Abstract
Variable (diversity) joining [V(D)J] recombination of immune gene loci proceeds in an ordered manner with D to J portions recombining first and then an upstream V joins that recombinant. We present evidence that the non-core domain of recombination activating gene (RAG) protein 2 is involved in the regulation of recombinatorial order. In mice lacking the non-core domain of RAG2 the ordered rearrangement is disturbed and direct V to D rearrangements are 10- to 1000-times increased in tri-partite immune gene loci. Some forms of inter-chromosomal translocations between TCRβ and TCRδ D gene segments are also increased in the core RAG2 animals as compared with their wild-type (WT) counterparts. In addition, the concise use of proper recombination signal sequences (RSSs) appears to be disturbed in the core RAG2 mice as compared with WT RAG2 animals.
INTRODUCTION
Variable exons of immunoglobulin (Ig) and T-cell receptor (TCR) genes are assembled during lymphoid development by a tightly regulated and highly ordered series of site-specific DNA recombination reactions known as V(D)J recombination (1). This process is essential for adaptive immunity. The first step in this reaction is performed by recombination activating gene (RAG) proteins 1 and 2 that specifically bind to a pair of recombination signal sequences (RSSs) that flank each variable (V), diversity (D) and joining (J) gene segment. RSSs consist of a conserved heptamer and nonamer separated by either 12 or 23 bp of non-conserved spacer DNA. Only gene segments flanked by dissimilar RSSs can rearrange with one another (the 12/23 rule). In the IgH and TCRβ loci that contain these three different types of gene segments, D to J rearrangement almost invariably occurs first, then a V gene segment rearranges to the initial DJ recombinant (Figures 1A and B) (2). The direct rearrangement of a V segment to an unrearranged D segment does occur but at a very low frequency (3). Where permissible (following the 12/23 rule) D-to-J rearrangement occurs by deletion far more often than by inversion. In the TCRβ locus, only deletions are permitted as the Dβ gene segments have 5′ RSS-12 and 3′ RSS-23 signals, and Jβ segments have RSS-12. In the IgH locus, however, DH gene segments have RSS-12 signals both 5′ and 3′ of the coding segment so that D–J or V–D rearrangements can occur either by a deletional or inversional mechanism.
Figure 1.
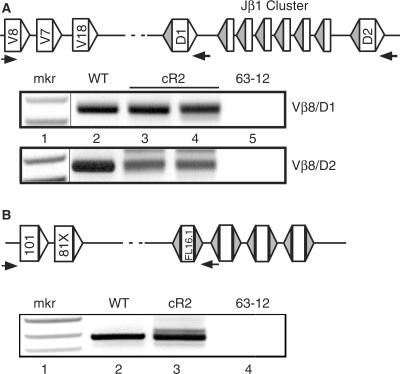
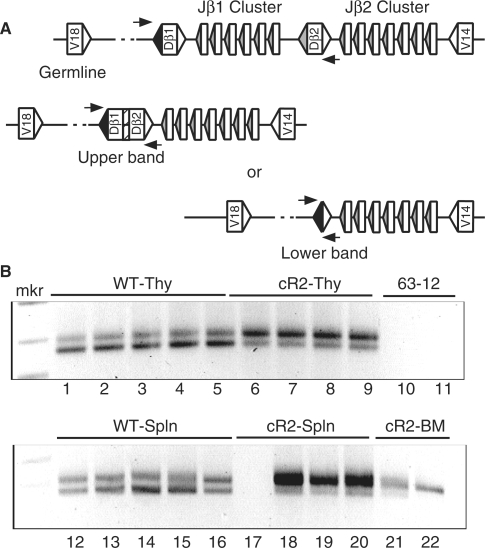
PCR amplification of direct V-to-D rearrangements obtained from splenocyte genomic DNA purified from WT and cR2 mice. (A) Diagram shows a portion of the TCRβ locus in its germline configuration with the positions of PCR primers (horizontal arrows) used to direct amplification of the products shown below (not to scale). Rectangles illustrate coding segments of V, D and J types. Triangles indicate RSS with either 23 bp (unfilled) or 12 bp (filled) spacers. Splenic DNA from individual WT (lane 2) or cR2 (lanes 3 and 4) mice was amplified with Vβ8 forward primers and either Dβ1 or Dβ2 return primers. The negative image of an ethidium-stained gel is shown. An MspI-digested pBR322 DNA marker (mkr) is shown as well as negative PCR control (lane 5, 62-12, RAG2 null cell line DNA). (B) Diagram shows the germline configuration of a portion of the IgH locus with the first two V gene segments followed by the first four D gene segments. Splenic (lanes 2 and 3) and control (lane 4) DNA samples were subjected to PCR with the primers indicated by arrows in the diagram. The negative image of an ethidium-stained gel is shown.
Mechanisms that enforce the ordered assembly of these tripartite loci remain unknown. One possibility is that V gene RSSs are inaccessible to the recombinase while D-to-J rearrangement is occurring. This cannot be the entire explanation, however, since V gene segments often bypass one or several intervening D gene segments while rearranging to a downstream DJ allele (Figure 1). Another hypothesis is that 3′ of D segment RSSs are inherently better or stronger signals, thus favoring the D-to-J events over V-to-D events. This too fails to explain why the V gene segments, which are fully accessible to the recombination mechanism, favor the rearranged DJ segments rather than one of the intervening unrearranged D gene segments. Conceivably, the 5′ of D RSS becomes available to the recombinase only after the 3′ RSS has been deleted by D-to-J rearrangement. It is also possible that V gene segments have no preference for rearrangement and recombine with either unrearranged or rearranged D segments, but that only V gene segments that rearrange with DJ recombinants are ever fixed into the cell's genome by IgH or TCRβ chain selection.
The RAG1 and RAG2 proteins are both required for V(D)J recombination. RAG1 contains the so-called DDE motif, the catalytic triad of amino acids used by recombinases of this type, as well as the RSS binding domain of the V(D)J recombinase (4). While RAG2 is necessary for recombinase activity, its precise role remains uncertain. The carboxy-terminal third of the protein is dispensable for catalytic activity both in vitro and in vivo, but is highly conserved through evolution. Recent reports suggest that it is capable of specific interaction with histone proteins leading these workers to suggest a role for RAG2 in regulation of recombination (5–7). To probe the function of the dispensable domain, we and others have engineered targeted mutant mice that express only the core domain of RAG2 (8,9). We reported previously that the core-RAG2 mutant (cR2) mouse shows a partial block in B- and T-cell development corresponding to a defect in V-to-DJ rearrangement in the IgH and TCRβ loci (9). Independent investigations revealed a defect in the precision of signal joint formation during V-to-DJ rearrangement (10) as well as increased hybrid joint formation (an inversion of the RSS-ended excised reaction byproduct) in the setting of a p53 DNA repair deficiency (11). Thus, it is possible that the non-core domain of RAG2 may be involved in the ordering of rearrangement in tripartite loci.
To begin to explore these issues, we undertook an investigation into the in vivo frequencies of direct V to (germline) D, D-to-J and complete VDJ rearrangements in cR2 and wild-type (WT) mice. Real-time quantitative PCR (qPCR) is ideal for determining the frequency of gene segment recombination events as the primers can be placed on different gene segments and only recombination of those specific segments permits amplification. Using this simple approach, we found that the frequency of direct V-to-D recombination events in WT mice was at least five orders of magnitude lower than the corresponding V-to-DJ event in the TCRβ and IgH loci in various tissues. Surprisingly, direct V-to-D rearrangement was 10- to 1000-fold more frequent in cR2 mice. In addition, we report that the VD coding joints formed in cR2 mice display abnormal sequence properties and can involve interchromosomal rearrangements between distinct antigen receptor loci. Our data suggest that the non-core domain of RAG2 is responsible for enforcing ordered rearrangements in the tripartite antigen receptor loci and suppressing chromosomal translocations.
MATERIALS AND METHODS
Animals and DNA collection
WT B6 and heterozygous and homozygous cR2 mice (backcrossed to B6 for more than 10 generations) were analyzed at approximately 1 month of age. Thymocytes were obtained by disrupting thymus glands, filtering the material through a 0.40 µm cell strainer, and washing in PBS. Splenocytes and bone marrow lymphocyte populations were obtained in a similar manner. DNA was purified by proteinase K digestion in the presence of 0.5% SDS, RNAse treatment, phenol:chloroform extraction and ethanol precipitation. Precipitated DNA was washed three times with 70% ethanol, air dried and then re-suspended in TE (10 mM Tris pH 8.0, 0.1 mM EDTA) at a final concentration of 200 ng/μl.
Conventional PCR
Nested primers were designed to amplify rearranging antigen receptor gene segments (Supplementary Table S1). PCR reactions (25 μl) contained 30 pmol of forward and reverse primers, 0.75 U JumpStart Taq polymerase (Sigma-Aldrich, St. Louis, MO) and up to 1000 ng of genomic DNA template. Primary reaction mixtures were heated to 94°C for 10 min and cycled 20- to 25-times (95°C for 20 s then 65°C for 1 min), with a final 7-min extension at 72°C. A secondary reaction, programmed with 1 μl of the completed primary reaction, used a set of nested primers and was cycled 25- to 30-times with an increased annealing/extension temperature of 68°C. Completed reactions were visualized on ethidium bromide-stained agarose gels.
Real-time qPCR
Primer and probe sets were manually designed following the design guidelines set by Applied Biosystems, Foster City, USA. In all cases, the forward primer and Taqman hydrolysis probe were complementary to groups of V gene segments. Reverse primers were complementary to sequences 3′ of various D gene segments, and thus were specific for direct V-to-D rearrangements. Other reverse primers anneal 3′ of various J gene segments and were used to detect complete VDJ rearrangements. A final set of reverse primers was designed to detect germline V gene segments in order to provide absolute standard curves using DNA from a non-rearranging RAG2 null cell line, 63-12. The V-to-D and V-to-DJ rearrangement frequencies were calibrated using those standard curves. DNA concentration and quality were verified using a real-time PCR assay that detects genomic DNA from the APRT locus. All rearrangement assay results were normalized by dividing by the amount of DNA determined by the APRT assay. The real-time PCR primers and probe for the various V, D and J segments for the TCRβ and the IgH segments are listed in Supplementary Table S1. Reactions followed the cycling conditions suggested by the qPCR device manufacturer (Opticon, Bio-rad, Hercules, CA).
Ligation-mediated PCR (LM-PCR) for signal end breaks
Intact nuclei from the thymocytes of three WT or cR2 mice were collected using standard protocols and fixed with a 4% paraformaldehyde solution in PBS. Fixed nuclei were briefly permeabilized with 0.25% Tween-20 and then stained with 20 μg/ml propidium iodide in the fixing solution. Stained nuclei were sorted by high-speed flow cytometry (MoFlo, Backman-Coulter; Fullerton, CA) and two discrete populations of cells were obtained reflecting either 2C or 4C DNA content. High molecular weight genomic DNA was then purified as above and LM-PCR was performed as previously described (12). PCR products were visualized on an ethidium bromide-stained gel.
RESULTS
Direct V-to-D rearrangement in coreRAG2 mice
To determine whether the ordered rearrangement of TCRβ and IgH gene segments is disrupted in cR2 mice, we designed PCR assays to detect and quantify direct V-to-D rearrangement. Using nested PCR primers located in Vβ8 gene segments and primers located just 3′ to a Dβ gene segment (Figure 1A), we amplified splenic DNA from individual WT (lane 2) and cR2 mutant (lanes 3, 4) mice, and genomic DNA from 63-12, a RAG2-null pro-B leukemia cell line. These PCR products were cloned and subjected to sequence analysis (Table 1). A similar PCR analysis (Figure 1B) was carried with nested VH101 primers and a primer located just 3′ of DFL16.1 in the IgH locus, and the products cloned and sequenced (Table 2). The sizes of the PCR products from the cR2 mice appeared more diverse than those from WT mice for both the IgH and TCRβ loci. Sequence analysis confirmed this difference in fragment diversity (Supplementary Table S2). This difference suggested that there may be a higher frequency of direct V-to-D rearrangement in cR2 compared with WT animals.
Table 1.
Sequence characterization of cloned PCR products from direct TCRβ V-to-D rearrangements
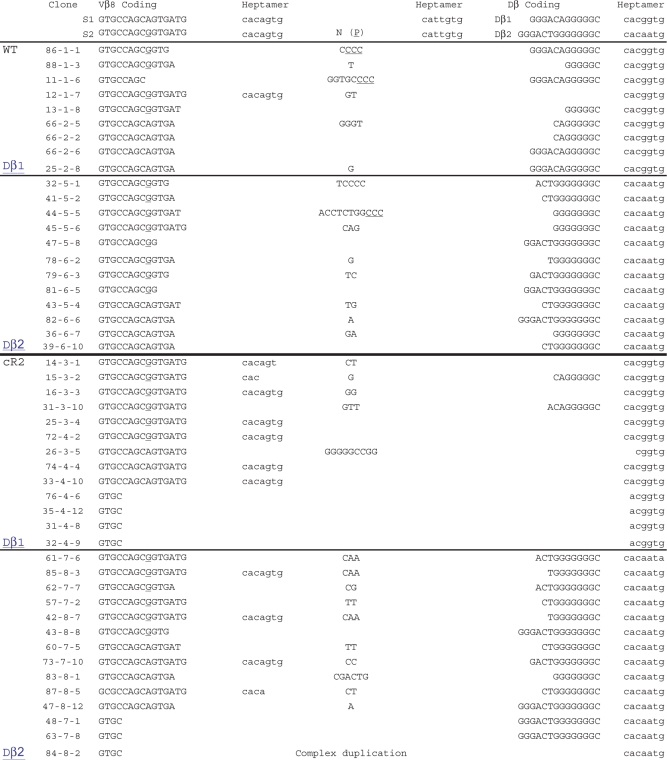 |
PCR products shown in Figure 1A were cloned and sequenced. Two of the three possible Vβ8 gene segments (a single underlined G differentiates them) were detected in rearrangements with intact Dβ1 or Dβ2 gene segments. The third Vβ8 gene segment is not amplified by these primers. The RSS heptamer adjacent to each of the rearranged gene segments is shown (lower case). Non-templated nucleotides (N) and associated P nucleotides (P, underlined) are shown. Recovered sequences span several hundred basepairs on either side of the coding joints (data not shown). Table includes sequences of cloned VD PCR products from several other animals.
Table 2.
Sequence characterization of cloned PCR products from direct IgH V-to-D rearrangements
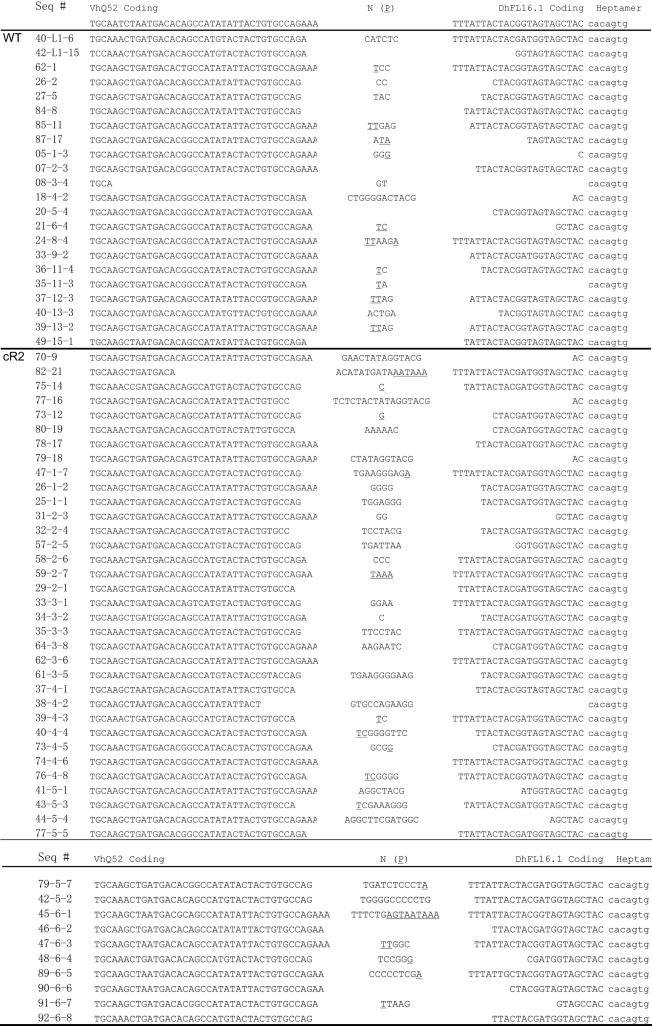 |
PCR products shown in Figure 1B were cloned and sequenced. The RSS heptamer 3′ adjacent to the D gene segment is shown (lower case). Non-templated nucleotides (N) and associated P nucleotides (P, underlined) are shown. Recovered sequences span several hundred basepairs on either side of the coding joints (data not shown). Table includes sequences of cloned V-to-D PCR products from several other animals.
Inclusion of Vβ8 heptamer sequences in VD rearrangements
Our sequence analysis revealed that the cR2 animals generate VD joints that frequently include the heptamer sequence adjacent to the Vβ8 gene segment (Table 1). A simple means test revealed that the cR2 animals (11/28 sequences) were 8-times more likely to create a VD joint that included this heptamer than their WT counterparts (1/21 sequences, P = 0.016). Limited sequencing of direct V-to-D rearrangement events involving other V gene segments failed to show such heptamer inclusions (data not shown). When examining V-to-D events that utilized the Dβ1 coding segment, we found that cR2 animals (11/13 sequences) were also 8 times more likely to delete the entire Dβ1 coding segment as compared with WT animals (1/9 sequences, P = 0.0013). Inclusion of the Vβ8 heptamer occurred more frequently in joints that also displayed deletion of Dβ1 in cR2 (6/13 sequences) but not WT (1/9 sequences) animals. When Dβ2 coding segments were rearranged, we found no inclusion of the Vβ8 heptamer in WT, but its inclusion was found four times in cR2 animals. In stark contrast to Dβ1, Dβ2 was never deleted in either WT or cR2 animals. It is possible that the cR2 recombinase is recognizing the Vβ8 RSS heptamer in an opposite orientation, using a nonamer-like DNA sequence within the coding region to form an RSS-12 (Supplementary Figure S1). This could result in an aberrant rearrangement event consisting of a signal joint containing this unusual RSS-12 and the RSS-23 3′ of Dβ1. We conclude that the cR2 recombinase is imprecise in its selection of cleavage sites, suggesting that the non-core domain of RAG2 is important for the fidelity of V(D)J recombination.
DNA sequence analysis of VH52-to-DH rearrangements reveals long N-regions in cR2 mice
We next characterized a set of direct V-to-D PCR products obtained from the spleens of either WT or cR2 animals using nested primers that amplify products of VHQ52-to-DFL16.1 joining (Figure 1B). DNA sequence analysis of 22 joints from WT animals and 44 joints from cR2 animals failed to reveal any V segment heptamer inclusion. We did note, however, that the N-regions were significantly longer in the cR2 animals. Direct VD joints in cR2 mice often (21/44 sequences) had seven or more N-nucleotides between the two coding segments as compared to the WT direct VD joints (4/22 sequences, proportions test P = 0.01). Additionally we noted that the extent of V gene segment sequence deletion (three or more basepairs) in the direct VD coding joints in cR2 animals (20/44 sequences) as compared with the WT animals (1/22 sequences) was significantly different (proportions test P = 0.0007).
An increased frequency of V-to-D rearrangements in cR2 mice
As our sequence analysis suggested a higher frequency of direct V-to-D rearrangements in cR2 mice, we designed a qPCR method to quantify various V(D)J rearrangements. Genomic DNA prepared from spleen and thymus of either cR2, heterozygous or WT mice was analyzed by qPCR for TCRβ rearrangements (Figure 2A). We found that the frequency of complete VDJ rearrangements utilizing the Vβ7 and Jβ2.7 gene segments is 5-fold higher in the thymus than in splenocytes, likely reflecting the unselected nature of the thymic repertoire (Figure 2B). Complete rearrangements in thymocytes from cR2 mice are just a few fold less frequent than in the WT animals while in the spleen they are slightly more frequent (Figure 2B).
Figure 2.
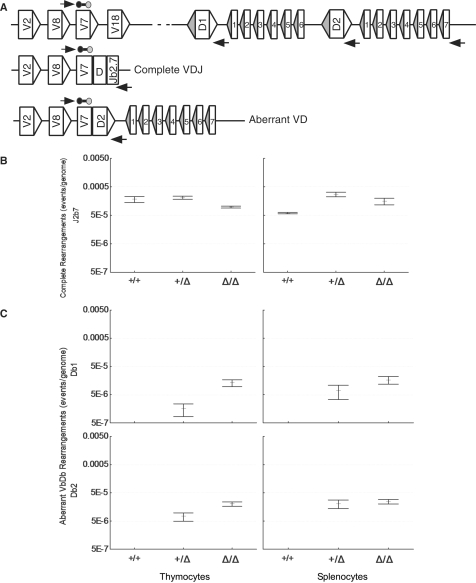
Frequency of complete Vβ7-to-Dβ-to-Jβ2.7 and direct Vβ7-to-Dβ rearrangements in cR2 and WT mice. (A) Diagram of the mouse TCRβ locus indicating germline configuration (top) as well as complete VDJ and direct VD rearrangements utilizing Vβ7. Real-time qPCR primers are shown as horizontal arrows and the probe as a line bounded by circles. DNA purified from the thymus or spleen of sets of four animals of each genotype was analyzed by real-time PCR for complete VDJ (B) and direct VDβ1 and VDβ2 (C) rearrangements and the mean values (bars) SEs (whiskers) are shown.
We went on to compare the frequencies of direct Vβ7 to germline Dβ1 or Dβ2 rearrangement in WT, heterozygous or homozygous cR2 animals (Figure 2C). The thymi and spleens of cR2 mice had readily detectable frequencies of direct VD rearrangements as compared with WT mice, where such events were undetectable with this qPCR assay (Figure 2C). Heterozygous mice had an intermediate frequency of these VD rearrangements. The frequency of complete V(D)J rearrangements involving Vβ7 and Jβ2.7 was about 20-times greater than the frequency of direct Vβ7-to-germline Dβ rearrangement in the cR2 mice. Since there are 14 total Jβ gene segments, we estimate that direct V-to-D rearrangement in cR2 mice is between 100- and 200-fold less frequent than V(D)J rearrangement. A similar quantitative analysis of direct rearrangements of Vβ2, Vβ8 and Vβ14 to germ-line Dβ gene segments revealed similar patterns (Supplementary Figure S2).
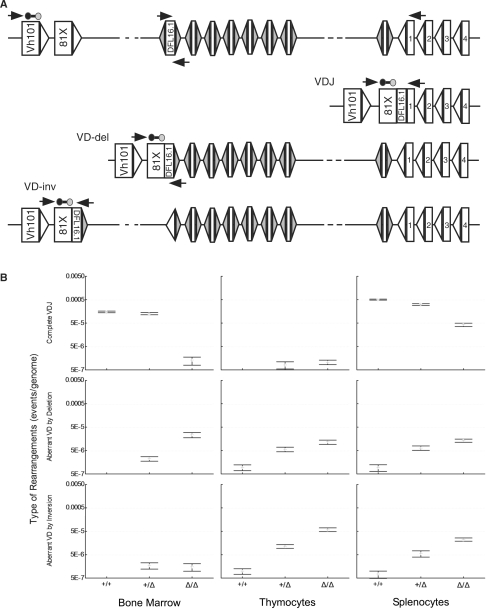
We next examined the IgH locus for direct V-to-D rearrangements. The heavy chain locus contains a suite of DH gene segments followed by a single grouping of four JH gene segments (Figure 3A). Each of the 13 heavy-chain DH segments is flanked by two similar RSS-12 sequences permitting rearrangements to occur by either deletion or inversion of the intervening sequences. The frequency of complete rearrangement of VH81X-to-DH-to-JH1 was measured in order to contrast with direct VH-to-DH rearrangement frequencies. Once again we observed that complete VDJ rearrangement was less frequent in the cR2 samples. In the thymus, where complete IgH rearrangements are thought not to occur in WT mice, we were able to detect such rearrangements in cR2 animals but not in their WT counterparts. The frequencies of direct V-to-D rearrangements that occur by deletion were examined in three tissues (Figure 3B). For all three tissues, the cR2 mice had at least 10-fold higher levels of direct VD rearrangement than did WT mice. For the bone marrow samples, more than a 1000-fold difference was observed. Again the heterozygous animals had an intermediate level of direct VD rearrangements. For each of the three tissues, the frequency of direct VH81X to DFL16.1 rearrangement occurring by inversion was assayed (Figure 3B). These results were nearly identical to those for deletional rearrangements; cR2 animals contain 10- to 100-fold more such rearrangements than their WT counterparts. To rule out the possibility that these observations might be influenced by the proximity of VH81X to the DH gene segment cluster, we looked at VH1S1 that represents a large family of VH gene segments (Supplementary Figure S3). We again observed the same pattern in the data with direct V-to-D rearrangement in cR2 animals ranging from 10- to 1000-fold higher than in WT animals.
Figure 3.
Frequency of complete VH-to-DH-to-JH and direct VH-to-DH rearrangements in cR2 and WT mice. (A) Diagram of a portion of the mouse IgH locus in its germline configuration (upper; only a subset of the total number of D segments bounded by RSS-12 are shown) as well as complete VDJ and direct VD rearrangements (VD-del and VD-inv, deletion and inversion) utilizing VH81x and DFL16.1. Real-time qPCR primers are shown as horizontal arrows and the probe (line bounded by circles). (B) DNA purified from the indicated tissues was analyzed for complete VDJ (upper) or direct VD joints (middle and lower; deletion or inversion). Hybrid joints (not illustrated) were excluded by placing the D segment primer just within that coding segment. Four animals from each of the three possible genotypes were examined and the mean values (bars) with SEs (whiskers) are shown.
IgH D-to-J rearrangements occur in T cells maturing in the thymus. Here again the DH segments can rearrange to downstream JH gene segments either by inversion or deletion. We found that DJ rearrangement by inversion is barely detectable in cR2 thymus (Supplementary Figure S4). DJ rearrangement by deletion was easily detectable in thymus and more frequent in cR2 mice. To determine if this pattern was occurring at other DH gene segments, we examined in the same manner DQ52 and sp2.9, and found similar results (Supplementary Figure S4).
Single D gene segment deletions
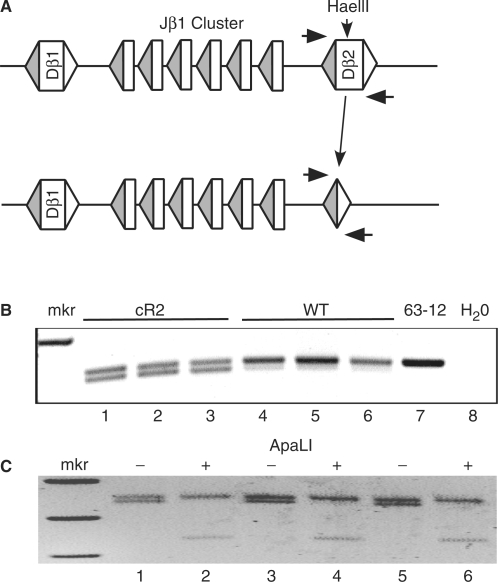
Sequence characterization of direct TCRβ VD joints revealed another unusual type of joint where the entire Dβ gene coding segment is removed and a perfect signal joint is left in its place. We devised PCR assays to specifically detect such events. The first assay made use of a HaeIII restriction site within the germline Dβ2 coding sequence (Figure 4A). PCR primers on either side of the Dβ segments were then used to amplify across the region. Two bands were detected with the largest originating from intact germline sequences (despite overnight digestion of the DNA). The smaller band reflects the formation of a signal joint between the 5′ and 3′ Dβ2 RSSs. cR2 mice were observed to contain Dβ2 coding segment deletions in the thymus more often than WT animals (Figure 4B). To confirm that this reaction generates a precise signal joint, some of the PCR reactions were digested with ApaLI that cuts perfect signal joints (GTG/CAC). Here, only the lower bands were digested (Figure 4C). Sequence characterization also confirmed that the faster migrating species represents precise signal joints (Supplementary Table S3, Part 1). Dβ1 gene segments also underwent deletion by this mechanism more frequently in cR2 than in WT mice (data not shown). We went on to examine the TCRδ locus which also contains two Dδ segments both of which are bounded by RSS-12 and RSS-23. We were able to detect Dδ segment deletion in this setting as well, occurring more frequently in cR2 than in WT animals (data not shown). Similar analyses of several of the IgH DH gene segments failed to reveal any such deletion events, suggesting that the causal mechanism is following the 12/23 rule, as DH gene segments are bounded by RSS-12.
Figure 4.
Dβ gene segments are frequently deleted in cR2 mice as compared with WT mice. (A) Diagram of a portion of the TCRβ locus showing the Dβ2 gene segment in its germline configuration (upper) and after a deletion event (lower). Horizontal arrows indicate the relative positions of a set of PCR primers that amplify the Dβ gene segment from HaeIII-restricted genomic DNA from either cR2 or WT thymus. (B) Negative image of agarose gel analysis of amplification products obtained from thymic DNA purified from six individual mice of the indicated genotypes (lanes 1–6) along with negative controls [reactions programmed with either DNA from 63-12, a RAG2-null cell line (lane 7) or H20 (lane 8)]. (C) The three cR2 PCR products (minus, lanes 1, 3 and 5) were restricted with ApaLI (plus, lanes 2, 4 and 6) which cleaves perfect signal joint sequences (GTG/CAC) remaining after deletion of the Dβ2 coding segment, releasing the expected fragments [341 and 106 bp (not visible)]. Sequence analysis of the PCR products further confirmed their nature (Supplementary Table S3).
It was surprising that RAG protein complexes could bind to one Dβ RSS-12 and then effectively capture the adjoining RSS-23 that is only 12 or 14 bp away without suffering steric hindrance. The possibility exists that such D gene segment deletions could be the result of inter-chromosomal recombination in which a Dβ RSS-12 on one chromosome is recombined with a Dβ RSS-23 on the other, effectively forming a chromosomal translocation that deletes a Dβ segment. To further explore this possibility, we used a mutant mouse that carries a deletion of the Dβ2–Jβ2 cluster of gene segments and crossed this to a WT mouse (13). The F1 generation had one chromosome 6 with the Dβ2 segment and the other without (Figure 5A). We devised a second, direct PCR assay that made use of the perfect signal joints resulting from Dβ gene segment deletions. Here, a downstream primer that spans the signal joint was paired with the upstream primer (Figure 5B). This arrangement effectively eliminates the larger band originating from undigested DNA sequences, making visualization easier. We observed that both the Dβ1 (data not shown) and Dβ2 (Figure 5C) gene segments still underwent deletion in the F1 mice, leading us to conclude that at least a fraction and perhaps all Dβ gene segment deletion events are intramolecular.
Figure 5.
Dβ2 coding segment deletion can occur via an intrachromosomal recombination reaction. (A) Diagram illustrating TCRβ locus structure of a heterozygous mutant in which a portion of the locus has been deleted by gene targeting resulting in the presence of only one copy of the Dβ2 gene segment. (B) Diagram showing a direct PCR strategy to detect perfect signal joints resulting from the deletion of Dβ2 gene segments. One of the PCR primers (horizontal arrows) overlaps the signal joint by several base pairs precluding the amplification of unrearranged gene segments. (C) Agarose gel analysis of PCR products obtained from the genomic DNA of several individual mice with either WT (Dβ2 +/+) or heterozygous Dβ2-deleted (Dβ2 +/KO) genotypes. The identities of PCR fragments were confirmed by DNA sequence analysis (Supplementary Table S3). Amplification of the β-globin locus serves as a DNA loading control. 63-12 is a RAG2-null cell line whose DNA served as a negative control.
Dβ1-to-Dβ2 deletions
Sequence from one of the cR2 direct VD rearrangement events revealed a Dβ1/Dβ2 joint adjacent to a Vβ gene segment. Previous reports have detailed the occasional rearrangement of Dβ1 to Dβ2 in WT mice (14). To examine these Dβ1/Dβ2 events further, we used nested primers in front of Dβ1 and behind Dβ2 (Figure 6A). Rearrangements between the 3′ Dβ1 RSS-23 and the 5′ Dβ2 RSS-12 would result in the two Dβ segments forming a coding joint. Alternatively, the 5′ Dβ1 RSS-12 could capture the 3′ Dβ2 RSS-23 effectively resulting in the loss of both Dβ segments to a coding joint circle and the formation of a signal joint on the chromosome. In WT thymocytes, both products were detected, but the smaller signal joint products appeared to be more prevalent in the five animals analyzed. In cR2 thymocytes, this situation was reversed with larger coding joint favored over smaller signal joint formation. This was also observed in splenocytes with the cR2 animals again showing a much higher level of the coding joint over the signal joint. We were unable to devise a qPCR assay to properly quantify these observations due to sequence constraints. Sequence analysis confirmed the identity of these joints and failed to reveal N-region length differences between joints in WT and cR2 mice (Supplementary Table S3, Part 2). We also looked for D–D joints in the TCRδ locus and again we found that the cR2 mice had a discernable increase in these deletion events (data not shown).
Figure 6.
Dβ1-to-Dβ2 coding joints in cR2 mice. (A) Diagrams showing two types of rearrangements between the Dβ segments in the TCRβ locus. Recombination targeting the Dβ1-RSS-23 (open triangle) and Dβ2-RSS-12 (filled triangle) results in a coding joint with N-nucleotides (hatched) and recombination targeting the Dβ1-RSS-12 and the Dβ2-RSS-23 results in a signal joint. Relative positions of the PCR primers (horizontal arrows) are shown. (B) A single round of PCR was used to amplify genomic DNA purified from the indicated tissues (Thy, thymus; Spln, spleen; BM, bone marrow) from multiple individual mice. An agarose gel analysis of PCR products is shown with the size of coding joints (upper band, 308 bp) and signal joints (lower band, 278 bp) shown. Joint structures were confirmed by DNA sequence analysis (Supplementary Table S3). 63-12 is a RAG2-null cell line whose DNA served as a negative control.
TCRβ-to-TCRδ D gene segment translocations
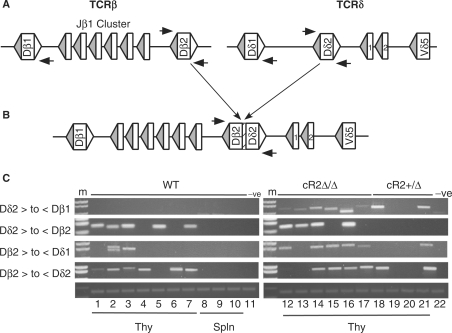
The non-core domain of RAG2 was shown by others to diminish the frequency of signal end transposition in vitro (15) and we recently reported that signal end insertions are greatly increased in cR2 mice as compared with their WT counterparts (16). Chromosomal translocations between antigen receptor loci and other targets have been associated with mis-regulated RAG activity (17). To further explore the possibility that the non-core domain of RAG2 might play a role in the suppression of interchromosomal recombination events, we compared the frequency of inter-locus Dβ-to-Dδ rearrangement in WT and cR2 animals. These loci are located on chromosome 6 and 14, respectively (Figure 7A). Genomic DNA from developing thymocytes was chosen as both these loci are accessible and undergoing RAG-mediated recombination in this tissue. Furthermore, the unselected repertoire would be available permitting the recovery of unbalanced translocation events. We chose to examine all the possible translocations that could occur between the Dβ2 and Dδ2 gene segments consistent with the 12/23 rule (Figure 7B). We found that WT mice are devoid of Dδ2-to-Dβ1 translocations, while in cR2 mice these are readily detectable in all six individuals examined (Figure 7C and Supplementary Table S3, Part 3). Two of the four heterozygous cR2 mice contained this type of translocation as well. Dδ2-to-Dβ2 translocation was detected in both WT and cR2 thymocytes. Dβ2-to-Dδ1 translocation occurred more frequently in the cR2 animals (5/6 versus 2/7, difference test P = 0.03), but Dβ2 to Dδ2 occurred with equal frequency in the two genotypes. These results suggest that the non-core domain of RAG2 might be involved in suppressing certain interchromosomal rearrangement events.
Figure 7.
Interchromosomal rearrangements (translocations) between TCRβ and TCRδ D gene segments. (A) Diagram of the germline configurations of the TCRβ (left) and TCRδ (right) loci. Rectangles represent coding gene segments (D's as marked and J's numbered) bounded by RSS-12 (filled) and RSS-23 (open) triangles. Nested amplifications were directed by primers (horizontal arrows) adjacent to the four D gene segments. (B) Diagram of a translocation between Dβ2 and Dδ2 resulting in a coding joint complete with N-region (hatched). (C) Agarose gel analyses of PCR products from reactions programmed with cR2 (homozygous or heterozygous) or WT genomic DNA purified from thymocytes (lanes 1–7 and 12–21) or splenocytes (lanes 8–10) from individual mice, or 63-12 (RAG2-null negative control) cell line DNA as indicated. PCR primer directions are given as forward (>) or reverse (<) for D gene segments indicated. Joint structures were confirmed by DNA sequence analysis (Supplementary Table S3).
The cR2 mutation alters the cell cycle regulation of RSS cleavage
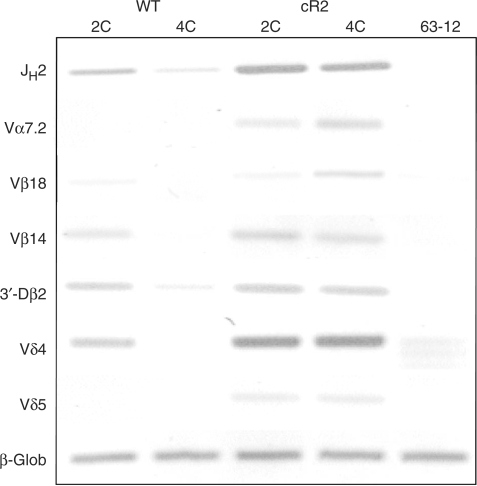
Previous studies have shown that V(D)J recombination is limited to the G1-phase of the cell cycle (12,18). This may be due, at least in part, to the cell cycle-dependent destruction of the RAG2 protein that depends upon a region within its non-core domain (18). Therefore, cR2 protein might persist after developing T or B cells re-enter the cell cycle resulting perhaps in various aberrant recombination events. To test whether the cR2 mouse shows abnormal cell cycle regulation of recombinase activity, we used LM-PCR to assay for double-stranded DNA RSS breaks in genomic DNA purified from thymocyte nuclei containing unreplicated (2C) or replicated (4C) genomes (19). We readily detected broken signal ends from a variety of immune receptor loci in G1 thymocyte DNA samples. We observed very few if any such breaks in WT cells during S/G2/M, whereas the corresponding fraction in cR2 mice had numerous breaks (Figure 8). Thus in cR2 mice, V(D)J recombination may persist through S-phase, perhaps affecting the mechanisms that dictate ordered rearrangement. Alternatively, signal ends generated in G1 may not have been converted to signal joints before the start of S-phase due to a defect in the core-RAG2 containing recombinase complex.
Figure 8.
Analysis of dsDNA RSS breaks in WT and cR2 thymocytes sorted based on DNA content (2C, 4C). Nuclei from several (pooled) 1-month-old mouse thymi were stained with PI and sorted into 2C (G0/G1) and 4C (G2/M) populations. Genomic DNA was subjected to LM-PCR to detect RSS breaks at the indicated antigen receptor gene segment RSSs. The negative image of an ethidium-stained agarose gel is shown. 63-12 is a RAG2-null cell line whose DNA served as a negative control. The direct amplification of β-globin serves as a DNA-loading control.
DISCUSSION
Most biochemical investigations of V(D)J recombination have been limited to truncated or core domains of the two RAG proteins. These core RAG proteins are more soluble than the full-length proteins, and hence are obtainable via traditional recombinant protein purification methods. While the core domain proteins are fully functional, one is left to wonder what the role of the highly conserved non-core domains might be. Biochemical studies have ascribed a transposition repression function to the non-core domain of RAG2 (15) and a transfection study showed a role in the suppression of hybrid joint formation in the setting of NHEJ deficiency (11). Recently, two groups reported that the PHD domain within the non-core region of RAG2 specifically binds to histone H3 trimethylated at lysine 4 (H3K4Me3) (5,6). These reports reinforced a previous work that had shown that the non-core domain of RAG2 binds to histones (7). Finally, Desiderio and co-workers (20) showed that the non-core domain of RAG2 contains a motif that results in the phosphorylation-dependent degradation of RAG2 upon S-phase entry.
To assess function in a more biological system, our group and another developed knock-in mice that express the core domain of RAG2 and found that the mice had defects in IgH and TCRβ gene assembly, most profoundly at the V-to-DJ step (8,9). A subsequent study found that cR2 mutant lymphocytes display a range of deletions at normally full-length signal joints and a spectrum of more subtle coding joint defects. These authors suggested a role for the non-core domain of RAG2 in the structure and stability of the post-cleavage complex (10). In the course of our analysis of V(D)J recombination in cR2 mice, we uncovered a role for the non-core domain at the pre-cleavage step during which the recombinase chooses pairs of RSSs for rearrangement.
D-to-J rearrangement almost invariably precedes V-to-DJ rearrangement in the IgH and TCRβ loci. We found that the frequency of direct V-to-D rearrangement in cR2 mice was increased to a level consistently 10- to 1000-fold that seen in WT mice. Heterozygous mice displayed an intermediate frequency of direct V-to-D rearrangement. In both loci, several different V gene segments were analyzed, and consistently the same results were found. These direct V-to-D rearrangements, while readily detectable in cR2 mice, were still very rare as compared with conventional V(D)J rearrangements. Through sequence analysis we also noted that the cR2 mice produce coding joints with much longer N-regions (>7 bp) as compared with WT RAG2 mice indicative of a joining phase defect. Sequence characterization of the Vβ8-to-Dβ rearrangements led us to discover that cR2 mice frequently included the D RSS heptamer sequence in direct V-to-D joints. This was due to the aberrant recognition the Vβ8 heptamer as part of a cryptic RSS in the inverted orientation leading to the formation of a chromosomal signal joint (Supplementary Figure S1). Thus, the non-core domain of RAG2 is involved in the proper selection of gene segments for V(D)J recombination and subsequent processing.
In addition, we used PCR to search for other possible recombinations that are rarely seen but permitted under the 12/23 rule. This rule stipulates that the recombinational machinery will only select RSS sequences with differing spacer sequences; 12 and 23 bp. In the TCRβ loci, the Dβ1 segment should be able to join the Dβ2 segment as each is flanked by a RSS-12 and RSS-23. This cannot occur in the IgH loci as the D segments are flanked by two RSS-12 signals. We found that cR2 mice make Dβ1-to-Dβ2 rearrangements at much higher frequencies than do WT mice in a fashion that leaves behind a perfect signal joint. Interestingly, this is similar to what we observed previously in an analysis V(D)J recombination in TCRβ enhancer-deleted mice (14). We also found that cR2 mice more frequently generate particular interchromosomal rearrangements between TCRβ and TCRδ.
The V(D)J recombinase has been shown to play a key role in certain chromosomal translocations associated with lymphoid malignancy. Such translocations usually involve rearrangements between Ig or TCR locus RSSs and ‘cryptic’ RSSs or random dsDNA breaks within various proto-oncogenes (17). We recently reported an additional source of recombinase-mediated genomic instability, signal circle insertion. We found that signal circles could undergo a V(D)J recombination-like events with cryptic RSSs leading to re-insertion into the chromosome (16). Remarkably, cR2 mice showed an approximately 7-fold increase in the frequency of such events. The overall conclusion from these observations is that the non-core domain of RAG2 is required for the fidelity of RSS recognition, the suppression of genomic instability and the appropriate regulation of V(D)J recombination during lymphoid development.
It is interesting to consider how the ability of the non-core domain of RAG2 to interact with H3K4me3 chromatin might be responsible for both pre- and post-cleavage functions. With regard to gene-segment choice, the non-core domain of RAG2 might provide some degree of restriction of the recombinase to RSSs within chromatin structure carrying the activating mark. As first suggested by others, the non-core domain might limit the activity of the recombinase in the absence of H3K4me3 binding (5,6). Arguing against this, however, is the several-fold greater activity of the intact as compared to the core recombinase in transient transfection studies where chromatin structure is less likely to play a role than with chromosomal loci (21,22). It is also possible that the different levels of H3K4me3 within chromatin might be involved in the bias for D-to-J rearrangement over V-to-D rearrangement. In the absence of the non-core domain, this bias may be diminished. The non-core domain might also be involved in the preference for rearrangement to occur in cis by promoting some kind of tracking mechanisms in which a RAG–RSS complex transiently interacts with H3K4me3 during the search for a partner RSS. Interestingly, heterozygous cR2 mice display an intermediate effect on the frequency of direct V-to-D rearrangement. This dominant phenotype might be explained by the multimeric nature of the RAG1/RAG2 complex with defective RAG2 subunits influencing overall recombinase activity (23,24). Further studies will be required to critically test these potential aspects of recombinase regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (AI40227 to M.S.S.); Funding for open access charge: NIH RO1 A140227.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Hector Nolla (UC-Berkeley) for assistance with preparative flow cytometry, Hong-Erh Liang for generating cR2 mice and Dan Huang (UC-Berkeley) for technical assistance.
REFERENCES
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Alt F, Yancopoulos G, Blackwell T, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.VanDyk LF, Wise TW, Moore BB, Meek K. Immunoglobulin D(H) recombination signal sequence targeting: effect of D(H) coding and flanking regions and recombination partner. J. Immunol. 1996;157:4005–4015. [PubMed] [Google Scholar]
- 4.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West KL, Singha NC, De Ioannes P, Lacomis L, Erdjument-Bromage H, Tempst P, Cortes P. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Akamatsu Y, Monroe R, Dudley DD, Elkin SK, Gartner F, Talukder SR, Takahama Y, Alt FW, Bassing CH, Oettinger MA. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc. Natl Acad. Sci. USA. 2003;100:1209–1214. doi: 10.1073/pnas.0237043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 10.Talukder SR, Dudley DD, Alt FW, Takahama Y, Akamatsu Y. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic Acids Res. 2004;32:4539–4549. doi: 10.1093/nar/gkh778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekiguchi JA, Whitlow S, Alt FW. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell. 2001;8:1383–1390. doi: 10.1016/s1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 12.Schlissel MS, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5'-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 13.Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, Lafaille JJ, Wang L, Ichikawa Y, Jaenisch R, Hooper ML, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 14.Hempel WM, Stanhope-Baker P, Mathieu N, Huang F, Schlissel MS, Ferrier P. Enhancer control of V(D)J recombination at the TCRbeta locus: differential effects on DNA cleavage and joining. Genes Dev. 1998;12:2305–2317. doi: 10.1101/gad.12.15.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai CL, Schatz DG. Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J. 2003;22:1922–1930. doi: 10.1093/emboj/cdg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curry JD, Schulz D, Guidos CJ, Danska JS, Nutter L, Nussenzweig A, Schlissel MS. Chromosomal reinsertion of broken RSS ends during T cell development. J. Exp. Med. 2007;204:2293–2303. doi: 10.1084/jem.20070583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marculescu R, Vanura K, Montpellier B, Roulland S, Le T, Navarro JM, Jager U, McBlane F, Nadel B. Recombinase, chromosomal translocations and lymphoid neoplasia: targeting mistakes and repair failures. DNA Repair. 2006;5:1246–1258. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 19.Constantinescu A, Schlissel MS. Changes in locus-specific V(D)J recombinase activity induced by immunoglobulin gene products during B cell development. J. Exp. Med. 1997;185:609–620. doi: 10.1084/jem.185.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin W-C, Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–959. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- 21.Cuomo CA, Oettinger MA. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadofsky MJ, Hesse JE, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mundy CL, Patenge N, Matthews AG, Oettinger MA. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanson PC. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 2002;22:7790–7801. doi: 10.1128/MCB.22.22.7790-7801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.