 |
Avg. % G opposite F |
Avg. % A opposite F |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′-N | 3′-N | 5′-N | 3′-N | 5′-N | 3′-N | ||||||
| A | 43 | A | 33 | A | 54 | A | 39 | A | 45 | A | 53 |
| C | 73 | C | 71 | C | 9 | C | 40 | C | 71 | C | 35 |
| G | 43 | G | 48 | G | 16 | G | 31 | G | 63 | G | 60 |
| T | 58 | T | 64 | T | 70 | T | 38 | T | 10 | T | 40 |
−1 Deletion mutations
The sequence contexts showing the greatest percentage of −1 deletion mutations were CFT, CFA, GFC and AFC (81, 77, 73 and 72%) (Table 1, Figure 3c). These deletion mutations can be explained by either a misinsertion-mediated misalignment mechanism (Figure 4d) and/or a misaligned primer-mediated extension mechanism (Figure 4e). Given that A and G were the two most frequently inserted nucleotides (84% total) opposite the abasic site in the substitution mutations, the misinsertion-mediated misalignment mechanism would be expected to favor 5′-PyFN-3′ sequences and would explain the origin of the CFT and CFA mutations. If an A or G opposite an abasic site is also more stabilizing than T or C, then a misaligned primer-mediated extension mechanism would be expected to favor 5′-NFPy-3′ sequences and would explain the high frequency of −1 deletion at GFC and AFC. In fact all of the −1 deletion mutations with a frequency greater 50% can be explained by either of these two mechanistic schemes. One cannot rule out a misaligned template-mediated extension (dNTP stabilized or lesion loop out) mechanism (Figure 4c) to explain the preference of −1 deletions at NFPy sequences if extension is facilitated by a primer terminating in a purine. The low yield (29%) of −1 deletion for the TFA sequence can be explained by a highly competitive insertion of G opposite F which cannot misalign and thus leads to the template strand TCA substitution mutation. With the exception of the TFA sequence, all other sequences with a low frequency of −1 deletion fall into the PuFPu sequence context, with no 3′-pyrimidine to template the addition of a purine to the primer or 5′-pyrimidine to facilitate misalignment by base pairing with a purine in the primer. The AFA, GFA, AFG and GFG contexts yielded only 16, 9, 32 and 36% of a −1 mutation.
Deletion mutation frequency with sequence-specific abasic sites
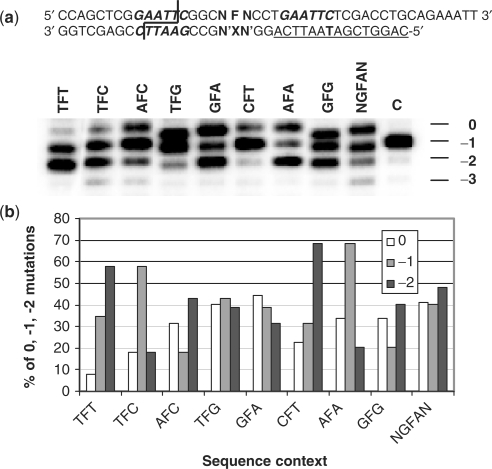
Because the SAMS assay was only conducted on the −1 and full-length products, we did not know if the under-represented sequences were the result of a slower rate of bypass, and/or due to significant formation of −2 mutations. To investigate these possibilities we carried out primer extension reactions on homogeneous templates containing F in 8 of the 16 sequence contexts (Figure 5a). We indeed found that primer extension opposite sequence contexts that were under-represented in the SAMS assay of −1 deletion and substitution mutations gave primarily −1 and −2 deletions and little substitution product, while over-represented sequences corresponded to those showing only −1 deletion and substitution mutations. For example, mutations arising from the TFT and TFC sequences which we found to be under-represented in the SAMS analysis of −1 and substitution deletions, can now be seen to have a high percentage of −2 deletion (Figure 5b). The percentage of −1 deletion relative to the −1 and substitution products for TFT and TFC (81% and 77%, Table 3) is virtually identical to that of 71% and 71% determined by the original SAMS assay (Table 1). Another sequence that was under-represented, but not as much as the two aforementioned sequences was AFA, which can also be seen to have a significant amount of −2 mutation (Figure 5b). The GFA and GFG sequences also showed a significant amount of −2 mutation but were not under-represented in the original SAMS data. On the other hand, the AFC and TFG sequences that we found to be over-represented contained mainly −1 and substitution mutations and very little −2 mutation. Another sequence that showed little −2 mutation, but was not as overly represented was CFT. In most cases the percentage of −1 relative to −1 and substitution mutations determined by the primer extension assay and SAMS assay were in very good agreement (Table 3). It differed substantially, however, for GFA and AFA (33% and 39% versus 9% and 16%, respectively) for some unknown reason.
Figure 5.
Formation of −1 and −2 mutations in specific sequence contexts. (a) Gel analysis of the full-length (0), and −1 and −2 deletion products in the bypass of homogeneous substrates in selected sequence contexts. C refers to the control sequence NN lacking the abasic site whose full-length product corresponds to a −1 deletion. The EcoRI sequence in the primer was mutated (G→T) to prevent cleavage at this site and retain the 5′-32P label on the bypass products. (b) Frequency of substitution, −1 and −2 mutations as a function of sequence context.
Table 3.
Percentage of −1/(−1 + substitution) mutations with homogeneous templates
| 5′-NFN-3′ | Gel assay | SAMS | |
|---|---|---|---|
| 1 | TFT | 81 | 78 |
| 2 | TFC | 77 | 71 |
| 3 | AFC | 69 | 90 |
| 4 | TFG | 51 | 60 |
| 5 | GFA | 33 | 9 |
| 6 | CFT | 76 | 81 |
| 7 | AFA | 39 | 16 |
| 8 | GFG | 50 | 36 |
| 9 | NGFAN | 50 | – |
We also carried out primer extension on NGFAN to see how flanking sequences one removed from the immediately flanking sequence affected the pattern of deletion mutations. In contrast to the large percentage of −2 deletion that was observed for sequence 5 (CGFAC), in the pool of all 16 possible NGFAN sequences, the −2 mutation was relatively minor, indicating an important role of sequences that do not immediately flank the damage site on −2 mutation frequency.
SAMS assay on −1 and −2 deletions
We then selected four sequence contexts showing different ratios of −1 and −2 mutations in the primer-extension assay for SAMS analysis. Both TFT and GFA showed a higher proportion of −2 to −1 mutations from the gel assay, which was accurately reflected in the SAMS assay (Table 4). Likewise, AFC and CFT showed a higher proportion of −1 than −2 deletions in the gel assay which was also accurately reflected in the SAMS assay. Sequence analysis revealed that the −1 deletion products for all four sequences retained both flanking nucleotides and could have arisen by one or more mechanisms. The −2 deletion mutations were all found to be exclusively complementary to the 3′-nucleotide flanking the abasic site.
Table 4.
Percentage of −2 deletions/(−1 and −2 deletions)
| Template | Gel assay | SAMS | 5′-NX′N-3′ | Number of mutants |
|---|---|---|---|---|
| 5′-TFT | 63 | 68 | GGCTTCCT | 58 |
| GGC TCCT | 121 | |||
| 5′-AFC | 22 | 17 | GGCACCCT | 70 |
| GGC CGCC | 14 | |||
| GGCA CCT | 0 | |||
| 5′-GFA | 64 | 81 | GGCGACCT | 24 |
| GGC ACCT | 98 | |||
| GGCG CCT | 0 | |||
| 5′-CFT | 8 | 8 | GGCCTCCT | 89 |
| GGC TCCT | 8 | |||
| GGCC CCT | 0 |
For TFT, the −1 product could have come from either extension of a misaligned template, also referred to as a dNTP stabilized misalignment (Figure 4c) (15), or a misinsertion-mediated misalignment (Figures 4d and 6a). The −2 mutation could have arisen via a misincorporation of G opposite the abasic site followed by misalignment to form a two base complementary stem and a 2-nt bulge loop (Figure 6a). Alternatively, the primer terminating in A opposite the 3′-T of the abasic site could have misaligned to yield a single nucleotide stem and a 2-nt bulge which was then extended. The higher frequency of the −2 deletion than the −1 deletion argues for the former pathway which proceeds through a more stable two base complementary stem. Two base pair stems with a template bulge have been shown to be stabilized by yeast pol η (16).
Figure 6.
Mechanisms to explain the sequence specificity of −2 deletion mutations. (a) Two possible misalignment-mediated pathways to explain the high frequency of the observed −2 deletion in the bypass of F flanked by T′s (5′-TFT-3′). (b) G misinsertion-mediated mechanism for all sequences.
The −2 deletion mutations observed for the other three sequence contexts, AFC, GFA and CFT, can all be explained by a misinsertion-mediated misalignment mechanism in which G is inserted opposite F followed by misalignment to form a G•C base paired terminus and a 2-nt bulge (Figure 6b). The −1 deletion for the AFC sequence can be explained by either a misaligned template or misaligned primer-mediated extension (Figure 4c and e). The −1 deletion produced in the GFA context can best be explained by a misaligned template-mediated extension (Figure 4c), but because the primer terminates in T, it occurs with much lower frequency than the −2 deletion. The −1 deletion for the CFT sequence can be explained any of these mechanisms.
DISCUSSION
The method that we have developed takes advantage of methodology to site-specifically incorporate a homogeneous DNA lesion into a random sequence library together with highly efficient sequencing of the bypass products through polymerization and cloning. Our method is an extension of methods that sequence clones containing the bypass products of an individual sequence context (7,17). Though in our example we only examined a library consisting of 16 sequence contexts, NFN, and sequenced 1000 bypass products, in principle our method could be extended to much larger sequence contexts with the currently available high-throughput sequencing technologies. A related strategy has been reported in which an 8-mer random library NNNNFNNNN was used to select sequence contexts that were most efficiently bypassed by T4 polymerase (18). Since only a small percentage of the sequence contexts were bypassed, and only about 100 bypass products were sequenced, only a very small part of the mutation spectrum was acquired. In our system, the polymerase is given sufficient time to bypass the majority of sequence contexts, and a large number of bypass products are sequenced to allow for a much more complete and accurate mutation spectrum to be obtained. Sequencing a large number of bypass products is facilitated by polymerizing the bypass products before cloning so that each sequenced clone has about 10 bypass products. We adopted this basic strategy of polymerizing the bypass products to increase sequencing throughput from the serial analysis of gene expression (SAGE) method (19), which we had also used in a method to select high-affinity antisense agents for native mRNA called serial analysis of antisense binding sites (SAABS) (20).
The high-throughput nature of the serial analysis method should also facilitate the mutation spectrum analysis of less mutagenic polymerases, where more bypass products will need to be sequenced. Alternatively, deletion and insertion mutants products can be isolated by PAGE prior to cloning and sequence analysis, and it may be possible to eliminate nonmutagenic bypass products through restriction digestion of appropriately engineered sequence contexts. To estimate the contribution of pre-existing and nontargeted mutations, the replication products of the identically synthesized template lacking the damage site by the polymerase of interest can be sequenced and compared to that of a high-fidelity polymerase. For DNA damage that can be directly reverted, such as dipyrimidine photoproducts or O6-methyguanine, the analysis can be carried out on the damaged template following repair.
Though yeast pol η is not the enzyme that has been found to bypass abasic sites in vivo, (21,22) it was chosen as a model polymerase because it is a DNA damage bypass polymerase, and its ability to bypass the tetrahydrofuran abasic site model (F) has been previously studied by steady-state kinetic methods (23). This abasic site model was also chosen because a commercially available phosphoramidite was available for its site-specific incorporation into DNA by automated DNA synthesis (14). There have also been a number of other studies of the bypass of the tetrahydrofuran model by other polymerases which could be used to compare similarities and differences in mechanism (15,18,24–26).
We found that A and G were preferentially inserted opposite F, with an overall preference for A, but with different sequence dependences. The previous study of nucleotide insertion specificity with pol η had found that G was preferred over A (1.9) in TFG sequence context. In that same local sequence context we also found that G was preferred over A but by a greater ratio (3.6). This difference may be the result of effects from more remote sequences or from a statistical difference due to an insufficient number of sequenced bypass products. We found no evidence for a misalignment-mediated insertion followed by realignment mechanism (Figure 4a) that was invoked to explain the incorporation of C opposite a C2-AP site by Pol II/IV opposite F(26) which would have led to a AAN′ or GGN′ sequence opposite TFN or CFN, respectively. Likewise, incorporation of A or G opposite F via misalignment followed by templated addition opposite the 5′-flanking N and realignment (Figure 4b) were not seen to predominate (N′AA or N′GG opposite NFG, NFC, respectively) as observed for human pol β (15). The comparable frequency of A and G insertion is unusual as most previously studied polymerases show a much higher preference for A relative to G. For Drosophila polymerase α the specificity of A to G varied from 6–11 depending on the 3′-nt to F, while human pol α selectivity ranged from 2.1 to 7 depending on the 3′-nt (15). In a study of the bypass of F by exo− KF and Dpo4 and human pol η by a reversion assay, insertion of A opposite F was highly favored by all enzymes in a TFG and AFC context, except for pol η, which showed a significant amount of G inserted in the AFC context (7).
We saw a significant amount of −1 deletion mutations which could be ascribed to either a misinsertion-mediated misalignment mechanism (Figure 4d) or misaligned primer-mediated extension mechanism (Figure 4e). In the first mechanism, misalignment takes place after the preferential insertion of A or G opposite the abasic site to form a Watson–Crick base pair with the base flanking the 5′-side of F, and in the second, misalignment takes place after templated insertion of A or G by the base flanking the 3′-side of F. Human pol β has been shown to misalign in preference to extend when the nucleotide opposite F is complementary to the 5′-flanking nucleotide with efficiencies that are similar for all four nucleotides (15). HIV reverse transcriptase also misaligns to give -1 deletions when the 5′-flanking nucleotide is G, and then with decreasing preference with C and T. Highly efficient misalignment-mediated insertion has also been observed for Dpo4 with an abasic site in a GFT context (17). In contrast yeast pol η appears in our study to prefer to misalign when the 5′-flanking nucleotide is C or T. In accord with our results, human pol η has been found to have a higher frequency of −1 deletions in a TFG sequence than in a AFC sequence context (7).
We also observed a significant amount of −2 deletions in specific sequence contexts. The −2 deletion could all be explained by misinsertion of G opposite the abasic site followed by misalignment to produce a 2-nt template bulge (Figure 6). This type of mechanism has also been proposed to account for the −2 deletion observed when exo− KF synthesized opposite TCFC in which A was misinserted opposite F following insertion of G, which then misaligned (24). The high yield of −2 deletions is consistent with the ability of pol η to stabilize and extend template-strand bulge loops (16). In the SAMS analysis of the homogeneous templates, we did not observe nontargeted mutations as has been reported for the bypass of an actual abasic site by Dpo4 (17), which is consistent with a misincorporation rate of about 1% for pol η.
CONCLUSION
We have developed a high-throughput method for determining the mutation spectrum for a DNA lesion as a function of sequence context which we call SAMS. The methodology allows one to focus on a particular class of mutations, such as substitution, and/or deletion or insertion mutations by excising the corresponding bypass products from a gel. The methodology is not limited to bypass products produced in vitro, but could also be used to assay bypass products produced in cell extracts or in vivo. One can also focus on specific sequences, or combinations of flanking nucleotides by synthesizing the appropriate sequences. By carrying out such a broad sweep of sequences in a single experiment, one can identify particularly interesting mutations for further study by SAMS, pre-steady-state kinetic experiments, or in vivo experiments. The methodology cannot, however, detect more complex mutations that involve substitutions in the random flanking sequences, or unambiguously identify the precursor sequence for deletion mutations, which would require a single homogeneous template. Using this methodology, we have found that A and G are inserted opposite the abasic site by yeast pol η in a sequence-dependent manner resulting in abasic site to T and C mutations. We have also discovered a high frequency of −1 mutation that can be attributed to misinsertion-mediated misalignment and misalignment-mediated insertion mechanisms. The high frequency of −2 mutations is consistent with a misinsertion mediated misalignment mechanism and the ability of pol η to stabilize template bulges. We believe that the SAMS methodology will greatly aid in assessing the role of sequence context in the mechanisms of mutagenesis and on the origin of mutation hotspots caused by DNA-damaging agents.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Institutes of Health (CA40463). Funding for open access publication charge: National Institutes of Health (CA40463).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. Washington, DC: ASM Press,; 2006. [Google Scholar]
- 2.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann AR. Replication of damaged DNA by translesion synthesis in human cells. FEBS Lett. 2005;579:873–876. doi: 10.1016/j.febslet.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 4.Seo KY, Jelinsky SA, Loechler EL. Factors that influence the mutagenic patterns of DNA adducts from chemical carcinogens. Mutat. Res. 2000;463:215–246. doi: 10.1016/s1383-5742(00)00047-8. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr. Top. Microbiol. Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 6.Canella KA, Seidman MM. Mutation spectra in supF: approaches to elucidating sequence context effects. Mutat. Res. 2000;450:61–73. doi: 10.1016/s0027-5107(00)00016-6. [DOI] [PubMed] [Google Scholar]
- 7.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 8.Fiala KA, Suo Z. Sloppy bypass of an abasic lesion catalyzed by a Y-family DNA polymerase. J. Biol. Chem. 2007;282:8199–8206. doi: 10.1074/jbc.M610719200. [DOI] [PubMed] [Google Scholar]
- 9.Delaney JC, Essigmann JM. Biological properties of single chemical-DNA adducts: a twenty year perspective. Chem. Res. Toxicol. 2008;21:232–252. doi: 10.1021/tx700292a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 11.Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem. Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 12.Ho SP, Britton DH, Stone BA, Behrens DL, Leffet LM, Hobbs FW, Miller JA, Trainor GL. Potent antisense oligonucleotides to the human multidrug resistance-1 mRNA are rationally selected by mapping RNA-accessible sites with oligonucleotide libraries. Nucleic Acids Res. 1996;24:1901–1907. doi: 10.1093/nar/24.10.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannistraro VJ, Taylor JS. DNA-Thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase eta. J. Biol. Chem. 2004;279:18288–18295. doi: 10.1074/jbc.M400282200. [DOI] [PubMed] [Google Scholar]
- 14.Ide H, Murayama H, Murakami A, Morii T, Makino K. Effects of base damages on DNA replication–mechanism of preferential purine nucleotide insertion opposite abasic site in template DNA. Nucleic Acids Symp. Ser. 1992:167–168. [PubMed] [Google Scholar]
- 15.Efrati E, Tocco G, Eritja R, Wilson S, Goodman MF. Abasic translesion synthesis by DNA polymerase beta violates the “A-rule.” Novel types of nucleotide incorporation by human DNA polymerase beta at an abasic lesion in different sequence contexts. J. Biol. Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 16.Cannistraro VJ, Taylor JS. Ability of polymerase eta and T7 DNA polymerase to bypass bulge structures. J. Biol. Chem. 2007;282:11188–11196. doi: 10.1074/jbc.M608478200. [DOI] [PubMed] [Google Scholar]
- 17.Fiala KA, Brown JA, Ling H, Kshetry AK, Zhang J, Taylor JS, Yang W, Suo Z. Mechanism of template-independent nucleotide incorporation catalyzed by a template-dependent DNA polymerase. J. Mol. Biol. 2007;365:590–602. doi: 10.1016/j.jmb.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatahet Z, Zhou M, Reha-Krantz LJ, Ide H, Morrical SW, Wallace SS. In vitro selection of sequence contexts which enhance bypass of abasic sites and tetrahydrofuran by T4 DNA polymerase holoenzyme. J. Mol. Biol. 1999;286:1045–1057. doi: 10.1006/jmbi.1998.2520. [DOI] [PubMed] [Google Scholar]
- 19.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 20.Fang H, Yue X, Li X, Taylor JS. Identification and characterization of high affinity antisense PNAs for the human unr (upstream of N-ras) mRNA which is uniquely overexpressed in MCF-7 breast cancer cells. Nucleic Acids Res. 2005;33:6700–6711. doi: 10.1093/nar/gki968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, Xie Z, Shen H, Wang Z. Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 2004;32:3984–3994. doi: 10.1093/nar/gkh710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pages V, Johnson RE, Prakash L, Prakash S. Mutational specificity and genetic control of replicative bypass of an abasic site in yeast. Proc. Natl Acad. Sci. USA. 2008;105:1170–1175. doi: 10.1073/pnas.0711227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haracska L, Washington MT, Prakash S, Prakash L. Inefficient bypass of an abasic site by DNA polymerase eta. J. Biol. Chem. 2001;276:6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- 24.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule.”. J. Biol. Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 25.Otsuka C, Sanadai S, Hata Y, Okuto H, Noskov VN, Loakes D, Negishi K. Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res. 2002;30:5129–5135. doi: 10.1093/nar/gkf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroeger KM, Kim J, Goodman MF, Greenberg MM. Replication of an oxidized abasic site in Escherichia coli by a dNTP-stabilized misalignment mechanism that reads upstream and downstream nucleotides. Biochemistry. 2006;45:5048–5056. doi: 10.1021/bi052276v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.