Abstract
The repair of DNA double-strand breaks by nonhomologous end-joining (NHEJ) is essential for maintenance of genomic integrity and cell viability. Central to the molecular mechanism of NHEJ is DNA ligase IV/XRCC4/XLF complex, which rejoins the DNA. During adenovirus (Ad5) infection, ligase IV is targeted for degradation in a process that requires expression of the viral E1B 55k and E4 34k proteins while XRCC4 and XLF protein levels remain unchanged. We show that in Ad5-infected cells, loss of ligase IV is accompanied by loss of DNA binding by XRCC4. Expression of E1B 55k and E4 34k was sufficient to cause loss of ligase IV and loss of XRCC4 DNA binding. Using ligase IV mutant human cell lines, we determined that the absence of ligase IV, and not expression of viral proteins, coincided with inhibition of DNA binding by XRCC4. In ligase IV mutant human cell lines, DNA binding by XLF was also inhibited. Expression of both wild-type and adenylation-mutant ligase IV in ligase IV-deficient cells restored DNA binding by XRCC4. These data suggest that the intrinsic DNA-binding activities of XRCC4 and XLF may be subject to regulation and are down regulated in human cells that lack ligase IV.
INTRODUCTION
DNA double strand breaks (DSBs) occur in mammalian chromosomal DNA as a result of normal cellular processes like immunoglobulin gene rearrangement and also as a result of environmental insults such as exposure to ionizing radiation. Unrepaired or inappropriately repaired DSBs may cause chromosomal aberrations, which can result in tumorigenesis. DNA DSBs may be repaired by nonhomologous end-joining (NHEJ) or by homologous recombination. NHEJ is a homology-independent process used for the repair of DSBs during G0, G1 and early S phases of the cell cycle and can be divided into three steps: (i) DSB detection—where exposed termini of double-stranded DNA (dsDNA) are detected and the repair pathway is initiated; (ii) NHEJ signal transduction and amplification—where the presence of a DSB is signaled to the ligation apparatus; and (iii) synapsis, processing and ligation—where the exposed DNA ends are brought into close proximity, modified to create ligatable ends and covalently re-joined.
In mammals, the ligation step of NHEJ is catalyzed by the ATP-dependent DNA ligase IV (ligase IV), which has been shown to form a functional complex with the DNA-binding proteins XRCC4 and XLF (1–7). Recent findings have demonstrated that the XRCC4 homodimer lies at the center of the ligase IV/XRCC4/XLF complex and directly contacts both ligase IV and XLF (8,9). In addition to the ligase IV/XRCC4/XLF complex, three other mammalian proteins directly participate in NHEJ. The Ku70/80 heterodimer (Ku) and the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) comprise the heterotrimeric DNA-dependent protein kinase (DNA-PK), which is required for NHEJ in vivo and in vitro (10,11). In addition to these six core factors, the Artemis protein has been shown to act as a substrate for DNA-PK and is important for both V(D)J recombination and genomic stability (12,13).
Much is known about mammalian NHEJ, yet the molecular mechanism driving this important biological process, in particular the roles of XRCC4 and XLF, remains unclear. The crystal structures of both proteins reveal N-terminal globular head domains with C-terminal coiled-coil stalks that mediate homo-dimerization (8,14,15). Recent studies have shown that the N-terminal globular head domains of XRCC4 and XLF interact directly (16), leaving the C-terminal coiled-coil domain of XRCC4 free for interaction with ligase IV. While the formation of a ligase IV/XRCC4 complex is necessary for stable expression and function of ligase IV (2), residues at the extreme N-terminus of XRCC4 that are required for its intrinsic DNA-binding activity are dispensable for stimulation of ligase IV in vitro (3,4,17). Interestingly, while N-terminally truncated DNA-binding mutants of XRCC4 are able to stimulate ligase IV in vitro, they are unable to participate in NHEJ in vivo (3,4,17). These observations suggest that the intrinsic DNA-binding activity of XRCC4 plays an important, yet uncharacterized, role in NHEJ in vivo. Because investigation of virus–host interactions frequently illuminates the mechanics of host cell processes, we are using adenoviral infection to study DSB repair by NHEJ in mammalian cells. Specifically, we are using adenovirus infection to study DNA substrate recognition by the ligase IV/XRCC4/XLF complex.
Wild-type human adenovirus type 5 (Ad5) encodes early proteins that inhibit NHEJ on a variety of substrates in infected cells (18–21). Previous work has revealed that a 55-kD protein encoded by adenovirus early region E1B (E1B 55k) and a 34-kD protein encoded by the open reading frame 6 of early region 4 (E4orf6; E4 34k) (19,20). E1B 55k and E4 34k act in concert with host proteins to form a virus-specific E3 ubiquitin ligase that targets several host proteins in infected cells, including p53, Mre11 and ligase IV for proteasome-mediated degradation (19,22–24). Degradation of these proteins inhibits a DNA damage response otherwise induced by adenovirus infection, inhibits p53-dependent apoptosis and prevents NHEJ-mediated end-to-end concatenation of intracellular viral DNA molecules (18,19,22–24). In addition to the E1B 55k/E4 34k ubiquitin ligase mechanism of NHEJ inhibition, there exists a second pathway capable of preventing end-to-end ligation of the viral genome that requires only the expression of an 11-kD protein product that is encoded by the open reading frame 3 of E4 region (E4orf3; E4 11k) (19). While the expression of either E1B 55k/E4 34k complex or E4 11k can prevent end-joining of the viral genome, expression of E4 11k does not inhibit other forms of NHEJ in vivo and fails to inhibit NHEJ in vitro (25). The distinct effect of E4 11k on adenoviral genome concatenation is most likely due to inactivation of Mre11 (26), which is required for genome concatenation (19).
In this communication, we show that E1B 55k/E4 34k-dependent degradation of ligase IV is accompanied by the unexpected loss of DNA binding by XRCC4. Direct assessment of the role of ligase IV in DNA substrate recognition by XRCC4 and XLF revealed that in mutant cells lacking functional ligase IV, XRCC4 and XLF fail to bind DNA. We found that while DNA binding by XRCC4 required the ligase IV polypeptide, binding occurred irrespective of the adenylation state of ligase IV. Finally, we determined that while XLF binds the globular head of XRCC4, which is responsible for XRCC4 DNA binding, formation of an XRCC4/XLF complex does not inhibit DNA recognition by these factors.
MATERIALS AND METHODS
Cell culture, viral infections and DNA transfection
Large-scale infections
HeLa cells (0.5 l) were grown in suspension to 106 cells/ml in Joklik's MEM with 5% Newborn Calf Serum, 100 units/ml penicillin and 100 μg/ml streptomycin (PenStrep), collected by centrifugation, resuspened in fresh medium at 5 × 106 cells/ml in a spinner flask and virus was added to 10 pfu/cell. Virus was allowed to adsorb for 2 h at 37°C with gentle agitation. Cells were collected by centrifugation, resuspended at 106 cells/ml in fresh culture medium and grown for 16 h postinfection (HPI). For small-scale infections, 293 cells were cultured as monolayers in EMEM supplemented with 10% fetal bovine serum (FBS) and PenStrep in 25 cm2 flasks. Small-scale infections in HeLa cells were done by culturing cells as a monolayer in 35 mm dishes supplemented with DMEM containing 10% FBS and Penstrep. For virus infection, culture media is aspirated and replaced with fresh FBS free media and the virus was added at 10 pfu/cell. Virus was allowed to adsorb for 2 h after which FBS free media is aspirated and replaced with fresh media containing 10% FBS. The infection was carried out for 18 h. 2V6.11 cells (27) were cultured as monolayers in EMEM with 10% FBS and PenStrep in 25 cm2 flasks. LB2304 cells were cultured in RPMI 1640 with 15% FBS, PenStrep and 10% sodium pyruvate in 10-cm dishes. NBS3703 cells cultured in DMEM with 15% FBS, PenStrep and 10% sodium pyruvate in 10-cm dishes. Nalm-6 cells were cultured in RPMI 1640 with 10% FBS with 50 μM β-mercaptoethanol in 25 cm2 flasks. Nalm-6 cell transfections were carried out by electroporation of 8 × 106 cells/ml with 20 μg of plasmid DNA at 250 V with five pulses each at 2 ms with 1 s between pulses. The electroporated cells were added back to the 25 cm2 flasks and grown for 60 h in RPMI 1640 with 10% FBS with 50 μM β-mercaptoethanol. pcDNA 3.1 plasmids expressing WT Ligase IV-his and R278H-his were a generous gift from Penny Jeggo (University of Sussex, UK).
Preparation of extracts
Suspension grown HeLa cells were harvested by centrifugation and whole cell extract (WCE) was prepared essentially as described by Baumann and West (28). Mini-whole cell extract (mWCEs) were prepared as previously described (25). Briefly, monolayer cells were harvested using a cell scraper, washed twice in phosphate-buffered saline (PBS), snap frozen on dry ice and stored at –80°C. Frozen cell pellets were resuspended in 70–90 μl of hypotonic lysis buffer [10 mM Tris (pH 8.0), 1 mM EDTA as described in (28)], incubated on ice for 20 min then subject to vigorous vortexing for 30 s. Nuclei were collected by gentle centrifugation (1300g, 2 min, and room temperature) and the supernatant was reserved. Nuclei were resuspended in 35–45 μl of nuclear extract buffer (25 mM Tris pH 8.0, 0.33 M KCl, 1.5 mM EDTA) and incubated on ice for 20 min. The reserved cytoplasmic extract was added back to the nuclei, cell debris was removed by centrifugation (16 500g, 10 min, 4°C) and the resulting supernatant was collected as mWCE. Protein concentrations were determined by Bradford (BioRad, California, USA) analysis prior to storage at –80°C.
Immunoprecipitaiton and adenylation assays
For co-immunoprecipitations (co-IP) HeLa cells were washed twice in PBS and lyzed in NP40 lysis buffer [50 mM Tris HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 10% glycerol, 1 mM DTT and 1% NP40] for 20 min at 4°C. Cellular debris was removed by centrifugation (13 000g, 4°C, 10 min) and the protein concentration of the resulting supernatant was determined by Bradford (BioRad) analysis. An extract of 200 μg was treated with 1:200 dilution of mouse polyclonal XRCC4 (Novus biologicals, Colorado, USA) for 2 h at 4°C with gentle end-over-end mixing. For control IP, FBS (1:200 dilution) was added to 200 μg of the extracts for 2 h at 4°C with gentle end-over-end mixing. After 2 h, 30 μl of protein A sepharose beads (Amersham) was added after being washed three times in NP40 lysis buffer. The mixture is then incubated for 1 h at 4°C with gentle end-over-end mixing. The samples were washed 5–8 times in NP40 lysis buffer to remove unbound proteins, proteins were resolved by SDS–PAGE, subject to western transfer and individual proteins were detected as indicated. Adenylation assays were performed as previously described (29).
In vitro NHEJ, dsDNA–cellulose fractionation and antibodies
Assays for in vitro NHEJ were carried out as described in (11,28). Briefly, reactions (10 μl) were carried out in 50 mM HEPES pH 8.0, 40 mM KOAc, 0.5 mM Mg(OAc)2, 1 mM ATP, 1 mM DTT, 0.1 mg/ml BSA, contained 2–4 μl (10–40 μg) of WCE and Hind III-linearized 5′-32P-labeled pBluescribe DNA (10 ng). Incubation was for 2 h at 37°C. 32P-labeled DNA products were deproteinized and analyzed by electrophoresis through 0.6% agarose gels followed by autoradiography. For dsDNA–cellulose fractionation, 20 μl of a 50% native DNA cellulose (Sigma, Montana, USA) slurry was added directly to 130 μl of 0.4 mg/ml extract, incubated for 3 h at room temperature with turning, collected by centrifugation, washed with 3 × 1 ml in HEK buffer [20 mM HEPES (pH 7.6), 0.1 M KOAc, 0.5 mM EDTA] and resuspended in 2× protein sample buffer. Samples were heated to 100°C for 5 min, resolved by SDS–PAGE and subject to western transfer. In quality control experiments, 32P end-labeled DNA incubated with extract for up to 4 h at 37°C showed no significant degradation (data not shown), and so we believe that DNA used in our DNA-binding experiment remains intact during the room temperature DNA-binding reaction as described above. XRCC4 was detected with anti-XRCC4 rabbit polyclonal antibodies (Serotec, North Carolina, USA, 1:3000). DNA-PKcs was detected with anti-DNA-PKcs rabbit polyclonal antibodies (Serotec, 1:2000). Ligase IV was detected with anti-ligase IV rabbit polyclonal antibody (Serotec, 1:1000). E4 34k was detected using anti-E4 34k rabbit polyclonal antibodies against C-terminal peptide (27,30) (1:1000). E1B 55kDa was detected using 2A6 mouse monoclonal antibody (1:5000) (31). Ku70 was detected using anti-Ku70 rabbit polyclonal antibody (1:1000) (32). Ku80 was detected using anti-Ku80 mouse monoclonal antibody (Serotec, 1:3000). V5-tagged XLF was detected using anti-V5 mouse monoclonal antibody (Serotec, 1:1000). WT his6-ligase IV and his6-R278H were detected using anti-his6 antibodies (GE, USA, 1:3000). Anti-XRCC4 antibodies (Serotec) were used to neutralize NHEJ in vitro at a 1:250 dilution. XLF antibodies were a generous gift from S. Jackson (Cambridge, UK).
Phosphocellulose, gel filtration and phosphopeptide-capture chromatography
HeLa WCEs were prepared and fractionated step-wise over phosphocellulose in L buffer [20 mM Tris (pH 8.0), 0.5 mM EDTA, 10% glycerol, 1 mM DTT] with step-elutions at 0.25 M, 0.6 M and 1 M KCl as previously described (11,28). Superdex-200 gel filtration was performed according to manufacturer's specifications in 25 mM Tris pH 8.0, 0.5 M KCl and 0.5 mM EDTA. An extract of 0.5 mg was fractionated and 0.6 ml fractions were collected. Molecular weight standards were purchased from GE. Phosphopeptide capture was carried out using the PhophoProtein Purification Kit (Qiagen, California, USA) according to manufacturer's instructions.
Cloning, coexpression and purification of GST-XLF and XRCC4-his6
To generate C-terminally his6-tagged XRCC4-his6: the XRCC4 cDNA was removed as an NcoI/EcoRV fragment from pcDNA3.1 XRCC4-V5-His [a generous gift from D. Durocher (33)]. pET28a(+) was digested with EcoRI, the 3′-overhang was Klenow filled and resulting DNA was cleaved with NcoI. Both the vector, pET28a(+) and insert, XRCC4, were gel purified, ligated, transformed into DH5α and plated on LB-Kan. Clones were selected by PCR amplification of the XRCC4 cDNA, positive clones were subject to direct DNA sequence analysis, found to be free of mutations, and transformed into Escherichia coli strain Rosetta 2 (Novagen, New Jersey, USA) to confirm production of XRCC4-his6. The resulting clone is called pET28a(+)XRCC4-his6. To produce N-terminally GST-tagged GST-XLF: pEX-4T-XLF, a generous gift from S. Jackson (Cambridge, UK), was subject to direct sequence analysis and found to be free of mutations. For protein expression, plasmids were transformed into Cam-resistant E. coli strain Rosetta 2. pET28a(+)XRCC4-his6 was plated on LB-Kan, Cam and pEX-4T-XLF plated on LB-Amp, Cam. For coexpression of XRCC4-his6 and GST-XLF, plasmids were co-transformed into Cam-resistant E. coli strain Rosetta 2 and plated on LB-Amp, Kan, Cam. Cells were grown at 37°C in LB broth with the appropriate antibiotics to OD600 = 1.0, induced with 0.25 mM IPTG and then cultured at 37°C for an additional 4.5 h before being harvested by centrifugation and stored at –80°C. For tandem affinity purification of XRCC4-his6 and GST-XLF, Ni-NTA (Qiagen) affinity chromatography was carried out according to manufacturer's instructions. Briefly, cells were lyzed in Ni-buffer [20 mM Tris (pH 8.0), 0.15 M NaCl] with 10 mM imidazole with 50 μg/ml lysozyme ncubated on ice for 15 min and PMSF was added to 1 mM final concentration. The resulting lysate was sonicated (3 × 20 s at 30% power) and cellular debris was removed by centrifugation (25 000g, 4°C, 30 min). A 50% Ni-NTA slurry of 0.8 ml was added to 10 ml of lysate (∼100 mg) and the sample was bound in batch at 4°C for 60 min with gentle end-over-end turning before being poured into a column. The lysate was allowed to flow through and the retained Ni-NTA resin was washed with 4 ml of Ni-buffer with 10 mM imidazole and eluted in Ni-buffer with 0.5 M imidazole. Peak fractions were identified by Bradford analysis, pooled, 4 M NaCl was added to bring the final NaCl concentration to 0.3 M and the resulting sample was applied to a 0.4 ml glutathione sepharose (GE) column prewashed with G-buffer (50 mM Tris pH 8.0, 0.3 M NaCl, 5 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.2 mM benzamidine). The sample was allowed to flow through the glutathione sepharose column, captured and re-applied, after which the column was washed in G-buffer eluted with 40 mM free glutathione. Peak fractions were identified by Bradford analysis, pooled and dialyzed into 20 mM Tris pH 8.0, 50 mM NaCl, 5% glycerol, 0.5 mM EDTA and 1 mM DTT. To purify XRCC4-his6 alone, Ni-NTA affinity chromatography was carried out as described above. To purify GST-XLF alone, glutathione sepharose affinity chromatography was carried out as described above. All protein concentration was determined by Bradford analysis and samples were stored at –80°C.
RESULTS
Inhibition of DNA binding by XRCC4 accompanied loss of ligase IV and inhibition of NHEJ in adenovirus-infected cells
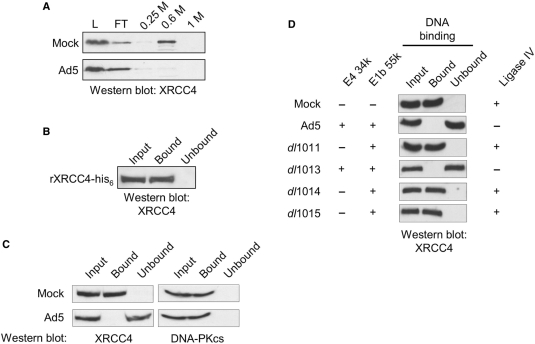
Previous studies have shown that NHEJ is inhibited in adenovirus-infected cells and that this is caused by the targeted degradation of ligase IV during infection (18–21,24,27). While the levels of other NHEJ factors remained unchanged during adenoviral infection (Supplementary Figure S1C) (24), phosphocellulose fractionation of extracts prepared from Ad5-infected HeLa cells revealed a change in the chemistry of XRCC4. Fractionation of extracts prepared from uninfected cells has shown that XRCC4 binds to phosphocellulose (Figure 1A) (28). In fractionating extracts prepared from Ad5-infected cells, we found that XRCC4 did not bind phosphocellulose, but quantitatively flowed through a phosphocellulose column (Figure 1A). While inhibition of NHEJ in Ad5-infected cells is undoubted due to virus-directed degradation of ligase IV, phosphocellulose fractionation of extracts from Ad5-infected cells showed that the chemistry of XRCC4 had changed, which may reflect changes of the entire ligase IV/XRCC4/XLF complex.
Figure 1.
XRCC4 DNA binding is inhibited in an E4 34k-dependent fashion in adenovirus-infected cells. (A) Partitioning of XRCC4 on phosphocellulose was altered by Ad5 infection. HeLa cells were either Mock- or Ad5-infected, extracts were prepared and subject to phosphocellulose chromatography. Load (L), flow-through (FT) 0.25, 0.6 and 1 M KCl washes were resolved by SDS–PAGE, subject to western transfer and probed for the presence of XRCC4. (B) Purified XRCC4 binds dsDNA cellulose. Recombinant XRCC4-his6 was expressed and purified as described in Materials and methods section and subject to dsDNA–cellulose fractionation at 1.4 pM XRCC4-his6. The input fraction (Input), DNA-bound fraction (Bound) and the sample following removal of DNA-bound XRCC4-his6 (Unbound) were resolved by SDS–PAGE, western transferred and probed for the presence of XRCC4. (C) mWCE (50 µg) prepared from 293 cells that were Mock- or Ad5-infected 293 cells were subject to dsDNA–cellulose fractionation. The input extract (Input), DNA-bound species (Bound) and the extract following removal of DNA-binding proteins (Unbound) were resolved by SDS–PAGE, subject to western transfer and probed for the presence of XRCC4 or DNA-PKcs, as indicated. (D) mWCE of 50 µg prepared from HeLa cells that were Mock-, Ad5-, dl1011-, dl1013-, dl1014- or dl1015-infected were treated as in (B) and probed for the presence of XRCC4. Expression of E3 34k, E1B 55k and Ligase IV is indicated. All extracts were prepared 18 HPI.
Like many of the factors involved in NHEJ, XRCC4 possesses an intrinsic DNA-binding activity and recognizes dsDNA in a sequence-independent manner (4,34). Because phosphate recognition is important in retention of a protein on phosphocellulose and in DNA binding, we chose to examine the DNA-binding activity of XRCC4 to determine if the observed lack of XRCC4 retention on phosphocellulose following adenovirus infection was the result of a change in the biologically relevant activity of XRCC4. Purified, recombinant his6-tagged XRCC4 was used as a reference and we determined the concentration of endogenous XRCC4 in mWCE to be ∼1.4 pM (data not shown). While XRCC4 is thought to have a relatively weak DNA-binding activity, we used native dsDNA cellulose fractionation to study the intrinsic DNA-binding activity of purified, recombinant XRCC4 at 1.4 pM. Western blot analysis was then used to assess the presence of XRCC4 in the input fraction (input), the DNA-bound fraction and the fraction following capture of DNA-binding proteins (unbound). We found that the recombinant XRCC4 bound dsDNA cellulose and could only be detected in the DNA-bound fraction (Figure 1B). These data show that DNA binding by XRCC4 can be examined using dsDNA cellulose fractionation.
To determine if adenoviral infection had any affect on DNA binding by XRCC4, we examined extracts prepared from mock- and Ad5-infected 293 cells and found that XRCC4 was present in similar amounts, despite viral infection. In extracts prepared from mock-infected 293 cells, we found XRCC4 to be capable of binding dsDNA and present in the DNA-bound fraction, which we had anticipated (Figure 1C). When dsDNA-cellulose fractionation was carried out with extracts prepared from Ad5-infected 293 cells, XRCC4 was not found in the DNA-bound fraction (Figure 1C). The lack of XRCC4 in the DNA-bound fraction was not due to degradation of XRCC4 during the DNA-binding reaction, as full-length XRCC4 was found in the unbound fraction (Figure 1C). These data suggest that adenoviral infection and expression of adenovirus early proteins directly or indirectly prevented DNA recognition by XRCC4.
In control experiments, we examined the effect of adenovirus infection on DNA binding by other NHEJ factors. DNA-PKcs was present in the input and DNA-bound fractions of mock- and Ad5-infected cells and was not detected in unbound fraction (Figure 1C). Similar results were obtained for the Ku heterodimer, which was found in the input and DNA-bound fractions of both mock- and Ad5-infected cells, but was not observed in unbound fractions (data not shown). These control experiments show that the dsDNA cellulose used in these experiments was available for recognition by DNA-binding proteins. In particular, the exposed DNA ends, which represent the physiological substrate for NHEJ factors, were accessible. These data indicate that inhibition of DNA binding was unique to XRCC4 and not caused by a general mechanism that affected all DNA-binding proteins.
Targeted degradation of ligase IV requires expression of viral proteins E1B 55K and E4 34k. To investigate the role of adenoviral protein E4 34k in the observed loss of DNA binding by XRCC4 in adenovirus-infected cells, we examined XRCC4 DNA recognition in extracts prepared from HeLa cells that had been infected with various early region 4 (E4)-deletion mutants. HeLa cells were infected with E4-deletion mutants shown in Supplementary Figure S1A, extracts were prepared and western blot analysis was used to assess the expression of E1B 55k and E4 34k (Supplementary Figure S1B). Native DNA cellulose was then used to capture DNA-binding proteins and western blot analysis to detect XRCC4 in the input extract, the DNA-bound fraction and in the extract following removal of DNA-binding proteins (unbound). Figure 1D shows that in extracts prepared from HeLa cells infected with E4-deletion mutants that did not express E4 34k, XRCC4 was detected in the DNA-bound fraction in amounts that were comparable to that observed with mock-infected cells. In extracts prepared from HeLa cells infected with wild-type Ad5 or with E4-deletion mutant dl1013, which retains E4 34k expression, XRCC4 was not detectable in the DNA-bound fraction but was found in the unbound fraction. Western blot analysis showed that loss of XRCC4 DNA-binding activity was coincident with expression of adenoviral proteins E1B 55k and E4 34k (Supplementary Figure S1B) and degradation of ligase IV (Supplementary Figure S1C). Taken together, data presented in Figure 1 and Supplementary Figure S1 show that inhibition of DNA recognition by XRCC4, like degradation of ligase IV, is dependent upon adenoviral infection and expression of both the E1B 55k and E4 34k proteins.
XRCC4 remains in large complexes in adenovirus-infected cells
Loss of DNA binding by XRCC4 in adenovirus-infected cells may reflect gross alteration of XRCC4 (i.e. changes in protein structure or protein–protein interaction) incurred during poly-ubiquinylation and degradation of the ligase IV that is bound to XRCC4. In Ad5-infected cells, the relocalization of Mre11 to cytoplasmic aggresomes is thought to interfere with the normal function of the Mre11-Rad50-Nbs1 (MRN) complex (26). XRCC4 is a nuclear factor and using in situ immunofluorescence we found that, unlike the MRN complex, XRCC4 remained nuclear in Ad5-infected cells (data not shown).
Having determined that XRCC4 was not re-localized in Ad5-infected cells, we went on to determine if interactions with other NHEJ factors were affected by adenoviral infection. First, we carried out gel filtration analysis on extracts prepared from mock- or Ad5-infected HeLa cells, then used western blot analysis to identify XRCC4-containing fractions to determine the average size of XRCC4-containing complexes. The average size of XRCC4-containing complexes was reduced in extracts prepared from Ad5-infected cells when compared with extracts prepared from mock-infected cells (Supplementary Figure 2A). While the loss of ligase IV might account for the observed decrease in the average size of XRCC4-containing complexes, these complexes are still relatively large (∼200–350 kDa), suggesting the continued participation of XRCC4 in multi-protein complexes. In addition to recognition of ligase IV that participates in a large protein complex, anti-ligase IV antibodies directed against the C-terminal domain of ligase IV also recognized a 100 kDa species that migrated near 100 kDa on gel filtration. While antibody recognition makes it reasonable to conclude that this species that was not degraded during Ad5 infection is an interesting sub-population of ligase IV, we cannot rule out the unlikely possibility that this species may be DNA ligase I or III. Our findings show that during adenoviral infection, XRCC4 protein levels, localization and participation in multi-protein complexes remained unchanged, which suggest that the XRCC4 protein was not grossly altered during adenoviral infection.
XRCC4 is known to interact directly with the DNA-PKcs, and it is thought that this interaction is important for NHEJ (35). We used co-IP assays to determine if this protein–protein interaction was maintained or lost in Ad5-infected cells. Immunoprecipitation (IP) of XRCC4 was carried out using extracts prepared from mock- or Ad5-infected HeLa cells and western blot analysis was used to determine if DNA-PKcs was co-immunoprecipitated with XRCC4. XRCC4 is associated with DNA-PKcs in mock-infected cells, and this interaction was unaffected by adenoviral infection (Supplementary Figure 2B). Our findings show that XRCC4 levels and localization remained unchanged and the ability to interact with DNA-PKcs was not altered, which suggest that the XRCC4 protein was not significantly altered during adenoviral infection.
E1B 55k and E4 34k are sufficient for inhibition of NHEJ and loss of XRCC4 DNA binding
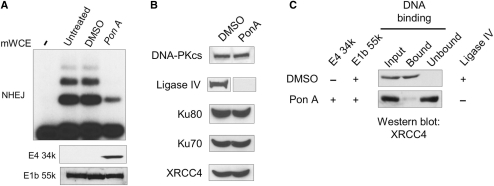
Data presented thus far show that E1B 55k and E4 34k expression are required to inhibit DNA recognition by XRCC4 and NHEJ in vitro. To determine if E1B 55k and E4 34k are sufficient to inhibit DNA binding by XRCC4, we examined the effect of expression of E1B 55k and E4 34k in the 2V6.11 cell line that had been engineered to carry E4 34k under Ponesterone A (Pon A) inducible control (27). 2V6.11 cells were derived from 293 cells, which constitutively express proteins from adenovirus early region 1 (E1), including E1B 55k, E1a 289R and 243R and E1B 19K (36,37). We found that, as previously described (27), the E1B 55k protein can be detected in extracts prepared from 2V6.11 cells and that Pon A treatment resulted in expression of E4 34k (Figure 2A). As shown in Figure 2A, extracts prepared from 2V6.11 cells that had been induced to express E4 34k showed reduced NHEJ activity when compared with extracts prepared from untreated or mock-induced (DMSO) controls. In control experiments, Pon A treatment of the parental 293 cells had no effect on in vitro NHEJ (data not shown). Western blot analysis showed that in 2V6.11 cells where E4 34k expression was induced, ligase IV was undetectable while levels of the other NHEJ factors were unchanged (Figure 2B). This is consistent with previous observations that of adenoviral gene products, E1B 55k and E4 34k are sufficient to ablate ligase IV, which presumably explains the reduction of NHEJ in 2V6.11 extracts in vitro.
Figure 2.
Expression of E1B 55k and E4 34k are sufficient for inhibition of NHEJ and loss of XRCC4 DNA binding. mWCEs were prepared from 2V6.11 cells that were untreated, DMSO or Pon A (2 µg/ml) treated for 48 h. (A) Expression of E1B 55k and E4 34k resulted in inhibition of in vitro NHEJ. (top) In vitro NHEJ assays were carried out using 40 µg of mWCE. ‘-’ denotes the lane where no mWCE was added during NHEJ reaction. mWCE of 50 µg was resolved on SDS–PAGE, subject to western transfer and probed for the presence of E4 34k or E1B 55k as noted. (B) Levels of NHEJ factors were assayed in 50 µg of mWCE prepared from DMSO- or Pon A-treated 2V6.11 cells. Extracts were resolved on SDS–PAGE, western transferred and proteins were detected as indicated. (C) Loss of XRCC4 DNA binding in extracts prepared from cells expressing E1B 55k and E4 34k. mWCE of 50 µg prepared from DMSO- or Pon A-treated 2V6.11 cells was subject to dsDNA–cellulose fractionation. The input extract (Input), DNA-bound species (Bound) and the extract following removal of DNA-binding proteins (Unbound) were resolved by SDS–PAGE, subject to western transfer and probed for the presence of XRCC4. Expression of E4 34k, E1B 55k and Ligase IV is indicated.
To determine if DNA binding by XRCC4 was inhibited by the expression of E1B 55k and E4 34k, we compared XRCC4 DNA binding in extracts prepared from Pon A-induced and mock-induced 2V6.11 cells. As shown in Figure 2C, while XRCC4 was clearly observed in the DNA-bound fraction of extracts prepared from mock-induced 2V6.11 cells, with extracts prepared from Pon A-induced 2V6.11 cells that expressed E4 34k, retention of XRCC4 on dsDNA–cellulose resin DNA binding by XRCC4 was greatly reduced. Thus, expression of E1B 55k and E4 34k and loss of ligase IV in 2V6.11 cells is coincident with loss of DNA binding by XRCC4. We note that these data do not rule out the possibility that another E1 protein participates in inhibiting DNA binding by XRCC4. Although ligase IV was undetectable in extracts prepared from Pon A-induced cells, low levels of in vitro NHEJ activity and XRCC4 DNA binding were detected. We attribute these observations to remaining ligase IV at levels below the limit of western blot detection.
Loss of ligase IV, not viral protein expression, correlated with loss of DNA binding by XRCC4
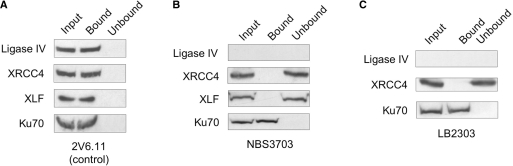
Thus far, our findings show that loss of ligase IV, through poly-ubiquitinylation and proteasome-mediated degradation, and the expression of viral proteins, in particular E1B 55k and E4 34k, correlate with the observed loss of DNA binding by XRCC4. To separate the effect(s) of loss of ligase IV from those of viral protein expression, we assayed for DNA binding by XRCC4 in extracts prepared from ligase IV-mutant human cell lines. In control DNA-binding experiments with extracts prepared from untreated 2V6.11 cells, both wild-type ligase IV and XRCC4 were found in the DNA-bound fraction (Figure 3A). Because this series of experiments focused on the substrate-recognition properties of the ligase IV/XRCC4/XLF complex, we also probed for the presence of XLF in the DNA-bound fraction. As shown in Figure 3A, XLF, like XRCC4 and wild-type ligase IV, was detected in the input and DNA-bound fractions of extracts prepared from untreated 2V6.11 cells. These data show that all three members of the ligase IV/XRCC4/XLF complex are retained on native DNA cellulose, which may reflect the intrinsic DNA-binding activities of the individual factors, or the DNA-binding activity of the complex as a whole.
Figure 3.
DNA binding by XRCC4 requires ligase IV. DNA binding by NHEJ factors in mWCEs prepared from untreated (control) 2V6.11 (A), NBS3703 (B) and LB2303 cells (C). 50 µg of mWCE was subject to dsDNA–cellulose fractionation. The input extract (Input), DNA-bound species (Bound) and the extract following removal of DNA-binding proteins (Unbound) were resolved by SDS–PAGE, subject to western transfer and individual factors were detected as indicated.
To investigate the role of ligase IV in DNA recognition by XRCC4 and XLF, we used cell lines NBS3703 and LB2304 that bear LIG4 nonsense mutations and consequently do not contain enough ligase IV to be observable by western blot (38,39). In NBS3703 cell extracts, we found that XRCC4 and XLF were detectable in the input extract and in the extracts following depletion of DNA-binding proteins (unbound fraction), but were not found in the DNA-bound fraction (Figure 3). Control experiments that tested for DNA binding by Ku (Figure 3) and DNA-PKcs (data not shown) demonstrated that the dsDNA used in these experiments was accessible and suitable for recognition by DNA-binding proteins. Similarly, we observed that in extracts prepared from LB2304 cells, XRCC4 was present but was not retained by the DNA cellulose, while Ku was detected in the DNA-bound fraction (Figure 3). To rule out the possibility that factors that inhibit DNA binding by the ligase IV/XRCC4/XLF complex may be present in extracts prepared from ligase IV mutant cells, we carried out mixing experiments in which extracts prepared from control untreated 2V6.11 or 293 cells were combined with native or heat-treated extracts prepared from LB2304 or NBS3703 cells, then assayed for DNA binding by XRCC4. Under these conditions, we observed no reduction in retention of XRCC4 or ligase IV, presumably contributed by the 2V6.11 extract, by dsDNA cellulose resin (data not shown).
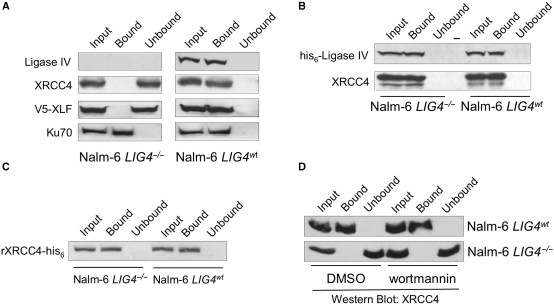
Data presented in Figure 3 show that in extracts prepared from cell lines that contain ligase IV nonsense mutations, XRCC4 and XLF were not retained by dsDNA cellulose, which indicates that the intrinsic DNA-binding activities associated with these factors have been inhibited. These findings rule out the participation of adenoviral proteins in inhibition of XRCC4 DNA binding and put forward the hypothesis that loss of ligase IV results in the observed loss of DNA binding by the remaining components of the ligase IV/XRCC4/XLF complex. To test this hypothesis, we examined retention of XRCC4 and XLF on native DNA cellulose in extracts prepared from Nalm-6 cells in which both copies of the LIG4 gene had been knocked out [Nalm-6 LIG4−/−, (40)]. Control dsDNA–cellulose fractionation experiments carried out with wild-type Nalm-6 cells (Figure 4A, right) showed that ligase IV, XRCC4 and Ku were detected in the input extract and DNA-bound fractions, but were not observed in extracts following depletion of DNA-binding proteins (Figure 4A, right). To assess DNA binding by XLF, V5-tagged XLF was transiently expressed in wild-type Nalm-6 cells. Western blot analysis using anti-V5 antibodies showed that V5-XLF was detected in the input extract and the DNA-bound fraction, but not in the unbound fraction (Figure 4A). In contrast, we found that in extracts prepared from LIG4−/− Nalm-6 cells (Figure 4A, left), the ligase IV protein was not detectable and while both XRCC4 and V5-XLF were detected in the input extract and in the extract following depletion of DNA-binding proteins, neither were detected in the DNA-bound fraction (Figure 4A). DNA accessibility was confirmed by the presence of Ku in the DNA-bound fraction, which suggests that occlusion of the DNA by other DNA-binding factors is not responsible for the lack of XRCC4 and V5-XLF retention by dsDNA cellulose.
Figure 4.
Loss of ligase IV protein correlates with inhibition of DNA binding by XRCC4. (A) DNA binding by NHEJ factors in extracts prepared from Nalm-6 cells. mWCE (50 µg) prepared from ligase IV-deficient (Nalm-6 LIG4−/−) and wild-type (Nalm-6 LIG4wt) cells was subject to dsDNA–cellulose fractionation. The input extract (Input), DNA-bound species (Bound) and the extract following removal of DNA-binding proteins (Unbound) were resolved by SDS–PAGE, subject to western transfer and individual factors were detected as indicated. For detection of XLF DNA binding: cells were transiently transfected with V5-tagged XLF, extracts were prepared 60 h after transfection, DNA-binding proteins were isolated and V5-XLF was detected using anti-V5 antibodies. (B) Expression of wild-type ligase IV restored DNA binding by XRCC4 in ligase IV-deficient cells. pcDNA3.1 expressing his-tagged wild-type ligase IV (his6-ligase IV) was transiently transfected into Nalm-6 LIG4−/− and Nalm-6 LIG4wt cells. Extracts were prepared 60 h after transfection and 50 µg of extract were fractionated on dsDNA–cellulose. XRCC4 was detected as described in (A) and ectopically expressed his6-Ligase IV was detected using anti-his6 antibodies. ‘-’ denotes empty lane. (C) Recombinant his6-tagged XRCC4 binds DNA in the presence of Nalm-6 cell extracts. Recombinant XRCC4-his6 was expressed and purified as described in Materials and methods section, then added to 100 µg of extract prepared from Nalm-6 LIG4−/− or Nalm-6 LIG4wt cells to a final concentration of 1.4 pM. Samples were incubated in the absence of DNA for 1 h, after which the samples were subject to dsDNA–cellulose fractionation. Recombinant XRCC4-his6 was detected using anti-his6 antibodies. (D) Nalm-6 cells (LIG4−/− and LIG4wt) were cultured in the presence of wortmannin (20 µM) or the vehicle (DMSO) for 24 h. mWCEs were prepared, samples were subject to dsDNA–cellulose fractionation and XRCC4 was detected as described in (A).
Thus far, our findings are consistent with the hypothesis that, in human cells, the ligase IV polypeptide is required for DNA binding by XRCC4 and XLF. To directly test this hypothesis, we used electroporation for high-efficiency transient expression of his-tagged wild-type ligase IV in wild-type and LIG4−/− Nalm-6 cells. As shown in Figure 4B, the his6-ligase IV cDNA was expressed in wild-type Nalm-6 cells and expression of this cDNA did not alter DNA binding by XRCC4. Importantly, we found that when his6-ligase IV was expressed in LIG4−/− Nalm-6 cells, XRCC4 was observed in the DNA-bound fraction (Figure 4B).
Ligase IV has been shown to interact with Ku, which is thought to recruit the ligase IV/XRCC4/XLF complex to exposed DNA ends (41–43). In the presence of ligase IV, binding of the ligase IV/XRCC4/XLF complex to DNA is facilitated by ligase IV/Ku interactions. In the absence of ligase IV, binding of abundant Ku to the dsDNA cellulose might prevent binding and recovery of XRCC4. To test this possibility, purified, recombinant XRCC4-his6 was added to extracts prepared from wild-type or LIG4−/− Nalm-6 cells at a final concentration of 1.4 pM, to match the concentration of endogenous XRCC4 in the extracts. The recombinant XRCC4-his6 was pre-incubated with the Nalm-6 extracts in the absence of dsDNA cellulose for an hour, after which DNA–cellulose fractionation was carried out to isolate DNA-binding proteins. Anti-his6 antibodies were used to detect the recombinant XRCC4-his6, apart from the endogenous XRCC4. As shown in Figure 4C, the recombinant XRCC4-his6 bound the DNA in the presence of both wild-type and LIG4−/− Nalm-6 extracts. These data show that the dsDNA cellulose used in these experiments is accessible to binding by XRCC4 and that lack of DNA binding by XRCC4 in LIG4−/− Nalm-6 extracts was not caused by occlusion of the DNA by Ku, or any other factor(s). Additionally, these data indicate that soluble factors are not likely to cause the observed inhibition of XRCC4 DNA recognition and implicate posttranslational modification and formation of stable protein–protein complexes as likely mechanisms for regulation of XRCC4 DNA binding.
Phosphorylation of recombinant XRCC4 by DNA-PK in vitro has been shown to inhibit XRCC4 DNA binding (4). DNA-PKcs phosphorylation sites were subsequently mapped to the C-terminal 100 amino acids of XRCC4 (44). Because the ligase IV binding site of XRCC4 lies within the C-terminal coiled-coil domain of XRCC4, it is plausible that these DNA-PK phosphorylation sites are made accessible when ligase IV is absent and that phosphorylation of XRCC4 results in inhibition of DNA binding by XRCC4. To test this hypothesis, we captured phosphopeptides from LIG4−/− Nalm-6 cell extracts and used western blot analysis to confirm that XRCC4 is a phosphoprotein in the absence of ligase IV (data not shown). We then treated wild-type and LIG4−/− Nalm-6 cells with wortmannin (20 μM) or the vehicle (DMSO) for 24 h, after which time extracts were prepared and DNA–cellulose fractionation was carried out to isolate DNA-binding proteins. As shown in Figure 4D, wortmannin treatment had no effect on the DNA-binding activity of XRCC4 in extracts prepared from wild-type Nalm-6 cells. XRCC4 in extracts prepared from LIG4−/− Nalm-6 cells remained unable to associate with DNA, despite wortmannin treatment. These data indicate that while XRCC4 is phosphorylated in the absence of ligase IV, phosphorylation dependent upon PI3K-related PK activity is not required for ablation of XRCC4 DNA binding activity.
Formation of the ligase-adenylate is not required for DNA binding by the ligase IV/XRCC4/XLF complex
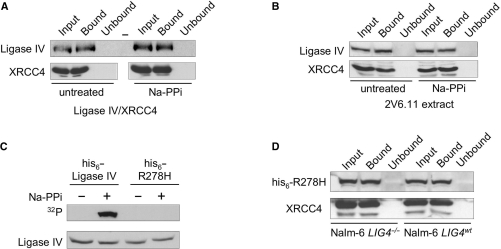
During ligation, an ATP-dependent ligase first reacts with ATP to form a ligase-adenylate. Some ligases, such as the Chlorella virus ligase, require formation of the ligase-adenylate for DNA recognition (45,46). Other ligases are capable of DNA recognition regardless of adenylation status. Recombinant ligase IV/XRCC4 exhibits a weak DNA-binding activity on short DNA substrates and it has been reported that formation of the ligase IV-adenylate is necessary for Ku-mediated binding of recombinant ligase IV/XRCC4 to short DNA substrates (47,48). Along these lines, we found that XRCC4 in cell line 180BRM, which expresses a mutant ligase IV with reduced ability to form the ligase IV-adenylate, fails to bind DNA (data not shown). These data suggest that formation of the ligase IV-adenylate may be required for DNA binding by the ligase IV/XRCC4/XCLF complex.
To directly assess the role of ligase IV-adenylate formation on binding to DNA cellulose, we chemically removed the adenosine from the ligase IV-adenylate, then assayed for DNA binding by the deadenylated ligase IV. To do this, we treated purified recombinant ligase IV/XRCC4 complex with inorganic pyrophosphate to disrupt preformed ligase-adenylates. Adenylation of untreated ligase IV/XRCC4 complex with 32PαATP confirmed that, as previously demonstrated (2,48), recombinant ligase IV was already in the ligase-adenylate form and could not be adenylated (data not shown). Adenylation of recombinant ligase IV following pyrophosphate treatment confirmed that ligase IV could form a 32P-labeled ligase IV-adenylate (data not shown). To determine what fraction of ligase IV-adenylate is disrupted by pyrophosphate treatment, we treated the 32P-labeled ligase IV-adenylate with pyrophosphate and were unable to detect 32P-labeled ligase IV-adenylate remaining in the sample (data not shown). These data indicate that treatment of ligase IV with pyrophosphate can disrupt the preformed ligase IV-adenylate. Following ligase-adenylate disruption, the pyrophosphate was removed from the reaction and native DNA cellulose was used to isolate DNA-binding proteins. We found that DNA binding by purified recombinant ligase IV/XRCC4 was unaffected by pyrophosphate treatment, which indicated that the formation of the ligase-adenylate was not required for DNA binding by this complex (Figure 5A). In similar experiments, extracts prepared from untreated 2V6.11 cells were treated with pyrophosphate to disrupt preformed ligase-adenylates. As observed for purified recombinant proteins, ligase IV and XRCC4 were detected in the input extracts and in the DNA-bound fractions, but were not detected in extracts following removal of DNA-binding proteins (Figure 5B). Thus, in the context of these treated human cell extracts, ligase-adenylates were not required for DNA binding.
Figure 5.
DNA binding by ligase IV/XRCC4 does not require ligase IV adenylation. (A) Disruption of the ligase IV-adenylate did not affect DNA binding by recombinant ligase IV/XRCC4 complex. Purified, recombinant ligase IV/XRCC4 complex (10 µg) was treated with 5 mM sodium pyrophosphate (NaPPi) for 15 min to disrupt the ligase–adenylate complex. Treatment was followed by removal of pyrophosphate and both NaPPi-treated and untreated samples were subject to dsDNA–cellulose fractionation. Input extract (Input), DNA-bound species (Bound) and the extract following removal of DNA-binding proteins (Unbound) were resolved by SDS–PAGE, western transferred and probed for the presence of Ligase IV and XRCC4 as indicated. Bottom panel was assembled from lanes from a single western blot. ‘-’ denotes empty lane. (B) Disruption of the ligase IV-adenylate did not affect DNA binding by ligase IV/XRCC4 in human cell extracts. An extract prepared from untreated 2V6.11 cells (50 µg) was treated with 5 mM NaPPi for 15 min, after which the sample was treated as described in (A). (C) Ligase IVR278H does not form the ligase IV-adenylate. pcDNA3.1 expressing wild-type or R278H mutant (his6-R278H) ligase IV was transiently transfected into Nalm-6 LIG4−/− cells. mWCEs were prepared 60 h after transfection, treated with 5 mM NaPPi for 15 min, after which pyrophosphate was removed. Ectopically expressed Ligase IV was co-immunoprecipitated from extracts using anti-XRCC4 antibodies and in vitro adenylated with 32PαATP as previously described (29). 32P-labeled ligase IV adenylate was detected by autoradiography (32P) and ligase IV was detected by western blot. (D) Expression of ligase IVR278H restored DNA binding by XRCC4 in ligase IV-deficient cells. pcDNA3.1 expressing his-tagged ligase IVR278H (his6-R278H) was transiently transfected into Nalm-6 LIG4−/− and Nalm-6 LIG4wt cells, extracts were prepared 60 h after transfection and 50 µg of mWCE were subject to dsDNA–cellulose fractionation. XRCC4 was detected as described in (A) and ectopically his6-ligase IVR278H was detected using anti-his6 antibodies.
In parallel experiments, we transiently expressed a mutant form of ligase IV (ligase IVR278H), which is severely impaired for formation of the ligase IV-adenylate (29), in Nalm-6 cells and assessed DNA binding by XRCC4. To confirm the adenylation defect of his6-ligase IVR278H, extracts prepared from LIG4−/− Nalm-6 cells expressing his6-ligase IVR278H were treated with pyrophosphate, or left untreated, and ligase IV immunoprecipitates were subject to adenylation with 32PαATP (Figure 5C). We found that, as previously described (29), neither untreated nor pyrophosphate-treated his6-ligase IVR278H was capable of forming a his6-ligase IVR278H-adenylate. In contrast, control reactions with extracts prepared from LIG4−/− Nalm-6 cells expressing wild-type his6-ligase IV showed that his6-ligase IV could be adenylated, but only after pyrophosphate treatment to disrupt the preformed his6-ligase IV-adenylate (Figure 5C). Detection of ligase IV showed that his6-ligase IV and his6-ligase IVR278H were expressed at comparable levels and that treatment with NaPPi and subsequent removal of NaPPi did not affect the amount of ligase IV present in the sample. DNA–cellulose fractionation of extracts prepared from wild-type NALM-6 cells expressing his6-ligase IVR278H showed XRCC4 and his6-ligase IVR278H in the input and DNA-bound fractions (Figure 5D, right). When the his6-ligase IVR278H protein was expressed in LIG4−/− Nalm-6 cells, XRCC4 and his6-ligase IVR278H were detected in the DNA-bound fraction (Figure 5D, left). These data show that restoration of the ligase IV polypeptide was sufficient to restore DNA binding by XRCC4 and that formation of the ligase-adenylate was not required for binding of long DNA substrates. Ligase IV in 180BRM cells has been describe as unstable (29), and we attribute our original observation that XRCC4 fails to recognize DNA in extracts prepared from 180BRM cells to the instability of ligase IV in these cells, and not to the inability of ligase IV in these cells to form the ligase IV-adenylate.
The XRCC4/XLF complex binds DNA
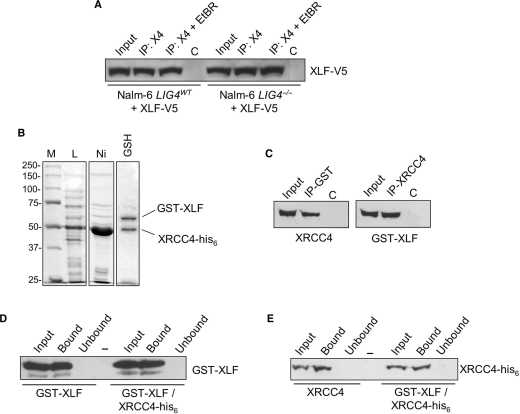
Investigation of the mode of interaction between XRCC4 and XLF has revealed that XLF binds to the N-terminal globular-head domain of XRCC4, which is also the DNA-binding domain (3,4,16,17). This observation suggests that binding of XRCC4 by XLF may inhibit DNA recognition by XRCC4. Consistent with this possibility, XLF co-immunoprecipitates (co-IP) with XRCC4 in extracts prepared from wild-type and LIG4−/− Nalm-6 cells (Figure 6A). V5-tagged XLF was transiently expressed in wild-type and LIG4−/− Nalm-6 cells and XRCC4 were immunoprecipitated (IP) to capture XRCC4-containing complexes. To rule out the possibility of DNA-mediated interactions, the XRCC4 IP was also carried out in the presence of ethidium bromide (EtBR), which can disrupt nonspecific protein–DNA interactions. As shown in Figure 6A, western blot analysis using anti-V5 antibodies shows that XLF is present in the XRCC4 co-IP, even in the presence of EtBR. These data show that in the absence of ligase IV, XLF binds XRCC4, and may interfere with XRCC4 DNA binding.
Figure 6.
DNA binding by the XRCC4/XLF complex. (A) Co-IP of XRCC4 and XLF from Nalm-6 cells. Cells were transiently transfected with V5-tagged XLF. Extracts were prepared 60 h after transfection and IP with anti-XRCC4 antibodies was used to collect XRCC4-containing complexes in the presence and absence of EtBr (50 µg/ml) to disrupt protein–DNA interactions (IP: X4 and IP: X4 + EtBR). Samples were resolved on SDS–PAGE, subject to western transfer and ectopically expressed V5-XLF was detected using anti-V5 antibodies. (B) Expression and purification of XRCC4-his6/GST-XLF complex. Escherichia coli strain Rosetta 2 was co-transformed with pET28a(+)XRCC4-his6 (KanR) and pEX-4T-XLF (AmpR) and protein expression was induced with IPTG for 4.5 h at 37°C. Crude lysate (L) was subject to tandem Ni-NTA (Ni), glutathione sepharose (GSH) affinity chromatography. Peak elution fractions are shown. (C) Co-IP of XRCC4-his6 and GST-XLF. Antibodies directed against XRCC4 (right) or GST (left) were used to IP XRCC4 and GST-XLF, respectively. The input fraction (Input), immunoprecipitated complexes (IP) and control IPs (C) of 10% were resolved on SDS–PAGE, subject to western transfer and XRCC4 and GST-XLF were detected as indicated. (D and E) DNA binding by the XRCC4-his6/GST-XLF complex. dsDNA–cellulose fractionation was carried out using glutathione sepharose eluate (GST-XLF/XRCC4-his6), GST-XLF alone and XRCC4-his6 alone. The input fraction (Input), DNA-bound species (Bound) and the sample following removal of DNA-binding proteins (Unbound) were resolved on SDS–PAGE, western transferred and XRCC4 and GST-XLF were detected as indicated. ‘-’ denotes empty lane.
To directly test the hypothesis that formation of an XRCC4/XLF complex inhibits DNA binding by XRCC4 and XLF, we coexpressed C-terminally his-tagged XRCC4 (XRCC4-his6) and N-terminally GST-tagged XLF (GST-XLF) in E. coli, used Ni-NTA and glutathione sepharose affinity chromatography in tandem to isolate complexes that contain XRCC4-his6 and GST-XLF, then assayed for DNA binding by these hetero-oligomeric complexes. Figure 6B shows the crude lysate and the Ni-NTA peak elution fraction, which was highly enriched for XRCC4-his6. Subsequent glutathione sepharose affinity chromatography captured those complexes also containing GST-XLF, while unbound XRCC4-his6 flowed through the column. The peak glutathione sepharose fraction shows two bands of equal intensity with molecular weights corresponding to those of XRCC4-his6 and GST-XLF. Co-IP and western blot analysis were used to confirm purification of an XRCC4-his6/GST-XLF complex (Figure 6B). Using native DNA cellulose, we found that both XRCC4-his6 and GST-XLF purified in this fashion were able to bind DNA (Figure 6D and E), which indicates that formation of an XRCC4/XLF complex does not inhibit the intrinsic DNA-binding activities of XRCC4 and XLF.
DISCUSSION
The details of cellular processes may be explored through the study of virus–host interactions. By using human adenovirus as a tool to study mammalian NHEJ, we have uncovered a novel aspect of NHEJ: in human cells, DNA recognition by XRCC4 and XLF requires the ligase IV polypeptide.
Expression of adenoviral E1B 55k and E4 34k proteins is required for proteasome-meditated degradation of ligase IV (24), which is the most likely cause of the observed inhibition of NHEJ in adenovirus-infected cells (19,21–25). We have observed that expression of E1B 55k and E4 34k also results in inhibition of DNA binding by XRCC4. Correlation of the loss of DNA binding by XRCC4 and XLF and degradation of ligase IV led us to take a closer look at the DNA-binding properties of XRCC4 in human cells that lacked ligase IV for other reasons. Using ligase IV-deficient mutants, we found that the lack of ligase IV correlated with loss of XRCC4 binding in uninfected cells, despite the documented intrinsic DNA-binding activity of XRCC4 (4). We then examined the behavior of another DNA-binding protein that interacts with XRCC4, the XLF protein (6,49). In human cells expressing defective ligase IV, we found that XLF, like XRCC4, failed to bind DNA. These data imply that substrate recognition by XRCC4 and XLF in vivo requires the participation of ligase IV. Furthermore, these data suggest the exciting hypothesis that the intrinsic DNA-binding activity of XRCC4, and possibly that of XLF, may be subject to regulation and is, in fact, down-regulated in cells lacking ligase IV.
Inhibition of XRCC4 DNA-binding activity was not due to degradation of XRCC4, as the absolute amount of XRCC4 in ligase IV mutant cell lines or in Ad5-infected cells was comparable to the wild-type and uninfected cells. Inhibition of XRCC4 DNA binding was not caused by a general mechanism affecting all DNA-binding proteins (such as inaccessibility of the DNA used in pull-down experiments or competition by DNA of host or viral genome origin in the extracts) because DNA binding by DNA-PKcs and Ku were unchanged by Ad5-infection, expression of a defective ligase IV or ligase IV deficiency.
Down-regulation of DNA binding by the XRCC4/XLF complex when ligase IV is absent would prevent assembly of unproductive, potentially mutagenic, complexes at exposed DNA termini. Our observation that XRCC4 and XLF, two intrinsic DNA-binding proteins, paradoxically fail to associate with DNA when ligase IV is absent supports this hypothesis. Related observations show that XRCC4 and XLF fail to associate with damaged chromatin in the absence of ligase IV (50,51). We found that when XRCC4 and XLF were coexpressed in E. coli to form an XRCC4/XLF complex, both proteins retained DNA-binding activity, which indicates that XRCC4 DNA-binding activity is not regulated through association with XLF.
Regulation of many biochemical activities is achieved by protein phosphorylation, and it has been shown that in vitro phosphorylation of recombinant XRCC4 by DNA-PK results in the loss of XRCC4 DNA binding (4). Subsequent mapping and genetic analysis of this phosphorylation event revealed that phosphorylation of XRCC4 by DNA-PK at the sites examined was dispensable for NHEJ in vivo (44). While these data argue that phosphorylation of XRCC4 by DNA-PK at these specific sites is of limited importance to NHEJ in vivo, they do not exclude the possibility that other sites of DNA-PK phosphorylation may be of biological importance. Using WCEs, we have observed that phosphorylation of XRCC4 by DNA-PKcs is coincident with accumulation of end-joining products in vitro (data not shown). Drouet et al. (51) have shown that in cells expressing ligase IV, recruitment of XRCC4 to damaged chromatin requires DNA-PK and is accompanied by DNA-PK-dependent phosphorylation of XRCC4. Consistent with this report, we failed to observe DNA binding by XRCC4 or ligase IV in extracts prepared from DNA-PKcs-deficient cells (data not shown). We also found that treatment of wild-type of LIG4−/− Nalm-6 cells with wortmannin to inhibit DNA-PKcs activity had no effect on DNA binding by XRCC4. Taken together, these observations indicate that the DNA-PKcs polypeptide plays an important role in assembly of NHEJ factors at exposed DNA termini, but the role of XRCC4 phosphorylation by DNA-PKcs and the molecular mechanism of XRCC4 DNA-binding regulation remain unresolved.
Whereas the possibility that DNA-PK may regulate substrate recognition by XRCC4 is of considerable interest, it is important to note that XRCC4 is also a substrate for the ubiquitous casein kinase II (CK2), which is thought to regulate interactions between the ligase IV/XRCC4-XLF complex and human polynucleotide kinase (PNK) (33,52). The regulation of DNA recognition by the ligase IV/XRCC4/XLF complex may require the concerted efforts of several PKs is a tantalizing possibility. Through western blot analysis, we have observed that XRCC4 from wild-type and LIG4−/− Nalm-6 cells have identical mobility on SDS–PAGE. Side-by-side comparison of XRCC4 in the DNA-bound and unbound fractions from wild-type and LIG4−/− Nalm-6 cells, respectively, also showed no detectable difference in XRCC4 mobility (data not shown). These data are consistent with 2D western blot analysis that showed no difference in the abundance or apparent charge of XRCC4 in extracts prepared from wild-type and ligase IV-deficient cells (data not shown). These observations suggest that XRCC4 carries the same number of phosphates in the presence and absence of ligase IV, but provides no information regarding the specific residues that are modified.
It is interesting to note that while all components of the ligase IV/XRCC4/XLF complex are DNA-binding proteins, the relative contributions of each factor to the process of substrate recognition have not been clearly defined. As previously described, XRCC4 N-terminal deletion mutants that fail to bind DNA remain capable of associating with ligase IV and stimulating ligation in vitro, suggesting that XRCC4 DNA-binding activity is, in fact, dispensable for ligase complex function (3,4,17). While these studies were limited to the ligase IV/XRCC4 complex, it is plausible that XRCC4 does not contribute significantly to substrate recognition by the ligase IV/XRCC4/XLF complex in mammalian cells and that the majority of protein–DNA contacts are formed by ligase IV. Our data support this hypothesis, as XRCC4/XLF complexes expressed in mammalian cells that lack ligase IV do not bind DNA. Further investigation into the regulation of DNA binding by XRCC4, and XLF, is expected to yield new insights into mechanisms that safeguard genomic stability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (GM070639-1 to L.A.H.), (5R01CA082127 to G.K.); Johns Hopkins University Bloomberg School of Public Health Faculty Research Initiatives Fund (to L.A.H. and G.K.); National Cell Culture Center (Minneapolis, MN), National Center for Research Resources, National Institutes of Health (U42 RR05991). Funding for open access charge: National Institutes of Health (GM070639-1).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Amy Baker, Joyce Cheung, Timra Gilson, B.T. Rantipole and Brenda Salerno for many thoughtful discussions. The authors thank Patrick Concannon (UVA) for the generous gift of LB2304 and NBS3703 cells and Steve Jackson for his generous gift of anti-XLF antibodies and pGEX-4T-XLF.
REFERENCES
- 1.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein Xrcc4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 2.Grawunder U, Wilm M, Wu XT, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with Xrcc4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 3.Grawunder U, Zimmer D, Kulesza P, Lieber MR. Requirement for an interaction of Xrcc4 with DNA ligase IV for wild-type V(D)J recombination and DNA double-strand break repair in vivo. J. Biol. Chem. 1998;273:24708–24714. doi: 10.1074/jbc.273.38.24708. [DOI] [PubMed] [Google Scholar]
- 4.Modesti M, Hesse JE, Gellert M. DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J. 1999;18:2008–2018. doi: 10.1093/emboj/18.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modesti M, Junop MS, Ghirlando R, van de Rakt M, Gellert M, Yang W, Kanaar R. Tetramerization and DNA ligase IV interaction of the DNA double-strand break repair protein XRCC4 are mutually exclusive. J. Mol. Biol. 2003;334:215–228. doi: 10.1016/j.jmb.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 6.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 7.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Chirgadze DY, Bolanos-Garcia VM, Sibanda BL, Davies OR, Ahnesorg P, Jackson SP, Blundell TL. Crystal structure of human XLF/cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ. EMBO J. 2007;27:290–300. doi: 10.1038/sj.emboj.7601942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu H, Pannicke U, Schwarz K, Lieber MR. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J. Biol. Chem. 2007;282:11155–11162. doi: 10.1074/jbc.M609904200. [DOI] [PubMed] [Google Scholar]
- 10.Smith GCM, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 11.Hanakahi LA, Bartlet-Jones M, Chappell C, Pappin D, West SC. Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell. 2000;102:721–729. doi: 10.1016/s0092-8674(00)00061-1. [DOI] [PubMed] [Google Scholar]
- 12.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 13.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, et al. Leaky scid phenotype associated with defective v(d)j coding end processing in artemis-deficient mice. Mol. Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 14.Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. Crystal structure of the XRCC4 DNA repair protein and implications for end joining. EMBO J. 2000;19:5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell. 2007;28:1093–1101. doi: 10.1016/j.molcel.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 16.Deshpande RA, Wilson TE. Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair (Amst) 2007;6:1507–1516. doi: 10.1016/j.dnarep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuta R, Cheng HL, Gao Y, Alt FW. Molecular genetic characterization of XRCC4 function. Int. Immunol. 1997;9:1607–1613. doi: 10.1093/intimm/9.10.1607. [DOI] [PubMed] [Google Scholar]
- 18.Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 20.Boyer J, Rohleder K, Ketner G. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology. 1999;263:307–312. doi: 10.1006/viro.1999.9866. [DOI] [PubMed] [Google Scholar]
- 21.Nicolas AL, Munz PL, Falck-Pedersen E, Young CS. Creation and repair of specific DNA double-strand breaks in vivo following infection with adenovirus vectors expressing Saccharomyces cerevisiae HO endonuclease. Virology. 2000;266:211–224. doi: 10.1006/viro.1999.0062. [DOI] [PubMed] [Google Scholar]
- 22.Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a cullin-containing complex. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchette P, Cheng CY, Yan Q, Ketner G, Ornelles DA, Dobner T, Conaway RC, Conaway JW, Branton PE. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell Biol. 2004;24:9619–9629. doi: 10.1128/MCB.24.21.9619-9629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker A, Rohleder KJ, Hanakahi LA, Ketner G. Adenovirus E1B 55k and E4 34k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 2007;81:7034–7040. doi: 10.1128/JVI.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smeaton MB, Miller PS, Ketner G, Hanakahi LA. Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining. Nucleic Acids Res. 2007;22:e152. doi: 10.1093/nar/gkm974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araujo FD, Stracker TH, Carson CT, Lee DV, Weitzman MD. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 2005;79:11382–11391. doi: 10.1128/JVI.79.17.11382-11391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohammadi ES, Ketner EA, Johns DC, Ketner G. Expression of the adenovirus E4 34k oncoprotein inhibits repair of double strand breaks in the cellular genome of a 293-based inducible cell line. Nucleic Acids Res. 2004;32:2652–2659. doi: 10.1093/nar/gkh593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumann P, West SC. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett CF, et al. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 30.Boyer JL, Ketner G. Genetic analysis of a potential zinc-binding domain of the adenovirus E4 34k protein. J. Biol. Chem. 2000;275:14969–14978. doi: 10.1074/jbc.M000566200. [DOI] [PubMed] [Google Scholar]
- 31.Sarnow P, Hearing P, Anderson CW, Halbert DN, Shenk T, Levine AJ. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J. Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanakahi LA. 2-Step purification of the Ku DNA repair protein expressed in Escherichia coli. Protein Expr. Purif. 2007;52:139–145. doi: 10.1016/j.pep.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 35.Hsu HL, Yannone SM, Chen DJ. Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair (Amst) 2002;1:225–235. doi: 10.1016/s1568-7864(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 36.Lewis JB, Mathews MB. Viral messenger RNAs in six lines of adenovirus-transformed cells. Virology. 1981;115:345–360. doi: 10.1016/0042-6822(81)90116-1. [DOI] [PubMed] [Google Scholar]
- 37.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 38.Ben-Omran TI, Cerosaletti K, Concannon P, Weitzman S, Nezarati MM. A patient with mutations in DNA ligase IV: clinical features and overlap with Nijmegen breakage syndrome. Am. J. Med. Genet. A. 2005;137:283–287. doi: 10.1002/ajmg.a.30869. [DOI] [PubMed] [Google Scholar]
- 39.O’Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer SE, Seidel J, et al. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 40.Iiizumi S, Nomura Y, So S, Uegaki K, Aoki K, Shibahara K, Adachi N, Koyama H. Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. Biotechniques. 2006;41:311–316. doi: 10.2144/000112233. [DOI] [PubMed] [Google Scholar]
- 41.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA. 2006;103:18597–602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costantini S, Woodbine L, Andreoli L, Jeggo PA, Vindigni A. Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair. 2007;6:712–722. doi: 10.1016/j.dnarep.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Yu Y, Wang W, Ding Q, Ye R, Chen D, Merkle D, Schriemer D, Meek K, Lees-Miller SP. DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair. 2003;2:1239–1252. doi: 10.1016/s1568-7864(03)00143-5. [DOI] [PubMed] [Google Scholar]
- 45.Sriskanda V, Shuman S. Chlorella virus DNA ligase: nick recognition and mutational analysis. Nucleic Acids Res. 1998;26:525–531. doi: 10.1093/nar/26.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Dell M, Sriskanda V, Shuman S, Nikolov DB. Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining. Mol. Cell. 2000;6:1183–1193. doi: 10.1016/s1097-2765(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 47.Kysela B, Doherty AJ, Chovanec M, Stiff T, Ameer-Beg SM, Vojnovic B, Girard PM, Jeggo PA. Ku stimulation of DNA ligase IV-dependent ligation requires inward movement along the DNA molecule. J. Biol. Chem. 2003;278:22466–22474. doi: 10.1074/jbc.M303273200. [DOI] [PubMed] [Google Scholar]
- 48.Marchetti C, Walker SA, Odreman F, Vindigni A, Doherty AJ, Jeggo P. Identification of a novel motif in DNA ligases exemplified by DNA ligase IV. DNA Repair. 2006;5:788–798. doi: 10.1016/j.dnarep.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Hentges P, Ahnesorg P, Pitcher RS, Bruce CK, Kysela B, Green AJ, Bianchi J, Wilson TE, Jackson SP, Doherty AJ. Evolutionary and functional conservation of the DNA non-homologous end-joining protein, XLF/Cernunnos. J. Biol. Chem. 2006;281:37517–37526. doi: 10.1074/jbc.M608727200. [DOI] [PubMed] [Google Scholar]
- 50.Wu PY, Frit P, Malivert L, Revy P, Biard D, Salles B, Calsou P. Interplay between Cernunnos-XLF and nonhomologous end-joining proteins at DNA ends in the cell. J. Biol. Chem. 2007;282:31937–31943. doi: 10.1074/jbc.M704554200. [DOI] [PubMed] [Google Scholar]
- 51.Drouet J, Delteil C, Lefrancois J, Concannon P, Salles B, Calsou P. DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J. Biol. Chem. 2005;280:7060–7069. doi: 10.1074/jbc.M410746200. [DOI] [PubMed] [Google Scholar]
- 52.Leber R, Wise TW, Mizuta R, Meek K. The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J. Biol. Chem. 1998;273:1794–1801. doi: 10.1074/jbc.273.3.1794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.