Abstract
The recently characterized NEIL1 and NEIL2 are distinct from the previously characterized mammalian DNA glycosylases (OGG1 and NTH1) involved in repair of oxidized bases because of the NEILs’ preference for excising base lesions from single-stranded DNA present in bubble and fork structures. OGG1 and NTH1 are active only with duplex DNA. This raises the possibility that NEILs function in the repair of base lesions during DNA replication and/or transcription. S-phase- specific activation of only NEIL1 suggests its preferential involvement in repair during DNA replication. Here we show that antisense oligonucleotides specific for human or Chinese hamster NEIL1 decreased in vivo NEIL1 levels by 70–80%, concomitant with increased oxidative damage in the genome. Moreover, NEIL1 downregulation enhanced spontaneous mutation in the HPRT locus by about 3-fold in both Chinese hamster V79 and human bronchial A549 cell lines. The mutant frequency was further enhanced (7- to 8-fold) under oxidative stress. The majority of both spontaneous and induced mutations occurred at A•T base pairs, indicating that oxidized A and/or T are NEIL1’s preferred in vivo substrates. NEIL1 thus plays a distinct and important role in repairing endogenous and induced mutagenic oxidized bases, and hence in maintaining the functional integrity of mammalian genomes.
Keywords: BER, HPRT, mutagenesis, NEIL1, oxidative stress
1. INTRODUCTION
Reactive oxygen species (ROS) generated both endogenously and exogenously induce oxidative DNA damage, together with oxidation of lipids and proteins. Sporadic mutations due to oxidized DNA base lesions and strand breaks have been implicated in the etiology of cancers, as well as in a variety of other pathophylogical states, and also in aging [1–3]. However, all organisms are able to repair DNA damage, and nearly all ROS-induced DNA lesions (except double-strand breaks) are repaired via the DNA base excision repair (BER) pathway [4,5]. The conserved BER process is initiated with excision of the damaged base by a DNA glycosylase, followed by cleavage of the DNA strand at the abasic site [4].
Three mammalian DNA glycosylases, involved in repair of oxidized bases, namely 8-oxoguanine DNA glycosylase (OGG1), endonuclease three homolog 1 (NTH1), and MutY homolog (MYH)- were previously identified and extensively characterized. OGG1 is the functional counterpart of Escherichia coli Fpg (MutM), which primarily removes oxidized purines from DNA. NTH1 (the functional counterpart of E. coli Nth) preferentially removes oxidized pyrimidines [6–8], while MYH removes the misincorporated A opposite 8-oxoG during DNA replication [9]. While OGG1, MYH and NTH1 were thought to be the three major enzymes for oxidized base repair in mammalian cells, individual null mutation of these genes in mice did not cause any major phenotype, suggesting that other DNA glycosylases are present in mammalian cells to repair oxidized bases [10–12].
We and others have subsequently identified two other DNA glycosylases, based on structural homology and reaction mechanism to E. coli Nei / MutM, which we named Nei-like 1 and 2 (NEIL1 and NEIL2) [13–17]. These two enzymes have been partially characterized, and we are currently exploring their distinct cellular functions.
The NEILs are functionally distinct from OGG1/NTH1 in preferentially excising substrate lesions from single-stranded DNA, whereas both NTH1and OGG1 are only active with duplex DNA substrates. This unusual feature of NEILs raises the possibility that they repair base lesions present in transient transcription bubbles or replication forks [18,19]. Futhermore, NEIL-initiated repair is dependent on polynucleotide kinase (PNK), whereas OGG1/NTH1-initiated repair is dependent on AP-endonuclease [20,21]. The physiological significance of the NEIL-initiated PNK-dependant repair pathway has not been elucidated in detail. Earlier studies have shown that inactivation or deficiency of DNA repair genes could induce a mutator phenotype in mammalian cell, as indicated by increased spontaneous mutation frequency [22]. Germline and/or somatic mutations in many DNA repair genes (including hMSH2, hMLH1, hMSH6, hPMS2, BRCA1, BRCA2, ATM and ATR), confer susceptibility to many cancers, notably nonpolyopsis colon cancer and breast cancer which arise from increase in instability and enhanced mutations in the genome [23–25]. Although NEIL1-deficient cells are sensitive to ionizing radiation [26], its role in preventing mutations has not previously been examined. Moreover, NEIL1 has been shown to have a wide range of DNA substrates by in vitro analysis; however, its major in vivo substrates have not yet been analyzed. Here we report that NEIL1 deficiency increases susceptibility to endogenous and ROS-induced mutations in mammalian genomes.
2. MATERIALS AND METHODS
2.1 Cell cultures
The A549 human bronchial epithelial tumor line was obtained from ATCC (Rockville, MD, USA) and cultured in Hank's F-12 medium containing 10% fetal bovine serum (FBS), streptomycin (100 µg/ml) and penicillin (100 IU/ml) at 37°C in 5% CO2. The Chinese hamster lung fibroblast line (V79), also from ATCC, was maintained in Eagle's minimum essential medium (EMEM; Invitrogen) containing 10% FBS supplemented with glutamine (292 mg/L), streptomycin (100 µg/ml), and penicillin (100 U/ml). Prior to the mutation screening, the cells were cultured in a medium containing 5 × 10−5 M hypoxanthine, 4 × 10−7 aminopterin, 5 × 10−6 M thymidine and stored in multiple replicate vials in liquid nitrogen [27].
2.2 Cloning of the Chinese hamster Neil1 gene
Human (NM_024608) and mouse (NM_028347) NEIL1 genes were aligned, and the primers were designed from homologous region for PCR amplification of the Chinese hamster (Cricetulus longicaudatus) Neil1 gene. Total RNA from Chinese hamster V79 cells was reverse-transcribed (Superscript III first strand synthesis kit, Invitrogen) and PCR-amplified using the forward (5’-TGGAGAAGTCCTCTGTCAGC C-3’) and reverse primers (5’-TGGAACCAGATGGTACGGTCATG-3’). The PCR product was sequenced in both directions and aligned to the mouse and human NEIL1 genes to confirm identity of the Chinese hamster NEIL1 gene.
2.3 NEIL1 downregulation by antisense oligonucleotide
V79 and A549 cells, grown to 90% confluency, were reseeded at105 cells/60 mm dish, and treated with 19 nt long NEIL1 sense (5′-G*A*A*GCTACAGCCCGCC*A*G*C-3’), or antisense oligonucleotide (5′-G*C*T* GGC GGG CTG TAG C*T*T* C-3’ (*, indicates a phosphorothioate bond). Oligonucleotidenucleotides (5µM) were added directly into the growth medium 4-times during a 30 h time period. The design of sense and antisense oligonucleotides was based on completely conserved sequences in the human and hamster Hprt genes. Total RNAs were isolated and reverse transcribed with Superscript III first strand synthesis kit (Invitrogen). The NEIL1 mRNA level was quantified by Q-PCR in duplicate using optimized NEIL1 probe (Applied Biosystems). Mammalian 18S RNA was used as the internal control. Cell extract was also prepared to monitor the level of NEIL1 expression by Western blot analysis.
2.4 Measurement of intracellular ROS level
Changes in intracellular ROS level due to glucose oxidase (GO) treatment were determined as described previously [28], [29]. Briefly, V79 cells were treated with increasing concentrations (10, 20, 40, 60, 80 and 100 ng per ml) of GO for 1h in culture medium and excess GO was removed by washing cells in PBS. Cells were then loaded with 5 µM of 5-(and-6) carboxy-2′, 7′-dichlorodihydro-fluorescein diacetate (H2DCF-DA; Molecular Probes) for 15 min at 37°C. As a control, we also treated the cells with the oxidized form of H2DCF-DA (5-and-6 carboxy-2’,7’-dichlorodihydro-fluorescein), and we observed a small increase in intracellular fluorescence (<5%) with or without GO treatment(data not shown), as we described previously [29]. The changes in DCF fluorescence of treated vs. mock-treated cells were determined by flow cytometry (Becton Dickinson FACScan). The mean fluorescence for ~12,000 cells, from three or more independent experiments were analyzed and expressed as ±SEM. Based on these studies V79 cells were treated with 20 ng per ml, while A549 cells received 100 ng per ml of GO in order to increase intracellular ROS levels by 2-fold. At these concentrations viability of cells (determined by Trypan blue extrusion) and cell proliferation was not affected by GO treatment.
2.5 Comet assay
The alkaline single cell gel electrophoresis (Comet assay) was performed with minor modifications as described earlier [28]. Briefly, V79 and A549 cells were trypsinized and cell suspensions (103 cells/ml) in low melting point agarose (1% in PBS, pre-warmed to 37 C) were applied to CometSlides™ (Trevigen, Gaithersburg, MD). DNA was released in the presence of NaI [30] after incubation of the agarose-embedded cells with a lysis buffer {2.5 M NaCl, 100 mM EDTA, 10 mM Tris–HCl (pH 7.8), 1% sodium lauryl sarcosinate and 0.01% Triton X-100}, washed and treated with E. coli formamidopyrimidine DNA glycosylases (Fpg, 1 µg/ml; New England Biolabs) in a digestion buffer (40 mM HEPES, pH 8.0, 100 mM KCl, 0.5 mM EDTA, 0.2 mg/ml bovine serum albumin) for 1h at 37°C [31]. The slides were then placed in a Bio-Rad submarine gel electrophoresis unit and it was then run in TBE, (pH 10, 89 mM Tris–HCl, 89 mM boric acid, 3 mM EDTA) at 4°C for 30 min at 1 V/cm. The samples were then neutralized (0.4 M Tris-HCl, pH 7.5) fixed in methanol and ethanol (5 min) at −20°C and dried overnight before staining with 10 ng/ml SyBr Green I (Molecular Probes Inc.). The comet images were recorded with a Nikon TE200 epifluorescence UV microscope photomicrographic system (Photometric CoolSNAP Fx camera). Randomly selected cells (30–50) were analyzed for each treatment by Euclid Comet Analysis software (Euclid Analysis, St. Louis, MO).
2.6 Screening of Hprt mutants
The ability of 6-thioguanine (6-TG)-resistant cells to replicate DNA and thus allowing cell multiplication in the presence of 6-TG is a reliable indicator of Hprt mutation [32, 33]. For mutant frequency analysis, the sense or antisense oligonucleotide-treated V79 cells were subcultured into 100-mm dishes (5×105 cells/dish) in the growth medium containing 6-TG (7 µg/ml). Experiments were repeated 4 to 5-times to identify presumptive mutants. Plating efficiencies were determined in the absence of the 6-TG [27, 32, 33]. In parallel experiments, sense or antisense oligonucleotide-treated cells were mock treated or treated with glucose oxidase (GO) to induce oxidative stress. GO concentration and time of treatment were optimized to induce a 2-fold increase in the ROS level as described above. Colonies of 6-TG-resistant cells grown for 6 to 8 days were fixed in 3.7% formalin, and stained with 0.1% crystal violet. The Hprt mutant frequency was calculated from the number of 6-TG-resistant colonies relative to the total number of colonies in non-selecting medium [32, 33].
A549 cells at 60% confluency (~3 × 106) were treated with NEIL1 antisense or sense oligonucleotide four times in a 30 h time period. Cells were trypsinized and plated on cover-slips (25 mm in diameter; 1 × 105 per cover-slip). Cells were then treated with 6-TG (10 µg/ml) for 6 additional days and pulse labeled for 3 h with 10 µM 5-bromodeoxyuridine (BrdU) in dark to prevent photolysis of BrdU-substituted DNA and stained for detection of BrdU incorporated into DNA as we described previously [34, 35]. After washing with PBS, the cells were fixed in 4% paraformaldehyde at 4°C for 15 min and placed into 0.1N HCl containing 100 µg/ml pepsin for 30 min at 37°C. For denaturation, the DNAs were treated with 1.5 N HCl for 15 min then Na-borate was added for 5 min. After washing, the incorporated BrdU was detected by immunostaining with monoclonal antibody to BrdU [28]. The HPRT mutant frequency for A549 cells was calculated by determining the number of BrdU positive cells relative to the total number of cells on the cover-slips.
The mutations were identified by sequencing the PCR product of Hprt cDNA from V79 cells as described earlier. Each cDNA was amplified with two Hprt primers (forward-5’-TTACCTCACCGCTTTCTCGTG-3’; reverse 5’-TGGCTGCAGAACTAGAATGCTTG-3’) flanking the Hprt cDNA and sequenced. Electropherograms were aligned with STADEN software to identify the mutations.
3. RESULTS
3.1 The humans and Chinese hamster NEIL1 genes are highly homologous
The Chinese hamster Neil1 gene was cloned as described in Materials and Methods. Sequence alignment showed that the Chinese hamster Neil1 has 80.4% and 87.8% identity to the human and mouse NEIL1 genes at the nucleotide level, and 74.2% and 82.7% identity at the amino acid level, respectively. Thus NEIL1 is a highly conserved protein.
3.2 The NEIL1 level is decreased in antisense oligonucleotide-treated cells
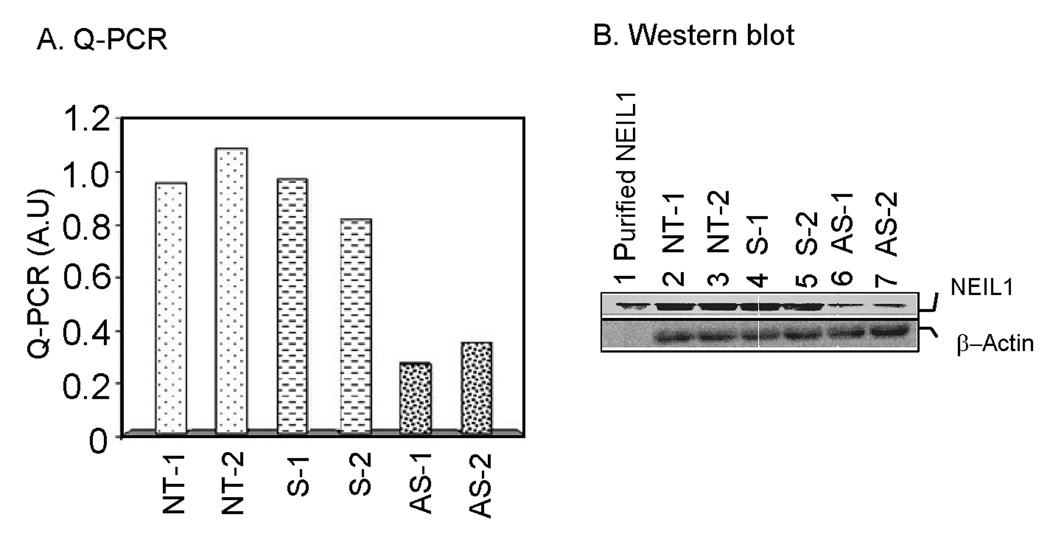
To downregulate intracellular NEIL1, V79 and A549 cells were treated with a NEIL1-antisense DNA oligonucleotide (along with a sense oligonucleotide in parallel as a control). We have tested NEIL1 downregulation in human cells using 4 different oligonucleotides; however the NEIL1 antisense oligonucleotide (common for both A549 and V79 cells) used in this study was optimized for downregulation of human A549 as well as Chinese hamster V79 cells. Quantitative RT- PCR analysis (in duplicate) showed that antisense-oligonucleotide treatment decreased the NEIL1 mRNA level by about 80%, while the sense oligonucleotide had no significant effect (Fig 1A). The NEIL1 protein level also showed a decrease using a similar amount of NEIL1 antisense oligonucleotide (Fig 1B). This indicates the specificity of NEIL1 downregulation due to antisense oligonucleotide treatment.
Figure 1. Antisense oligonucleotide-mediated downregulation of NEIL1 in V79 cells.
A. Q-PCR. Total RNA was isolated from cells treated in two independent experiments with antisense (AS-1, AS-2) or sense (S-1, S-2) oligonucleotide or from non-treated cells (NT-1, NT-2), and NEIL1 mRNA levels were measured by Q-RTPCR (in duplicate). A.U., arbitrary units. Other details are given in Materials and Methods. B. Western blot analysis. Total cell lysate (150 µg) was used for immunoblot analysis with anti-NEIL1 or anti-β actin antibody (Santa Cruz Biotech) using the ECL system (GE-health care) for detection.
3.3 Elevated DNA damage in NEIL1-downregulated cells
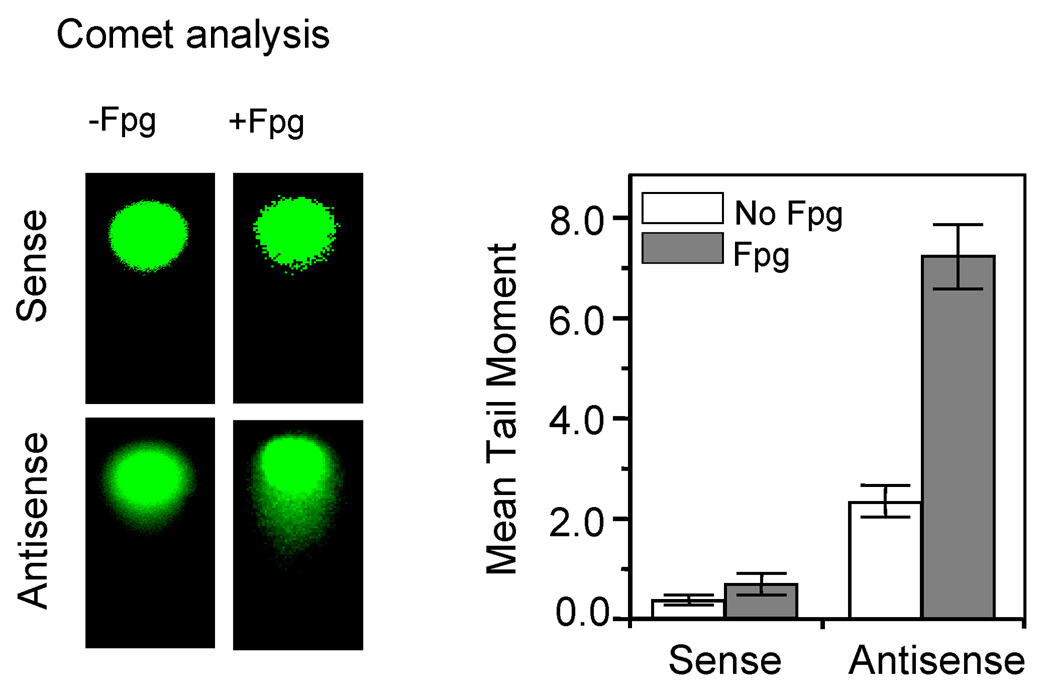
To test whether NEIL1 downregulation causes an increase in DNA base damage due to endogenous ROS, we used alkaline Comet assays in combination with E. coli Fpg treatment. The alkaline Comet assay detects both single- and double-strand breaks in the DNA. Because this procedure separates DNA single-strand fragments and also cleaves the alkali-labile sites including AP sites, this assay measures total alkali-labile lesions. Furthermore, treating the cellular genome prior to Comet analysis with DNA glycosylases, such as E. coli Fpg, which has broad substrate range for oxidized bases and robust DNA strand cleavage activity after base excision, allows analysis of most oxidized base lesions as well [36]. We therefore compared the effect of Fpg and DNA strand breaks in control and NEIL1-dowregulated cells. Fig 2 showed that NEIL1 downregulation significantly increased the tail moment of DNA from V79 cells after Fpg treatment compared to the sense oligonucleotide-treated cells, suggesting accumulation of spontaneous oxidative damage in the genomic DNA. Similar results were obtained with the A549 cells (data not shown). Taken together, these results suggest that NEIL1 is necessary to maintain genomic integrity by repairing basal genomic damage.
Figure 2. Accumulation of endogenous DNA damage due to NEIL1-downregulation.
Alkaline Comet analysis of endogenous DNA damage in V79 cells treated with AS or S oligonucleotides as before. The details of Fpg treatment are described in Materials and Methods. The Comet images were taken with a Nikon TE200 epifluorescence UV microscope equipped with a Photometric CoolSNAP Fx camera (Left panel). Tail moments of randomly selected cells (≥50) were analyzed for each treatment with Euclid Comet Analysis software (Euclid Analysis, St. Louis, MO). Data points are the mean (±S.E) of at least three independent experiments (Right panel).
3.4 Increased mutation frequency in NEIL1-downregulated cells
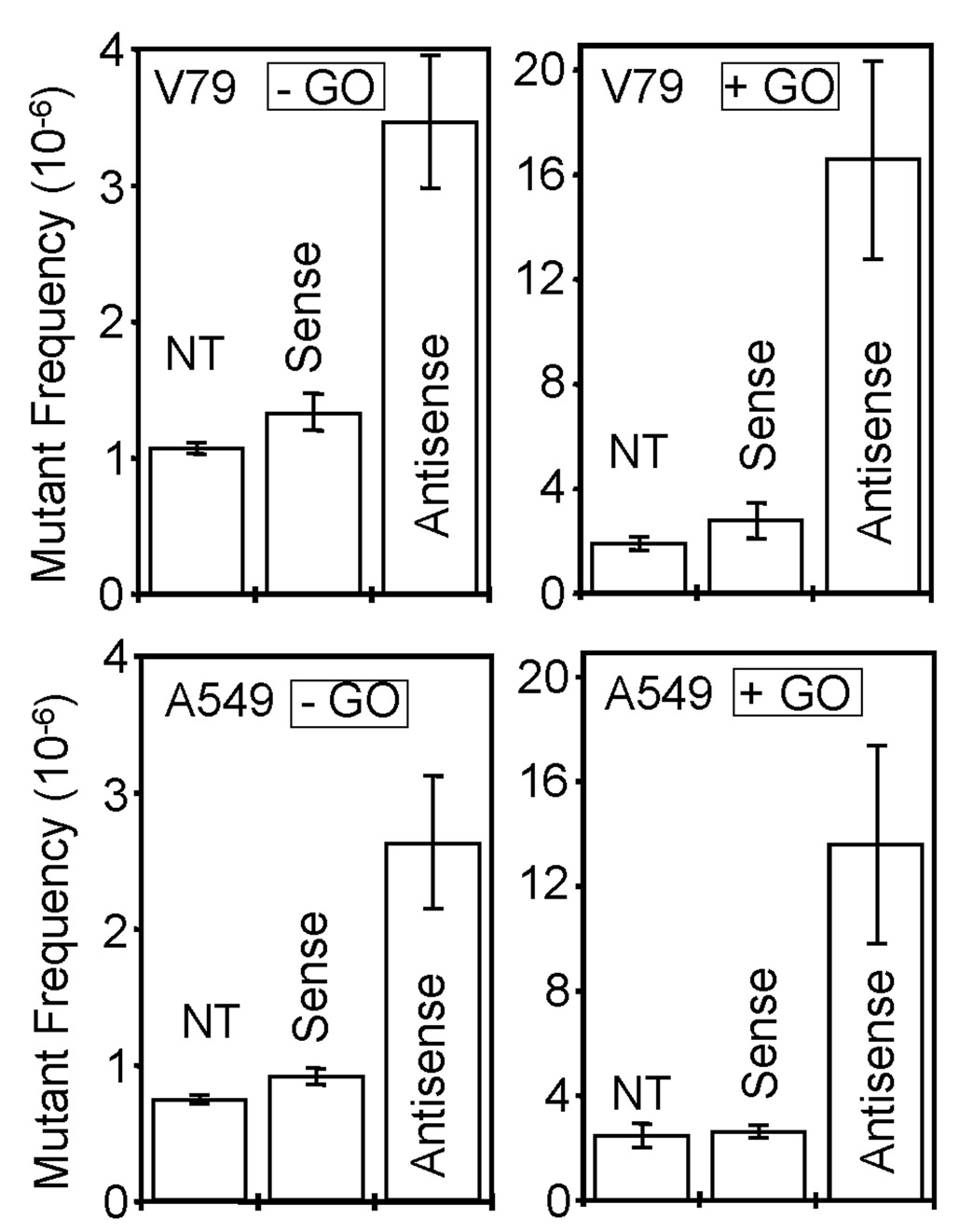
To test whether NEIL1 helps to prevent mutations due to endogenous oxidatively damaged bases, we screened for mutations in the Hprt gene. Because of the ease in scoring its forward mutations [32,33] the single copy, X-linked Hprt locus has been widely used to evaluate the genotoxicity of various chemical and physical agents, as well as the impact of altered levels of DNA repair proteins, [37,38]. Fig 3 shows that NEIL1 downregulation increased the HPRT mutant frequency by about 3-fold in both V79 and A549 cells relative to treatment with the sense (control) oligonucleotide, or to the untreated cells. We then compiled the mutational spectra in the Hprt locus of V79 after PCR amplification and sequence analysis. Table 1 shows that the majority of the mutations occurred at A•T base pairs (75%) in NEIL1 antisense oligonucleotide-treated cells. Interestingly, we have detected a mutational hot spot at nucleotide position 359 (359A>T) of the V79 Hprt gene. Similar analysis of the mutation spectrum in oxidatively-stressed V79 cells was also carried out (Table 2). It is evident that oxidative stress significantly increased the mutant frequency (~8 fold for V79 and 6 fold for A549 over the control cells), while maintaining similar base pair-specific distribution, i.e., with preference for the A•T pairs (79%). However, 359A>T base substitution was moderate, not a mutational hot spot under oxidatively stressed condition.
Figure 3. Increased HPRT mutant frequency in NEIL1-deficient cells.
V79 (upper panel) and A549 cells (lower panel) were nontreated (NT) or treated with sense or antisense oligonucleotides to NEIL1, then treated with (+) or without (−) glucose oxidase (GO) for 1h to induce oxidative stress. NT, non-treated; S, sense oligonucleotide, AS, antisense oligonucleotide. The HPRT mutants were scored in 6-TG-containing HAT medium. The bar graphs represent the means ± standard error from 4 independent experiments.
Table-1.
Nature of spontaneous mutations in NEIL1 sense or antisense oligonucleotide-treated V79 cells
| SENSE | ANTISENSE | |||||||
|---|---|---|---|---|---|---|---|---|
| Position | Mutation | Occurrence | AA change | Position | Mutation | Occurrence | AA change | |
| −51 | G>T | 1 | – | −16 | INS-C | 1 | – | |
| +10 | C>G | 2 | R>G | −24 | C>T | 1 | – | |
| +58 | G>T | 1 | D>Y | +1–505 | Rearranged | 1 | Rearranged | |
| +118 | G>T | 1 | G>stop | +2 | T>G | 1 | M>R | |
| +121 | G>A | 1 | V>M | +10 | C>G | 1 | R>G | |
| +133 | A>T | 2 | D>V | +21 | C>T | 2 | S>S | |
| +139 | C>T | 2 | T>I | +28 | INS-T | 1 | Frameshift | |
| −52 to +91 | Del 144 bp | 1 | Frameshift | +47 | G>C | 1 | G>A | |
| +190 | G>C | 1 | A>P | +61 | A>G | 1 | E>E | |
| − 43 to +299 | Del 342 bp | 1 | Frameshift | +82 | T>C | 1 | Y>H | |
| −32 to +280 | Del 312 bp | 1 | Frameshift | +88 | G>A | 1 | D>K | |
| +287 | C>A | 1 | T>N | +133 | A>T | 2 | D>V | |
| +300 | C>G | 1 | I>M | +165 | A>G | 1 | K>K | |
| +359 | A>T | 4 | D>V | +230 | A>G | 1 | D>G | |
| +379 | G>A | 1 | G>R | +236 | T>C | 1 | L>P | |
| +425 | C>A | 1 | T>K | +248 | A>G | 1 | K>R | |
| +442 | T>C | 1 | S>P | +251 | C>T | 1 | A>V | |
| + 460 | A>C | 1 | N>T | +259 | A>G | 1 | R>G | |
| +505–514 | Del-9 bp | 2 | Missing AA | +272 | G>A | 1 | R>K | |
| +505–522 | Del-18 bp | 3 | Missing AA | +274 | T>C | 1 | S>P | |
| +569 | G>C | 1 | G>A | +278 | T>C | 1 | I >P | |
| +602 | A>T | 1 | D>V | +291 | G>C | 1 | D>H | |
| +299 | T>C | 1 | I >C | |||||
| +359 | A>T | 16 | D>V | |||||
| +360 | T>A | 2 | D>A | |||||
| +391 to 407 | Del 17 bp | 2 | Frameshift | |||||
| +413 | A>G | 1 | D>G | |||||
| +421 | A>G | 1 | A>G | |||||
| +427 | A>G | 1 | M>V | |||||
| +430 | C>T | 1 | Q>stop | |||||
| +437 | T>C | 1 | L>P | |||||
| +459 | Del-G | 1 | Frameshift | |||||
| +488 to stop | Rearranged | 1 | Frameshift | |||||
| +493 | G>C | 1 | V>L | |||||
| +504 to 513 | Del-9 bp | 1 | Missing AA | |||||
| +505 to 522 | Del-18 bp | 3 | Missing AA | |||||
| +563 | Del-C | 1 | Frameshift | |||||
| +590 | A>T | 1 | A>V | |||||
| +580 | G>C | 1 | D>H | |||||
| +633 | A>G | 1 | L>R | |||||
| +657 | Del-T | 1 | Frameshift | |||||
| Total | 31 | 62 | ||||||
| Base substitutions | 23 | 49 | ||||||
| A.T | 9 (39 %) | 37 (75.5 %) | ||||||
| G.C | 14 (61 %) | 12 (24.5 %) | ||||||
| Others | 8 | 13 | ||||||
| Not amplified | 8 | 11 | ||||||
Table 2.
Nature of mutations in NEIL1 sense or antisense oligonucleotide-treated V79 cells after GO treatment.
| SENSE | ANTISENSE | |||||||
|---|---|---|---|---|---|---|---|---|
| Position | Mutation | Occurrence | AA change | Position | Mutation | Occurrence | AA change | |
| +21 | C>T | 5 | S>S | 1 to 31 | Rearranged | 1 | Frameshift | |
| +37 | G>C | 1 | D>H | 1 to 505 | Rearranged | 1 | Frameshift | |
| +57 | Del-A | 1 | Frameshift | −30 to +187 | Rearranged | 1 | Frameshift | |
| +72 | C>T | 1 | D>Q | +8 | INS-C | 1 | Frameshift | |
| +118 | G>C | 1 | G>R | +21 | INS-G | 1 | Frameshift | |
| +128 | T>C | 2 | M>T | +21 | C>T | 1 | S>S | |
| +209 | G>A | 1 | G>R | +31 | A>G | 3 | S>G | |
| +289 | G>C | 1 | V>L | +34 | G>A | 1 | D>N | |
| +382 | A>C | 2 | K>Q | +94 | T>G | 1 | L>V | |
| +389 to 405 | Del 17 bp | 2 | Frameshift | +108 | A>T | 1 | I >F | |
| +391 to 407 | Del-17 bp | 3 | Frameshift | +133 | A>T | 4 | D >V | |
| +403 | G>T | 1 | D>Y | +191 | C>T | 1 | A>V | |
| +413 | A>C | 1 | D>A | +202 | C>T | 1 | L>L | |
| +455 | Del-C | 1 | Frameshift | +224 | T>C | 1 | F>L | |
| +459 | Del-G | 1 | Frameshift | +269 | A>G | 1 | D>G | |
| 505–522 | Del-18 bp | 10 | Missing AA | +273 | A>C | 1 | R>S | |
| +464 | Del-C | 1 | Frameshift | +308 | A>G | 1 | K>R | |
| +555 | Del-C | 1 | Frameshift | +355 | G>A | 1 | G>D | |
| +591 | G>A | 1 | E>E | +359 | A>T | 3 | D>V | |
| +559 | T>G | 2 | F>V | +389 to 407 | Del 17 bp | 2 | Frameshift | |
| +599 | G>A | 2 | R>K | +449 | T>C | 1 | V>I | |
| +648 | T>G | 1 | Y>D | +457 | T>A | 1 | Y>N | |
| +648 | T>A | 1 | Y>N | +459 | Del-G | 1 | Frameshift | |
| +657 | Del-T | 1 | Frameshift | +465 | A>C | 1 | K>Q | |
| +505 to 522 | Del-18 bp | 4 | Missing AA | |||||
| Total | 44 | 36 | ||||||
| Base substitutions | 23 | 24 | ||||||
| A.T | 9 (39 %) | 19 (79 %) | ||||||
| G.C | 14 (61 %) | 5 (21 %) | ||||||
| Others | 21 | 12 | ||||||
| Not amplified | 9 | 11 | ||||||
Chi-square analysis also shows that the number of mutations at the A•T base pairs was significantly higher than the number of mutations at the G•C base pairs in antisense oligonucleotide-treated cells. For the antisense data, all of the Chi-square tests yielded significant results at the 95% significance level. The χ2 statistic for the antisense-treated V79 cells (without GO) was 7.17, with GO treatment was 4.99, and for the combined data was 12.03. Each value was higher than the critical value of the χ2 test at the 95% significance level with 1 degree of freedom.
These results strongly suggest that oxidized adenine or thymine is the preferred substrates of NEIL1. Downregulation of NEIL1 is thus associated with enhanced spontaneous as well as oxidative stress-induced mutations in the HPRT gene.
4. DISCUSSION
Human NEIL1 is one of two recently characterized oxidized base-specific mammalian DNA glycosylases with broad substrate specificity. However, its specific role in BER in general, and its contribution to the maintenance of genomic integrity in particular are not well understood. Stephen Lloyd and his collaborators have recently generated NEIL1-null mice, which show an unexpected phenotype. These mice are diabetic, develop fatty livers and also accumulate higher level of mitochondrial DNA damage, although no significant increase in the spontaneous mutation frequency has been reported [39]. However, Rosenquist et al. showed that mouse embryo fibroblasts with siRNA-mediated NEIL1 downregulation were sensitive to ionizing radiation [26], which induces oxidative base damage and DNA strand breaks. Their results thus support the protective function of NEIL1 for oxidative DNA damage. In this study, we have shown that the decrease in the NEIL1 level induced accumulation of oxidative DNA damage, and significantly increased the endogenous mutant frequency. The mutant frequency increased even further under oxidative stress, as expected.
Epidemiological linkage of gastric cancer to NEIL1-inactivating mutations has recently been reported, which is also consistent with an antimutagenic role for NEIL1 as directly demonstrated in our studies [40]. We have found a significant increase in the mutant frequency to NEIL1-deficient cells, and a large majority of the mutations we have characterized are due to endogenous DNA damage subject to repair by NEIL1. It is also reassuring that most of the mutations are single base changes, as expected from misreplication of oxidized DNA bases. Importantly, that the base pair bias was not affected by exogenous oxidative stress is consistent with the similarity in the genotoxic effect of endogenous vs. exogenous oxidants. The majority of mutations induced by NEIL1 deficiency occurred at A•T base pairs in both untreated and GO-treated cells. Thus about 75–80% of mutations occurred at A•T base pairs in NEIL1-deficient cells, compared to about half that many in control cells. We have recently analyzed the HPRT mutational spectra in NEIL2-deficient cells, where the majority of mutations were generated at C•G base pairs (data not shown). This is consistent with the substrate preference of NEIL2 for mutagenic oxidation products of C and G (including 8-oxoguanine) [14, 41]. We and others have shown that NEIL1 has broad substrate specificity in that it preferentially excises formamidopyrymidine (Fapy)-adenine (FapyA), Fapyguanine (Fapy G), thymine glycol, 5-hydroxyuracil (5-OHU) and several other pyrimidine derivatives [13,15,16] in vitro. Our results here suggest that oxidation products of A and T, are among NEIL1’s preferred substrates in vivo, and are likely to be the lesions leading to mutations identified in the HPRT locus [13, 42]. NEIL1 was recently shown to efficiently remove 5-formyluracil and 5-hydroxymethyluracil, oxidation products of thymine that are present at a significant level in mammalian genomes [43]. It has also been suggested that oxidation of the methyl group of T by hydroxyl radicals could frequently occur in the free nucleotide pool [43]. If mammalian DNA polymerases incorporate oxidized dNTPs into the nascent DNA, the cells would face a serious threat of genomic instability without repair of these incorporated lesions. We have previously shown that NEIL1 expression is cell cycle-dependent, with a higher level in the S-phase [13]. We have recently shown that NEIL1 interacts with PCNA, a replication-associated protein [44], and so is likely to be involved in replication-associated repair. Whether NEIL1 removes the incorporated oxidized bases in the nascent DNA strand warrants further investigation.
ACKNOWLEDGEMENTS
This work was supported by USPHS grants RO1 CA102271 (TKH), R01 CA 81063 (SM), P01 AG021830 (SM, IB) and P30 ES 06676 that supports the NIEHS Center Cell Biology Core and Molecular Genomics Core of UTMB’s NIEHS Center for DNA sequencing. We thank David Konkel for critically reading the manuscript and Wanda Smith for expert secretarial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Amit K. Maiti, Department of Biochemistry and Molecular Biology and Sealy Center for Molecular Medicine, University of Texas Medical Branch, Galveston; TX 77555
Istvan Boldogh, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston; TX 77555.
Heidi Spratt, Bioinformatics Program, University of Texas Medical Branch, Galveston; TX 77555.
Sankar Mitra, Department of Biochemistry and Molecular Biology and Sealy Center for Molecular Medicine, University of Texas Medical Branch, Galveston; TX 77555.
Tapas K. Hazra, Department of Biochemistry and Molecular Biology and Sealy Center for Molecular Medicine, University of Texas Medical Branch, Galveston; TX 77555.
REFERENCES
- 1.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radical Biology and Medicine. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 3.Gotz ME, Kunig G, Riederer P, Youdim MB. Oxidative stress: free radical production in neural degeneration. Pharmacology & Therapeutics. 1994;63:37–122. doi: 10.1016/0163-7258(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 4.Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 5.Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes(1,2) Free Radic Biol Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and charcterization of human hNTH1, a homolog of Escherichia coli endonuclease III: Direct identification of Lys-212 as the active nucleophilic residue. Journal of Biological Chemistry. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 7.Lu R, Nash HM, Verdine GL. A DNA repair enzyme that excises oxidatively damaged guanines from the mammalian genome is frequently lost in lung cancer. Current Biology. 1997;7:397–407. doi: 10.1016/s0960-9822(06)00187-4. [DOI] [PubMed] [Google Scholar]
- 8.Aspinwall R, Rothwell DG, Roldan-Arjona T, Anselmino C, Ward CJ, Cheadle JP, Sampson JR, Lindahl T, Harris PC, Hickson ID. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc Natl Acad Sci U S A. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slupska MM, Baiialov C, Luther WM, Chiang J-H, Wei Y-F, Miller JH. Cloning and sequencing a human homolog ({IhMYH}) of the {IEscherichia coli mutY} gene whose function is required for the repair of oxidative DNA damage. Journal of Bacteriology. 1996;178:3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, Itoh M, Takano H, Hippou Y, Aburatani H, Masumura K, Nohmi T, Nishimura S, Noda T. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci U S A. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ocampo MT, Chaung W, Marenstein DR, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Targeted deletion of mNth1 reveals a novel DNA repair enzyme activity. Mol Cell Biol. 2002;22:6111–6121. doi: 10.1128/MCB.22.17.6111-6121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and Characterization of a Novel Human DNA Glycosylase for Repair of Cytosine-derived Lesions. J Biol Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- 15.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA repair. 2002:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 16.Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takao M KS, Kobayashi K, Zhang QM, Yonei S, Van Der Horst GT, Yasui A. A back-up glycosylase in Nth1-knockout mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002 doi: 10.1074/jbc.M206884200. in press. [DOI] [PubMed] [Google Scholar]
- 18.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 19.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, Wilson SH, Mitra S, Hazra TK. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair (Amst) 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Nakabeppu Y, Sakumi K, Sakamoto K, Tsuchimoto D, Tsuzuki T, Nakatsu Y. Mutagenesis and carcinogenesis caused by the oxidation of nucleic acids. Biol Chem. 2006;387:373–379. doi: 10.1515/BC.2006.050. [DOI] [PubMed] [Google Scholar]
- 23.Riccio A, Aaltonen LA, Godwin AK, Loukola A, Percesepe A, Salovaara R, Masciullo V, Genuardi M, Paravatou-Petsotas M, Bassi DE, Ruggeri BA, Klein-Szanto AJ, Testa JR, Neri G, Bellacosa A. The DNA repair gene MBD4 (MED1) is mutated in human carcinomas with microsatellite instability. Nat Genet. 1999;23:266–268. doi: 10.1038/15443. [DOI] [PubMed] [Google Scholar]
- 24.Duval A, Rolland S, Compoint A, Tubacher E, Iacopetta B, Thomas G, Hamelin R. Evolution of instability at coding and non-coding repeat sequences in human MSI-H colorectal cancers. Hum Mol Genet. 2001;10:513–518. doi: 10.1093/hmg/10.5.513. [DOI] [PubMed] [Google Scholar]
- 25.Loukola A, Vilkki S, Singh J, Launonen V, Aaltonen LA. Germline and somatic mutation analysis of MLH3 in MSI-positive colorectal cancer. Am J Pathol. 2000;157:347–352. doi: 10.1016/S0002-9440(10)64546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 27.Bacsi A, Aguilera-Aguirre L, German P, Kruzel ML, Boldogh I. Colostrinin decreases spontaneous and induced mutation frequencies at the hprt locus in Chinese hamster V79 cells. J Exp Ther Oncol. 2006;5:249–259. [PubMed] [Google Scholar]
- 28.Boldogh I, Roy G, Lee MS, Bacsi A, Hazra TK, Bhakat KK, Das GC, Mitra S. Reduced DNA double strand breaks in chlorambucil resistant cells are related to high DNA-PKcs activity and low oxidative stress. Toxicology. 2003;193:137–152. doi: 10.1016/j.tox.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Das A, Hazra TK, Boldogh I, Mitra S, Bhakat KK. Induction of the human oxidized base-specific DNA glycosylase NEIL1 by reactive oxygen species. J Biol Chem. 2005;280:35272–35280. doi: 10.1074/jbc.M505526200. [DOI] [PubMed] [Google Scholar]
- 30.Hamilton ML, Guo Z, Fuller CD, Van Remmen H, Ward WF, Austad SN, Troyer DA, Thompson I, Richardson A. A reliable assessment of 8-oxo-2-deoxyguanosine levels in nuclear and mitochondrial DNA using the sodium iodide method to isolate DNA. Nucleic Acids Res. 2001;29:2117–2126. doi: 10.1093/nar/29.10.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trzeciak AR, Nyaga SG, Jaruga P, Lohani A, Dizdaroglu M, Evans MK. Cellular repair of oxidatively induced DNA base lesions is defective in prostate cancer cell lines PC-3 and DU-145. Carcinogenesis. 2004;25:1359–1370. doi: 10.1093/carcin/bgh144. [DOI] [PubMed] [Google Scholar]
- 32.Bradley MO, Bhuyan B, Francis MC, Langenbach R, Peterson A, Huberman E. Mutagenesis by chemical agents in V79 chinese hamster cells: a review and analysis of the literature. A report of the Gene-Tox Program. Mutat Res. 1981;87:81–142. doi: 10.1016/0165-1110(81)90029-4. [DOI] [PubMed] [Google Scholar]
- 33.Albrecht T, Fons MP, Deng CZ, Boldogh I. Increased frequency of specific locus mutation following human cytomegalovirus infection. Virology. 1997;230:48–61. doi: 10.1006/viro.1997.8467. [DOI] [PubMed] [Google Scholar]
- 34.Boldogh I, Milligan D, Lee MS, Bassett H, Lloyd RS, McCullough AK. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res. 2001;29:2802–2809. doi: 10.1093/nar/29.13.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobrovolsky VN, Casciano DA, Heflich RH. Development of a novel mouse tk+/− embryonic stem cell line for use in mutagenicity studies. Environ Mol Mutagen. 1996;28:483–489. doi: 10.1002/(SICI)1098-2280(1996)28:4<483::AID-EM26>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 36.Blaisdell JO, Wallace SS. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc Natl Acad Sci U S A. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malkhosyan S, McCarty A, Sawai H, Perucho M. Differences in the spectrum of spontaneous mutations in the hprt gene between tumor cells of the microsatellite mutator phenotype. Mutat Res. 1996;316:249–259. doi: 10.1016/s0921-8734(96)90007-7. [DOI] [PubMed] [Google Scholar]
- 38.Richards B, Zhang H, Phear G, Meuth M. Conditional mutator phenotypes in hMSH2-deficient tumor cell lines. Science. 1997;277:1523–1526. doi: 10.1126/science.277.5331.1523. [DOI] [PubMed] [Google Scholar]
- 39.Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci U S A. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinmura K, Tao H, Goto M, Igarashi H, Taniguchi T, Maekawa M, Takezaki T, Sugimura H. Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis. 2004;25:2311–2317. doi: 10.1093/carcin/bgh267. [DOI] [PubMed] [Google Scholar]
- 41.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 42.Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 43.Zhang QM, Yonekura S, Takao M, Yasui A, Sugiyama H, Yonei S. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA Repair (Amst) 2005;4:71–79. doi: 10.1016/j.dnarep.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]