Abstract
In mammals, the silencing step of the X-chromosome inactivation (XCI) process is initiated by the non-coding Xist RNA. Xist is known to be controlled by the non-coding Xite and Tsix loci, but the mechanisms by which Tsix and Xite regulate Xist are yet to be fully elucidated. Here, we examine the role of higher order chromatin structure across the 100-kb region of the mouse X-inactivation center (Xic) and map domains of specialized chromatin in vivo. By hypersensitive site mapping and chromosome conformation capture (3C), we identify two domains of higher order chromatin structure. Xite makes looping interactions with Tsix, while Xist makes contacts with Jpx/Enox, another non-coding gene not previously implicated in XCI. These regions interact in a developmentally specific and sex-specific manner that is consistent with a regulatory role in XCI. We propose that dynamic changes in three-dimensional architecture leads to formation of separate chromatin hubs in Tsix and Xist that together regulate the initiation of X-chromosome inactivation.
Keywords: X-chromosome inactivation, Xist, Tsix, Xite, chromosome conformation capture, higher order structure, non-coding RNA
INTRODUCTION
X-chromosome inactivation (XCI) evolved to compensate for gene dosage in sexually dimorphic species. In mammals, XX females and XY males achieve dosage compensation by the transcriptional silencing of one X chromosome in each diploid female cell (Lyon, 1961). In the embryo proper, the choice of which X chromosome is to be silenced occurs randomly, with both the maternal and paternal X chromosome having an equal chance of being inactivated. However, in the extra-embryonic tissues, a parent-of-origin effect is observed where the paternal X chromosome is preferentially silenced. Both mechanisms result in one active X chromosome (Xa) and one inactive X chromosome (Xi), which are then stably maintained through subsequent cell divisions.
The molecular players in XCI have been defined over recent years and many are located within a 100-kb region on the X chromosome, known as the X-inactivation center (Xic) (Lee et al., 1999b). Non-coding elements within the Xic are responsible for the three steps of random XCI, including counting of the X-to-autosome ratio, choice of which X chromosome to inactivate, and silencing of the Xi (reviewed by (Plath et al., 2002)). The Xist gene encodes a large, non-translated RNA that is transcribed from the inactive X chromosome and ‘coats’ the Xi in cis, inducing chromosome-wide gene silencing (Brockdorff et al., 1992; Clemson et al., 1996; Penny et al., 1996). Xist is negatively regulated by its antisense partner, Tsix (Lee, 2000; Lee et al., 1999a; Sado et al., 2001), also a non-coding, nuclear RNA. Tsix is in turn regulated by Xite, an upstream locus that produces several non-coding transcripts and harbors several strong DNase I hypersensitive sites (DHS) (Ogawa and Lee, 2003).
Murine embryonic stem (ES) cells have provided an excellent model system for the investigation of XCI and the roles of Xite, Tsix and Xist. In this system, Xist and Tsix RNAs are transcribed at low levels from both X chromosomes in undifferentiated ES cells. Upon differentiation, Tsix expression is extinguished on the future Xi (Lee et al., 1999a). Conversely, Xist expression is up-regulated in cis during the same developmental phase, suggesting that Tsix antagonism is responsible for Xist regulation. Tsix expression persists on the Xa until slightly later in development and Xist expression is prevented in cis. However, the exact mode of action of Tsix on Xist is currently still unclear. A number of possibilities exist whereby the process of antisense transcription through the Xist locus may prevent Xist expression in undifferentiated cells, including promoter blocking, transcriptional interference or alterations in chromatin structure. Recent evidence argues for a role of Tsix RNA-directed chromatin change (Navarro et al., 2005; Sado et al., 2005; Sun et al., 2006), in which Tsix RNA, coupled to anti-parallel transcription through the Xist promoter, results in recruitment of specific regulatory factors and chromatin modifications to the Xist promoter.
Thus, while long-range interactions between Tsix and Xist clearly exist, the three-dimensional (3-D) nature of such interactions has yet to be addressed. Higher order chromatin interplay has been implicated in other epigenetic systems, such as β-globin (Patrinos et al., 2004; Tolhuis et al., 2002), TH2 and interleukin genes (Lee et al., 2005), and H19/Igf2 loci (Kurukuti et al., 2006; Murrell et al., 2004), where looping interactions among locus control regions (LCR), enhancers, and co-clustered genes determine the pattern of gene expression. At the Xic, only two enhancers have been identified so far. DNase I hypersensitive site (DHS) mapping and transient transfection enhancer assays showed that one such enhancer resides at Xite (Ogawa and Lee, 2003). Current models posit that the Xite enhancer enables Tsix to persist allele-specifically on the future Xa. A second enhancer lies within the 5′ end of Tsix itself and is essential for high level expression of Tsix in ES cells (Stavropoulos et al., 2005). Although Xist is also developmentally regulated, transcriptional enhancers have not yet been described for this gene. How these regulatory elements interact with Tsix and Xist in 3-D space is not known.
The mode of action of enhancers or LCR elements on gene promoters located at large distances is the subject of intense debate. The ‘tracking’ model postulates that proteins bound to LCRs and enhancers track linearly along the DNA helix and scans surrounding sequences for promoter elements (Li et al., 1999). By contrast, the ‘linking’ model proposes that enhancer-bound proteins recruit additional protein factors, which then sequentially extend towards the promoter. Finally, the ‘looping’ model posits that random collisions between regions of the flexible chromatin fiber enable an enhancer to come into direct contact with a gene promoter in three-dimensional space, thereby looping out all intervening DNA sequences (Bulger and Groudine, 1999). The ‘chromosome conformation capture’ (3C) technique (Dekker et al., 2002) has lend strong support to the concept of looping chromatin in cis (Kurukuti et al., 2006; Lee et al., 2005; Murrell et al., 2004; Patrinos et al., 2004; Tolhuis et al., 2002), as well as to the idea of interaction in trans between chromosomes (Ling et al., 2006; Lomvardas et al., 2006; Spilianakis et al., 2005; Xu et al., 2006). In principle, an enhancer or an LCR element at the Xic might employ one or several of these mechanisms.
Here, to determine how critical elements interact during the process of XCI, we combine DHS identification and the 3C technique to map higher order chromatin structures at the Xic. We find several domains of interactions between Xist, Tsix and Xite that are regulated in a developmentally specific manner. We thus provide a first glimpse of dynamic three-dimensional architectural changes that accompany the onset of XCI.
MATERIALS AND METHODS
Cell lines and culture
Mouse embryonic stem cell lines J1 (40XY, 129Sv/J)(Li et al., 1992) and 16.7 (40XX, 129 × [M.castaneus × 129] (Lee and Lu, 1999)) were maintained on gelatin-treated tissue culture flasks with a feeder layer of γ-irradiated mouse embryonic fibroblasts (obtained from d13.5 embryos) in Dulbecco modified Eagle medium plus 15% fetal bovine serum (heat inactivated) and 500U/ml Leukemia Inhibitory Factor (LIF). Embryoid bodies were differentiated by suspension culture for 4 days without LIF and maintained thereafter under adherent conditions for 6 days. Primary fibroblasts were obtained from d13.5 embryos and maintained in DMEM plus 10% FBS.
DNase I Hypersensitive Site (DHS) Mapping
DHS mapping was performed essentially as previously described (Ogawa and Lee, 2003). Briefly, 1 × 108 trypsinized cells were washed with PBS, pelleted, and resuspended in 10 ml ice-cold Nuclear Isolation Buffer (NIB) (0.32 M sucrose, 3 mM CaCl2, 2 mM MgOAc, 10 mM Tris-HCl (pH7.5), 0.1 mM EDTA, 1 mM DTT). One-twentieth volume of NIB + 0.3% NP-40 was added and the suspension was subsequently dounced 15 times with pestle B. Nuclei were centrifuged at 400 × g and resuspended in ice-cold NIB at 0.2 mg/ml as determined by total nucleic acid content. For ES cells, DNase I (Worthington) was added to aliquots at a final concentration of 0 – 10 μg/ml, and samples were subsequently incubated at 37°C for 2 min. For fibroblasts, final concentration of DNase I was 0 – 80 μg/ml and samples were incubated for 5 min. Digestions were terminated by adding an equal volume of 2 ×stop solution (1% SDS, 0.6 M NaCl, 20 mM Tris-HCl (pH7.5), 10 mM EDTA, 0.2 mg/ml Proteinase K) and incubated overnight at 37°C. DNA was precipitated with 1.25 M NH40Ac, resuspended, and digested for Southern analysis.
For Southern blotting, panels A, C and D were digested with EcoRI; panels B and D with BamHI. Probes used were positions a) 108150–108354, b) 113059–113266, c) 114507–114718, d) 124506–124723 and e) 91198–91433 from GenBank AJ421279.
Chromosome conformation capture
Chromosome conformation capture experiments were carried out as described in Splinter et al (Splinter et al., 2004) with minor modifications and appropriate controls (Dekker, 2006). Briefly, 107 cells were cross-linked with 2% formaldehyde at room temp for 10 minutes. Cross-linking reactions were terminated by addition of glycine to the final concentration of 125 mM. Nuclei of crossed linked cells were purified by incubating the cells with lysis buffer (10 mM Tris 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.5% NP-40 with protease inhibitor cocktail) twice on ice for 4 minutes, then washed with 5 ml of ice-cold PBS. The purified nuclei were resuspended in 760 μl restriction buffer consisting of 80μl of 10X restriction buffer H (GE Healthcare) and 680 μl H2O, and incubated with mild agitation at 37°C for 1 hour with 12 μl 20% SDS. Reactions were treated with 18 μl of Triton-X100, incubated at 37° C for 1 hour and subjected to restriction enzyme digestion with 240U of BamHI and 200U of BglII at 37°C for 2 hours. An additional 120U of BamHI and 100U of BglII was carried out at 37°C overnight. The digested samples were treated with 40 μl of 20% SDS at 65°C for 20 min to inactivate restriction enzymes. Samples were transferred to 9.2 ml 1X ligase buffer containing 1% triton X-100 and incubated at room temperature for 1 hour and then incubated with 1600U of T4 DNA ligase at 16°C for 4.5 hours and additional 0.5 hour at room temperature. Ligated samples were treated with 3 μl of proteinase K (20 mg/ml) at 65°C overnight to reverse cross-linking, and then treated with 2 μl RNaseA (10 mg/ml) at 37°C for 1 hour. Genomic DNA was extracted with phenol/chloroform and precipitated with NaOAc and isopropanol. Precipitated DNA was washed with 1 ml 75% ethanol and resuspended with 600μl H2O. To estimate genomic DNA concentration, 5 μl of each sample were analyzed on a 0.8% gel and visualized by EtBr staining. About 40 ng of genomic DNA was used for each 25 μl PCR reaction.
Positive control templates were made by digesting pπJL2 with BamHI and BglII and then random re-ligation of the restriction fragments. To mimic the complexity of the genomic DNA obtained from 3C experiments, the controls PCR reactions were performed with the mixture of 2.0 pg of positive control template (see below) and 40 ng of control genomic DNA that was processed in parallel with other 3C samples, except that the formaldehyde crosslinking step was omitted. Amplification of specific PCR products was strictly formaldehyde-cross-linking and ligation-dependent (data not shown). All PCR reactions were performed with hot-start Taq DNA polymerase (AmpliTaq, Applied Bioscience) and with 34–36 cycles of amplification, where all the amplifications were in the exponential range. PCR reactions amplifying un-rearranged genomic DNA were also carried out as described above but with only 26 cycles of amplification. To obtain quantitative information, amplifications were performed in the exponential phase as confirmed by decreases in PCR product levels as the concentrations of templates decreased by successive 3-fold dilutions (Fig. 2B, 3B, and data not shown). The relative crosslinking efficiency was calculated at least from duplicate PCR analyses of 2 independent 3C preparations (average of at least 4 sets of PCR analyses). PCR primer sequences used for 3C analysis are available on request.
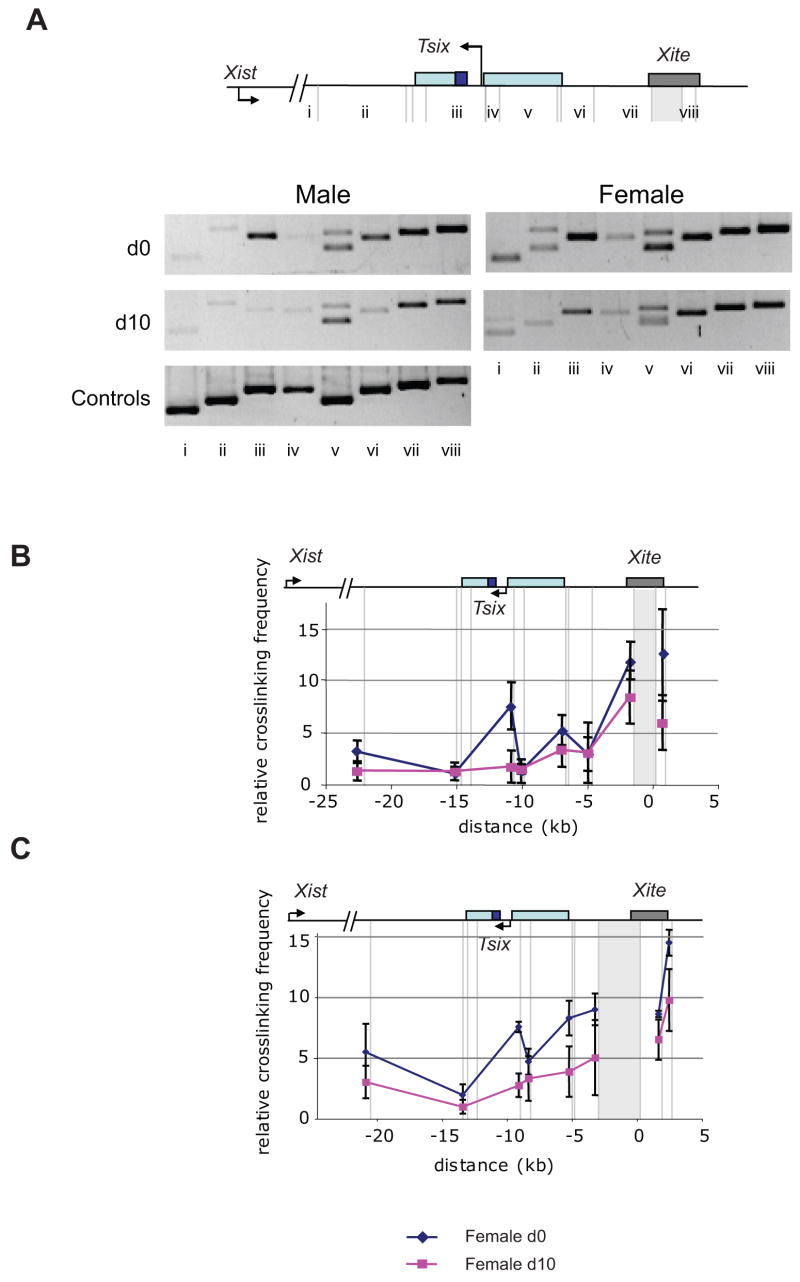
Figure 2. Xist and Jpx form an interaction domain within the Xic.
3C analysis of chromatin from female (EL16.7) and male (J1) ES cells. Grey dashed lines indicate BamHI and BglII digestion sites and each restriction fragment tested is numbered below. Note that the smallest BglII-BamHI fragments are not represented because of their size. Control PCRs are shown for all of the restriction fragments tested using the digested P1 construct. (A) PCR assays determine relative intra-chromosomal crosslinking efficiency of the GAPDH locus on chromosome 7, as well as inter-chromosomal cross-linking between chromosomes 7 and X, of undifferentiated (d0) and differentiated (d10) female ES cells. (B) Semi-quantitative PCR analyses of selected intra-chromosomal interactions. Interaction between Xist promoter and Jpx promoter (iii), the immediate upstream fragment of the Xist promoter (vii), and the end of the Xist gene (xi) were further analyzed by PCR (37 cycles) with a series of 3-fold dilution of input templates. The concentration of X chromosome was measured by PCR (26 cycles) using the same templates with primers that amplified a region in Xic free of BamHI and BglII restriction sites. (C) Anchor primer within the fragment containing the Xist promoter is indicated by the grey shaded region and a red arrow. The end of the Xist gene domain is indicated with a red dashed line. Gray triangles indicate specific PCR products. Multiple PCR bands in some of the lanes were due to incomplete digest of restriction enzymes. (D) Anchor primer within the fragment containing the Jpx promoter is indicated by the grey shaded region and a red arrow. The interaction with the Xist promoter fragment (vii) is boxed. The size of the control fragment (iv) is slightly smaller due to the presence of a cryptic restriction enzyme site within the P1 construct.
Figure 3. Xite and Tsix form a second interaction domain within the Xic.
3C analysis chromatin from female (EL16.7) and male (J1) ES cells. A schematic of the Xic region tested is shown above each panel. BamHI and BglII-digested restriction fragments are shown as grey lines and the anchor primer regions as a grey box. (A) Representative gel image of 3C analysis using the anchor primer localized to the DNA fragment bearing the Tsix promoter and the rest of the Xic. (B) Semi-quantitative nature of the PCR demonstrated by 3-fold serial dilutions of template for selected intra-chromosomal interactions, as indicated. (C) Average relative cross-linking frequencies of female ES cells between the anchor fragment containing the Tsix promoter and DXPas34 repeat region and the rest of the Xic. Blue and red lines indicate the average relative crosslinking efficiency of undifferentiated (d0) and differentiated (d10) female ES cells, respectively. (D) Comparison of changes in chromosomal conformation in male and female ES cell upon differentiation. Blue and red lines indicate the change in crosslinking efficiency of male ES cells and female ES cells upon differentiation, respectively.
To create a pool of all possible interacting products as control, we digested an 80-kb P1 construct, πJL2 (Lee et al., 1999b), with BamHI and BglII and randomly ligated the resulting fragments. To mimic the complexity of the mammalian genome during the PCR step, we digested female fibroblast genomic DNA with BamHI and Bgl This control was included in all 3C experiments. As a control for cross-linking efficiency, we carried out PCR using primers that amplify the GAPDH locus (cloned, digested, and randomly ligated to create all pairwise combinations as above) using primers GAPDH-a (5′-ACACAGGCAAAATACCAATG-3′ ) and GAPDH-b (5′-GAATGCTTGGATGTACAACC-3′) and found that the cross-linking efficiency of various samples was similar (Fig. 2A). Positive controls for inter-chromosomal (Ch7:ChX) were prepared by mixing the PCR fragments of GAPDH locus and X chromosome, digesting with BglII and BamHI, and then randomly ligating the restriction fragments. No detectable inter-chromosome crosslinking were detected after 40 cycles of PCR amplification.
The relative cross linking frequency between the anchor fragment and the fragment of interest (f(x)) was normalized to the cross linking frequency between the anchor fragment and its adjacent 5′ fragment. The relative cross linking frequency can be expressed as
Where IS(X:A) is the intensity of PCR products amplified from ligation products between the anchor fragment A and any given fragment X in the rearranged genomic DNA of ES cells; IC(X:A) is the intensity of PCR products amplified from ligation products between the anchor fragment A and any given fragment X in the positive control template; [IS(Y:A) is the intensity of PCR products amplified from ligation products between the anchor fragment A and the fragment (fragment Y) that yield the fewest ligation products in the rearranged genomic DNA of ES cells; IC(Y:A) is the intensity of PCR products amplified from ligation products between the anchor fragment A and the fragment Y in the positive control template.
RESULTS
Multiple clusters of DNAse I hypersensitive sites within the Xic
Previous studies showed that clustering of regulatory elements tend to occur at or around DHS (Tolhuis et al., 2002). At the Xic, DHS have previously been located at the Xist promoter (Hendrich et al., 1997; Navarro et al., 2005; Sado et al., 2005; Sheardown et al., 1997), at the 5′ end of Tsix (Stavropoulos et al., 2005), and all across Xite (Ogawa and Lee, 2003). To identify additional DHS within the Xic, we carried out DHS assays across regions that were not previously studied. We performed DHS assays in male and female ES cells in the pre-XCI state of the undifferentiated cells (d0) and the post-XCI state represented by differentiated ES cells (d12).
The analysis revealed multiple new DHS (Fig. 1). Within Xist, we found several clustered DHS. At the promoter, we observed three sites, two of which showed developmental-and sex-specific regulation (Fig. 1A), consistent with previous analysis (Hendrich et al., 1997; Navarro et al., 2005; Sado et al., 2005; Sheardown et al., 1997). While DHS-a, DHS-b and DHS-c were present in d0 in both male and female cells, DHS-b and DHS-c only persisted in the d12 female cells. In addition, we found novel DHS within exon 1 (Fig. 1B), the first intron (Fig. 1C), and the last intron (Fig. 1D).
Figure 1. Specialized chromatin structures within the Xic.
(A–E) A schematic of the location of DHS identified is shown above each set of panels. Southern blots show chromatin digested, from left to right, with DNase concentrations 0, 0.5, 1, 2.5, 5, 10 μg/ml. Unless stated otherwise in the text, identical results were generated for female cells (data not shown). The restrictions enzyme sites are marked as black lines and the positions of the probes shown in red. Constitutively active DHS are shown by dark grey arrowheads, DHS observed in undifferentiated ES cells are indicated by green arrowheads. (F) Summary of DHS described in this study and previously. Within cluster 2, DHS ‘a’ corresponds to HS3, DHS ‘b’ corresponds to HS1 and DHS ‘c’ to HS5 (Sado et al., 2005; Sheardown et al., 1997). (G) MAR predication software shows four major peaks of MAR potential greater than 0.6. MAR criteria analyzed included origin of DNA replication, AT- and TG-richness, curved and kinked DNA and topoisomerase II recognition. The window width was set to 300, the slide distance was 50 and the run length was set to 3. Asterisks indicate conservation to human sequence. A schematic of the Xic is shown below to specify the MAR locations.
The analysis also revealed another hypersensitive site approximately 10 kb upstream of the Xist promoter (Fig. 1E). This DHS is located at the 5′ end of another non-coding gene known as Jpx or Enox (Chureau et al., 2002; Johnston et al., 2002) and appears to be constitutive in differentiating male and female ES cells (Fig. 1E). Its constitutively hypersensitive profile is consistent with the gene’s biallelic status in female cells and constitutive expression profile during development (Chureau et al., 2002; Johnston et al., 2002). Combined with previous analyses, these data showed that the 100 kb region of the mouse Xic has multiple DHS in all non-coding loci, many of which are clustered (Fig. 1F). Analysis by the MAR-Wiz (v1.5) prediction software revealed that several DHS clusters lay close to potential matrix attachment regions (MARs) (Fig. 1G), two of which were predicted to be conserved from mouse to humans. At Jpx, the MAR-like sequence lay immediately downstream of the promoter and DHS. At Xist, one coincided with DHS cluster 2 and another with the intronic cluster 4. At Tsix, the putative MAR lay in the major promoter and coincided with DHS cluster 6. Thus, although the existence of MARs is an in silico prediction, the MAR and DHS data together suggest that specialized chromatin reside in discrete domains within the Xic.
3C analysis reveals an Xist-Jpx interaction domain
To look for 3-D interactions among Xic domains in vivo, we utilized the ‘chromosome conformation capture’ (3C) technique, which identifies two regions of interacting DNA by cross-linking chromatin in vivo (Dekker et al., 2002; Tolhuis et al., 2002). Prior to analysis of samples, multiple control experiments were performed to test for cross-linking efficiency, the quantitative nature of PCR, and equal PCR efficiencies of the primer pairs used (Dekker, 2006). As a control for cross-linking efficiency, we carried out PCR using primers that amplify the rearranged GAPDH locus (Spilianakis and Flavell, 2004) and found similar cross-linking efficiencies among all samples, whereas no cross-linking was seen in the negative control PCR between Gapdh and Xist or Tsix (an example is shown in Fig. 2A). For analysis of test samples, cells were treated with formaldehyde to cross-link interacting loops of DNA, the DNA was digested by BamHI and BglII to release separate complexes that were not cross-linked, and the resulting fragments diluted to promote intra-molecular reactions during ligation. We then carried out semi-quantitative PCR analysis on ligation products to determine whether two non-contiguous Xic sequences interact by 3-D looping. The quantitative nature of the PCR was ensured by carrying out the analysis in the exponential phase of amplification, as confirmed by appropriate decreases in product levels in serial dilutions of template (an example is shown in Fig. 2B; described in full in the Methods section). To create a pool of all possible interacting products as control, we digested an 80-kb P1 construct, πJL2 (Lee et al., 1999b), with BamHI and BglII, and randomly ligated the resulting fragments (Fig. 2C, panel e). For all 3C analyses below, at least 2 PCR analyses have been carried out for each of 2–3 independent cross-linking experiments. Representative results are shown throughout.
To look for Xist promoter interactions, we carried out 3C with an Xist promoter primer as the invariant ‘anchor’ (Fig. 2C, grey shading, red arrow) and a second, variable primer situated in distinct restriction fragments across the Xic region. We found that the overall interaction pattern in undifferentiated (d0) male and female cells was very similar, as all PCR amplifications with the Xist promoter anchor primer yielded similar band intensities across the Xist locus (Fig. 2C, compare panels a to c). However, the interaction frequency seemed to decrease 3′ of the last Xist exon (exon 8), as the PCR bands from the Tsix/Xite region were consistently less intense (separated by a dotted red line). In contrast, the control amplification performed in parallel was essentially equal across all Xic regions (Fig. 2C, panel e). These results suggested that, in d0 ES cells, chromatin within the Xist gene body and the immediate upstream region is in close contact as compared to chromatin lying downstream in the Tsix/Xite region.
We next looked for interactions that showed dynamic changes in a sex- or developmentally specific manner and found several noteworthy changes. The close contacts in the region in between Jpx and Xist varied during cell differentiation and between XX and XY cells. In male ES cells, differentiation led to a significant and reproducible loss of the interactions on d4 (data not shown) and d10 (Fig. 2C panel d). In female cells, differentiation also resulted in a reproducible decrease in interactions, though not to the same extent as seen in the male cells (Fig. 2C panel b). These observations indicated that the region spanning Jpx and Xist forms a compact structure in pre-XCI cells (d0), a time when Xist is at a basal transcriptional state but is poised to undergo full transcriptional activation (Navarro et al., 2005; Sado et al., 2005; Sun et al., 2006). The structure becomes relaxed on the single Xa in differentiated male cells, correlating with Xist expression. The partial loss of interactions in post-XCI female cells suggested differences between Xa and Xi, with the compact structure maintained on one but not the other X. Alternatively, a partial loss of compaction may occur on both Xs.
Given the chromosome conformation in the Jpx-Xist domain, we looked for specific contact points in this region and noticed further interactions that showed differentiation- and sex-specific dynamics. In particular, the contact between the Xist promoter and fragment iii, which carries the 5′ end of Jpx, occurred in both d0 male and female cells; however, while the interaction was reproducibly sustained in d10 female cells, the contact was specifically lost in d10 male cells (Fig. 2C, fragment iii: b versus d).
To verify these results, we carried out the reciprocal 3C analysis by anchoring a primer at the 5′ end of Jpx, just downstream of the promoter (Fig. 2D). A relatively high crosslinking frequency was seen for adjacent fragments as expected for closely linked regions, but the frequency dropped further upstream at fragment vi. Significantly, however, the frequency increased again at fragment vii, the fragment that spans the Xist promoter. This is a reproducible pattern observed in two PCR tests on two independent cross-linking experiments. Therefore, the 5′ ends of Jpx and Xist make contact. We furthermore found that, while the contact was preserved in female cells on d4 and d10 and in fibroblasts (Fig. 2D), the contact was lost in male cells on d4, d10 and in fibroblasts. Together, these data demonstrated that an interaction between the 5′ end of Jpx and the Xist promoter takes place only in cells that trans-activate Xist or are poised to do so. A loss of the Jpx-Xist interaction strictly correlated with repression of the Xist gene on Xa.
Tsix and Xite form a distinct interaction domain
Because the above analysis indicated that Tsix and Xite do not participate in the Xist-Jpx interactions, we next asked whether Tsix and Xite formed their own interaction domain. Indeed, 3C examination showed a similar ‘domain’ structure spanning Xite and Tsix using quantitative PCR assays (Fig. 3A,B). In d0 female ES cells, significant peaks of interaction could be observed throughout Xite and the 5′ half of Tsix (Fig. 3A). The interaction abruptly ceased at the last exon of Xist (Fig. 3A). Two interactions yielded 10- to 30-fold higher cross-linking frequencies above background, one involving the Tsix promoter and the 1.2 kb Xite enhancer, and the other involving the Tsix promoter and the 3′ end of Xist (Fig. 3C,D).
To confirm the interaction between the Tsix promoter and Xite, we anchored a primer in Xite and carried out the reciprocal 3C analysis (Fig. 4A). Because the Xite DHS span several kilobases (Ogawa and Lee, 2003; Stavropoulos et al., 2005), we tested anchors in two BamHI-BglII fragments and obtained similar results for each (Fig. 4B,C). As expected, the reciprocal analysis identified an interaction between the Xite enhancer and the Tsix promoter in the case of both tested anchors. The developmentally specific nature of these interactions suggests that the contacts occur in vivo and are not artifacts of the 3C technique.
Figure 4. Dynamic interaction between Xite and Tsix.
(A) Representative gel images of 3C analysis using the anchor primer localized to the DNA fragment bearing the Tsix promoter and the rest of the Xic.
(B) Relative cross-linking frequencies between the anchor fragment containing the 1.2kb Xite enhancer and the minimal Xic region. (C) Relative cross-linking frequencies between the anchor fragment containing fragment 5′ to the Xite 1.2 kb enhancer and the minimal Xic region.
Current models postulate that the Xite enhancer acts on Tsix only at the onset of XCI and not in post-XCI cells (Ogawa and Lee, 2003; Stavropoulos et al., 2005), as Tsix expression is extinguished shortly after XCI takes place. Consistent with this idea, the contact between the Xite enhancer and the Tsix promoter occurred in d0 and d4 cells, but was sharply reduced on d10 (Fig. 3,4, and data not shown). These results were also consistent with the loss of the Tsix promoter DHS (Stavropoulos et al., 2005) and the weakening of multiple DHS in Xite (Ogawa and Lee, 2003) in differentiated ES cells. Thus, an interaction between Xite and Tsix during XCI may enable one X-chromosome to persist as Xa. Indeed, the same interaction was observed for the single Xa of male ES cells (Fig. 3D). Combined, these data showed that Xite makes critical contacts with Tsix in cells where the antisense gene is highly expressed and that the contacts are lost once the window of XCI is past. Because the contact occurs in 3-D between discontiguous elements, we propose that the Tsix promoter and the Xite enhancer engage each other through ‘looping’ interactions, coming together in space to form a ‘chromatin hub’ which enables the antisense gene to persist briefly on the future Xa.
Comparative analysis of 3C profiles for the Jpx-Xist and Xite-Tsix domains highlight a striking interaction boundary within the last exon of Xist. A sharp boundary near exon 8 was easily discerned when viewed either proximally (Fig. 2) or distally (Fig. 3). When the anchor primer was placed in the Tsix promoter, strong interactions could be observed across Tsix and Xite, but all interactions abruptly decreased beyond Xist exon 8 (Fig. 3A and C). When an anchor primer was placed in the Xist promoter, the interactions likewise ceased beyond Xist exon 8 (Fig. 2C). The fact that this chromatin border coincided with DHS5 (Fig. 1D,F) raised the possibility that the 3′ end of Xist could harbor a distinct physical structure to separate the Jpx-Xist domain from that of Xite-Tsix.
Curiously, this border region demonstrated strong contacts with both the Xist promoter and the Tsix promoter. In undifferentiated male and female ES cells, the Tsix promoter consistently showed robust interaction with the Xist exon 8 fragment (Fig. 3C). The Xist promoter similarly made contacts with Xist exon 8 (Fig. 2C). Also notable was the fact that these interactions were lost during and after XCI. The region around Xist exon 8 may therefore function as an interaction boundary between the Xite-Tsix and Jpx-Xist chromatin domains.
Chromosome conformation in Tsix mutants
We observed some of the most dynamic changes in the fragment containing the Tsix major promoter and the repeat element, DXPas34 (Courtier et al., 1995). DXPas34 has been noted for its multiple Ctcf-binding sites (Chao et al., 2002), its differentially methylated domains in sperm and oocytes (Boumil et al., 2006), and its role in X-chromosome counting and choice (Cohen et al., 2007; Lee, 2005; Vigneau et al., 2006). In transcription assays in vitro, DXPas34 is absolutely required to enhance Tsix transcription (Stavropoulos et al., 2005). To determine whether DXPas34 may mediate the Tsix-Xite interactions, we analyzed two recently generated alleles of Tsix (Cohen et al., 2007). First, we tested the ΔDXPas34 allele, which specifically deletes the repeat element without affecting the Tsix major promoter and surrounding sequences (Fig. 5A). In female cells carrying a targeted deletion of DXPas34, the mutated X-chromosome down-regulates Tsix and is favored to be inactivated (Cohen et al., 2007). Male ES cells were used in order to examine the mutated Xic in isolation. The ΔDXPas34 allele retains all the restriction sites of the wild-type allele, thus allowing us to compare data obtained from mutant and wild-type cells. By anchoring the 3C assay at the Tsix promoter (Fig. 5B) or the Xite enhancer (Fig. 5C), we found that deleting DXPas34 did not significantly alter the chromosome conformation profile anywhere within the Xic at any time of differentiation. Therefore, DXPas34 is not absolutely required for the formation of the developmentally-regulated clustering of Xite with the Tsix promoter. Nor is DXPas34 necessary for Tsix’s interaction with the interaction boundary.
Figure 5. Chromosome conformation in a Tsix loss-of-function and a gain-of-function mutant.
3C analysis of chromatin from male (J1) mutant ES cells. A schematic of the Xic region tested is shown above each panel. BamHI and BglII-digested restriction fragments are shown as grey lines and the anchor primer regions as a grey box. (A) Schematic of the genomic region analyzed in the wild-type and ΔDXPas34 deletion male cell lines. (B) Relative cross-linking frequencies between stable fragment containing the Tsix promoter and DXPas34 repeat region and the rest of the Xic. (C) Relative cross-linking frequencies between stable fragment containing the 1.2kb Xite enhancer and the minimal Xic region.
We next tested the gain-of-function allele, ΔPneo, an allele that carries a Neo insertion in place of a deleted Tsix promoter. This deletion/insertion allele unexpectedly resulted in Tsix hyperactivity and the constitutively active state of the linked X-chromosome (Cohen et al., 2007) – a neomorphic allele believed to stem from the ectopic enhancer activity conferred by the Pgk1-driven Neo cassette. Heterozygous females carrying this mutation show non-random inactivation of the wild-type X, thereby providing an excellent opportunity to test how the Tsix-Xite interaction is altered either as a cause or consequence. We carried out allele-specific analysis of the ΔPneo mutation in female heterozygotes – an assay made possible by two adjacent BamHI sites created by the Neo insertion (Fig. 6A). Intriguingly, while the overall 3C profile was relatively unchanged (Fig. 6B), the allele-specific 3C assay at position ii showed that Xite contacted both Tsix alleles on d0 but preferentially engaged the ΔPneo allele upon differentiation (Fig. 6C). This correlated with the gain-of-function in Tsix and constitutive activity of the ΔPneo chromosome in differentiating ES cells. By contrast, the wild-type allele of the ΔPneo/+ mutant could not sustain its interaction with the Xite enhancer during differentiation, correlating with the inability of the wildtype X to remain active. These results demonstrate a correlation between positive Xite-Tsix interactions and the choice of the linked X-chromosome to be Xa. They therefore raise the possibility that the 3D interactions may mediate the stimulation of Tsix by Xite.
Figure 6. Chromosome conformation in a gain-of-function mutant.

3C analysis of chromatin from female (EL16) mutant ES cells. A schematic of the Xic region tested is shown above each panel. BamHI and BglII-digested restriction fragments are shown as grey lines and the anchor primer regions as a grey box. (A) Schematic of the wild-type allele and ΔPneo gain-of-function allele. The blue line of the wild type allele indicated the restriction site present only in the wild type allele. The red lines in the ΔPneo allele indicated the additional BglII and BamHI restriction sites in the mutant alleles. The anchor primer used is shown as grey shaded areas. (B) Ethidium bromide-stained gels showing PCR products representing interaction between the anchor fragment and various DNA fragments as indicated in (A). Wild-type and ΔPneo cells were analyzed at d0 (before XCI) and d10 (after XCI). The red rectangles indicated the interactions between the anchor fragment and the fragment bearing either the wild type sequence or exogenous Neo promoter. Note that the doublet reflects interaction with two alleles at position ii. (C) Higher resolution agarose gel analysis of allele-specific PCR products ii amplified from undifferentiated (d0) and differentiated (d10) mutant ES cells.
DISCUSSION
Long-range interactions have previously been demonstrated in several mammalian loci with complex patterns of gene regulation. In mouse and human, adult β-globin genes interact with DHS of the LCR located 40–60kb away by looping out the intervening sequence (de Laat and Grosveld, 2003; Patrinos et al., 2004; Tolhuis et al., 2002). These interactions regulate gene expression by the formation of an ‘active chromatin hub’ (ACH), a permissive higher order structure for developmentally-specific expression states (Tolhuis et al., 2002). Similar ACH formation has been reported for the mouse α-globin locus (Zhou et al., 2006). Here, we propose that the XCI regulation by the non-coding genes also involves formation of ACH (Fig. 7). According to our model, the Xic would be partitioned into two compact hubs prior to XCI. ACH-1 would encompass Xist and Jpx (Fig. 2A) and define a hub that directs silencing on the future Xi. ACH-2 would encompass all of Xite and the 5′ half of Tsix (Fig. 3A) and define a hub that blocks silencing on the future Xa. The transition between ACH-1 and -2 is sharp, perhaps made possible by an interaction boundary comprising specialized chromatin. It appears that this interaction boundary makes contacts with elements on both sides -- the Xist promoter proximally (Fig. 2) and with the Tsix promoter distally (Fig. 3).
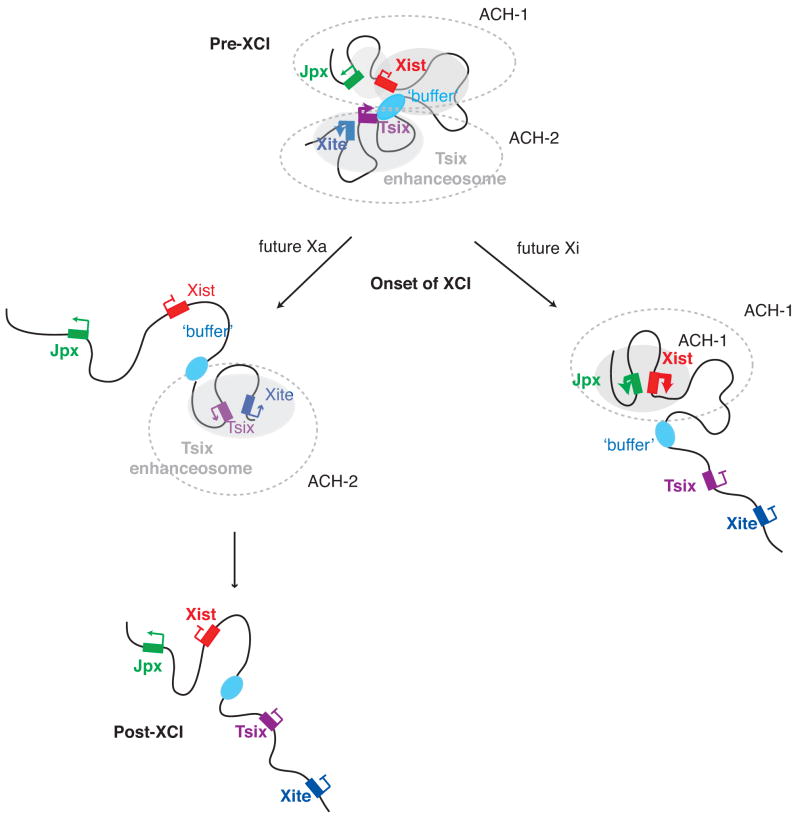
Figure 7. Dynamic chromosome conformation changes at the Xic during XCI.
Model depicting changes in the structure of Xic during X chromosome inactivation. Prior to XCI the Tsix and Xite form an active chromatin hub that promotes Tsix transcription, which in turn prevents the full activation of Xist. During this time, Jpx makes contact with Xist in a state that is “poised” for activation. Upon cellular differentiation, Tsix-Xite interactions disintegrate on the future Xi, resulting in the loss of Tsix expression and enabling productive contacts between Xist and Jpx. Xist is then fully transactivated. On the future Xa, the Xite-Tsix interaction persists, thereby enabling continued expression of Tsix and permanent downregulation of Xist (Ogawa and Lee, 2003). The Jpx-Xist interaction is lost. After the establishment of XCI, the Xite-Tsix interaction is also lost, coincident with downregulation of Tsix expression on Xa.
Each ACH structure would be a physical framework in which specific looping interactions can occur. We suggest that Jpx may be a positive regulator of Xist, consistent with previous analysis showing that a 30-kb region upstream of Xist is required to transactivate Xist (Lee et al., 1999b). In ACH-1, the Xist promoter contacts Jpx in the pre-XCI state when the Xist gene is poised for transcriptional activation but has not been stimulated in full. Events after cell differentiation would then allow a more productive Jpx-Xist interaction for full Xist expression. In males where XCI is absent, ACH-1 and the Xist-Jpx interaction are lost upon cell differentiation. We suggest that ACH-2 forms the basis of contact between the Xite enhancer and Tsix promoter described previously (Ogawa and Lee, 2003). These contacts occur before XCI (d0, Fig. 3A) and persists briefly at the onset of XCI (d4). The significance of these interactions is supported by the gain-of-function ΔPneo allele of Tsix, in which Xite becomes constitutively associated with the 5′ end of Tsix (Fig. 6) and the neomorphic Tsix allele constitutively represses Xist and XCI.
An intriguing observation is the occurrence of an interaction boundary at the 3′ end of Xist, a region of the Xic that has not yet been scrutinized. For some time, Xist was thought to terminate in exon 7 (formerly exon 6) (Brockdorff et al., 1992), but later characterization showed that Xist extends an additional 3 kb or so and includes a small exon 8 (Hong et al., 1999). However, the function of the last intron and exon has been unclear, as truncating Xist after exon 7 does not apparently affect the ability of Xist to initiate X-chromosome silencing (Clerc and Avner, 1998). Therefore, while intron 7 and exon 8 are not required for the silencing event, they may be involved in the upstream regulation of Xist. The interaction boundary may prevent chromatin from one ACH from looping or oozing into the adjacent ACH. We do not currently know whether the interaction boundary is congruent to the chromatin boundary elements and insulators defined in other systems (Bell et al., 1999; Ohlsson et al., 2001). At the Xic, such a chromatin boundary may function by preventing the oozing of active chromatin from Xite-Tsix to Xist-Jpx, and vice versa. It may function additionally by sequestering Xist and Tsix and releasing them for productive contacts only upon cell differentiation. The boundary structure raises the question of whether any direct communication can occur between the Jpx-Xist and Xite-Tsix domains. Using 3C, we failed to detect any significant interaction between them, but such looping contacts may too transient to be detected by the 3C method. We also cannot exclude the possibility that interactions may occur by tracking or linking, rather than looping. Finally, the existence of two distinct structure-function domains raises the possibility of a ‘global control region’ (GCR), one which controls multiple domains within a cluster of genes, as has been proposed for the HoxD/Evx2 gene clusters (Spitz et al., 2003).
Specific proteins responsible for mediating cis-interactions at the Xic have yet to be identified. One candidate is Ctcf, a factor that has been implicated in formation of an active chromatin hub (ACH) at the H19/Igf2 imprinted locus (Kurukuti et al., 2006; Ling et al., 2006) and the β-globin locus (Splinter et al., 2004). At the murine Xic, Ctcf binds multiple sites near the major Tsix promoter (Chao et al., 2002; Donohoe et al., 2007) within the fragment defined by 3C as a point of contact for the Xite enhancer. Furthermore, Ctcf binds the Xist promoter (Pugacheva et al., 2005) in the domain of contact for Jpx. Removing DXPas34 -- the major Ctcf-binding elements in Tsix -- does not affect the formation of the interaction between the Tsix promoter and the Xite enhancer. However, as Ctcf sites in Tsix also lie outside of DXPas34, the residual Ctcf sites may be sufficient for looping interactions.
In the future, we must consider the relationship of looping interactions in cis to chromosome-pairing interactions in trans. The Tsix/Xite domain that defines ACH-2 is known to be critical for homologous chromosome pairing (Bacher et al., 2006; Xu et al., 2006). Significantly, the timing of pairing interactions coincides with the appearance and disappearance of the Tsix-Xite hub. Futhermore, the pairing interaction is now known to involve Ctcf (Xu et al., 2007). Therefore, one interesting possibility is that the Tsix-Xite hub may first promote and then prevent pairing events during the initiation phase of XCI. The delineation of higher order chromatin structure at the Xic is therefore a critical step towards elucidating how the noncoding elements of the Xic regulate many aspects of XCI.
Acknowledgments
We thank all lab members for their input and critical discussion. This work was supported by NIH grant R01 (GM38895) and MGH Fund for Medical Discovery Post-doctoral Fellowship (R.K.R). J.T.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006 doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–96. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Boumil RM, Ogawa Y, Sun BK, Huynh KD, Lee JT. Differential Methylation of Xite and CTCF Sites in Tsix Mirrors the Pattern of X-Inactivation Choice in Mice. Mol Cell Biol. 2006;26:2109–17. doi: 10.1128/MCB.26.6.2109-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–26. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–77. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–7. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, Eggen A, Avner P, Duret L. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 2002;12:894–908. doi: 10.1101/gr.152902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–75. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Avner P. Role of the region 3′ to Xist exon 6 in the counting process of X-chromosome inactivation. Nat Genet. 1998;19:249–53. doi: 10.1038/924. [DOI] [PubMed] [Google Scholar]
- Cohen DE, Davidow LS, Erwin JA, Xu N, Warshawsky D, Lee JT. The DXPas34 repeat regulates random and imprinted X inactivation. Dev Cell. 2007;12:57–71. doi: 10.1016/j.devcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Courtier B, Heard E, Avner P. Xce haplotypes show modified methylation in a region of the active X chromosome lying 3′ to Xist. Proc Natl Acad Sci U S A. 1995;92:3531–5. doi: 10.1073/pnas.92.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11:447–59. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- Dekker J. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Hendrich BD, Plenge RM, Willard HF. Identification and characterization of the human XIST gene promoter: implications for models of X chromosome inactivation. Nucleic Acids Res. 1997;25:2661–2671. doi: 10.1093/nar/25.13.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YK, Ontiveros SD, Chen C, Strauss WM. A new structure for the murine Xist gene and its relationship to chromosome choice/counting during X-chromosome inactivation. Proc Natl Acad Sci U S A. 1999;96:6829–34. doi: 10.1073/pnas.96.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Newall A, Brockdorff N, Nesterova T. Enox, a Novel Gene That Maps 10 kb Upstream of Xist and Partially Escapes X Inactivation. Genomics. 2002;80:236. doi: 10.1006/geno.2002.6819. [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–9. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat Immunol. 2005;6:42–8. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Lee JT. Regulation of X-chromosome counting by Tsix and Xite sequences. Science. 2005;309:768–71. doi: 10.1126/science.1113673. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999a;21:400–4. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N, Han Y. Genetic analysis of the mouse X inactivation center defines an 80-kb multifunction domain. Proc Natl Acad Sci U S A. 1999b;96:3836–41. doi: 10.1073/pnas.96.7.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li Q, Harju S, Peterson KR. Locus control regions: coming of age at a decade plus. Trends Genet. 1999;15:403–8. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–72. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–13. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–3. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–93. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: implications for X-chromosome inactivation. Genes Dev. 2005;19:1474–84. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Lee JT. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell. 2003;11:731–43. doi: 10.1016/s1097-2765(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–7. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Patrinos GP, de Krom M, de Boer E, Langeveld A, Imam AM, Strouboulis J, de Laat W, Grosveld FG. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004;18:1495–509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–7. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of x chromosome inactivation. Annu Rev Genet. 2002;36:233–78. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- Pugacheva EM, Tiwari VK, Abdullaev Z, Vostrov AA, Flanagan PT, Quitschke WW, Loukinov DI, Ohlsson R, Lobanenkov VV. Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and preemptive choices of X chromosome inactivation. Hum Mol Genet. 2005;14:953–65. doi: 10.1093/hmg/ddi089. [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–65. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 2001;128:1275–86. doi: 10.1242/dev.128.8.1275. [DOI] [PubMed] [Google Scholar]
- Sheardown SA, Newall AE, Norris DP, Rastan S, Brockdorff N. Regulatory elements in the minimal promoter region of the mouse Xist gene. Gene. 1997;203:159–68. doi: 10.1016/s0378-1119(97)00507-6. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–27. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–17. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Splinter E, Grosveld F, de Laat W. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- Stavropoulos N, Rowntree RK, Lee JT. Identification of developmentally specific enhancers for Tsix in the regulation of X chromosome inactivation. Mol Cell Biol. 2005;25:2757–69. doi: 10.1128/MCB.25.7.2757-2769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–28. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vigneau S, Augui S, Navarro P, Avner P, Clerc P. An essential role for the DXPas34 tandem repeat and Tsix transcription in the counting process of X chromosome inactivation. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0602381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet. 2007;39:1390–6. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. Transient Homologous Chromosome Pairing Marks the Onset of X Inactivation. Science. 2006 doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC. Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol Cell Biol. 2006;26:5096–105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]