Abstract
Quantitative RT–PCR and overexpression studies of two Dicer-like proteins, MoDcl1 and MoDcl2, in Magnaporthe oryzae indicated that the functional diversification of the MoDcl1 and MoDcl2 proteins in RNA-mediated gene silencing pathways was likely to have arisen from both transcriptional control and protein specialization.
DICER is a member of the RNase III superfamily of bidentate nucleases that process long dsRNA precursors into shorter units (21–30 nt) that subsequently act as specificity determinants in various RNA-mediated gene silencing pathways. Genome sequencing projects have revealed that Dicer-like (DCL) proteins are evolutionarily conserved in a wide range of eukaryotic genomes although the number of DCL proteins in the genome varies among different organisms. Mammals have only one DCL protein, which participates in at least two different types of gene silencing pathways, namely, siRNA and miRNA pathways (Billy et al. 2001; Hutvagner et al. 2001). In contrast, multiple DCL proteins have been found in other organisms such as Drosophila melanogaster and Arabidopsis thaliana. The multiple DCL proteins are shown to mainly function in discrete RNA silencing pathways with some overlap in specificity (Jaskiewicz and Filipowicz 2008). It is of interest to know how the functional diversification of paralogous Dicer proteins has been achieved along with the development of various RNA silencing pathways during evolution.
In the model filamentous fungus Neurospora crassa, two different forms of RNA-mediated gene silencing, namely, quelling (Romano and Macino 1992) and meiotic silencing by unpaired DNA (MSUD) (Aramayo and Metzenberg 1996; Shiu et al. 2001), have been identified. Quelling operates during the vegetative phase while MSUD acts during the meiotic phase. Quelling belongs to the broad category of gene silencing pathways exemplified by RNAi, in which a specific mRNA is targeted for degradation via an siRNA-mediated pathway (referred to as “RNAi pathway” hereafter). MSUD silences the expression of such genes that cause unpaired DNA during meiosis in the zygote. Quelling has been shown to be mediated by qde-1 (RdRP) (Cogoni and Macino 1999), qde-2 (Argonaute) (Cogoni and Macino 2000), and two redundantly functioning Dicer proteins, dcl-1/sms-3 and dcl-2 (Galagan et al. 2003; Catalanotto et al. 2004), with a probable greater contribution of dcl-2 to the pathway (Catalanotto et al. 2004; Maiti et al. 2007). Interestingly, MSUD uses paralogous proteins of the quelling components such as sad-1 (RdRP) (Shiu et al. 2001) and sms-2 (Argonaute) (Lee et al. 2003). Recently, DCL-1/SMS-3 protein but not DCL-2 protein was shown to be required for MSUD and to colocalize with SAD-1 and SMS-2 in the perinuclear region (Alexander et al. 2008).
In the rice blast fungus Magnaporthe oryzae, two DCL genes, designated MoDcl1 and MoDcl2 (formerly Mdl1 and Mdl2, respectively), have been identified (Kadotani et al. 2004). MoDcl1 and MoDcl2 are orthologous to Neurospora dcl-1/sms-3 and dcl-2, respectively. We previously reported that, in Magnaporthe, MoDcl2 was solely responsible for the RNAi pathway in mycelia (Kadotani et al. 2004). Therefore, unlike Neurospora, no redundancy between the two DCL proteins was observed, suggesting a clear functional diversification of the M. oryzae Dicers. Here we show that this functional diversification likely stems from a combination of transcriptional control and protein specialization.
First, we examined the expression levels of MoDcl1 and MoDcl2 mRNAs in the vegetative mycelia of the M. oryzae isolate Br48 (a wheat-infecting strain) by quantitative RT–PCR (qRT–PCR). Three independent qRT–PCR analyses indicated that 689.1. ± 97.8 copies of MoDcl1 and 9267.7 ± 935.8 copies of MoDcl2 transcripts on average were detected in 25 ng of total RNA from mycelia (Table 1). Therefore, the average copy number of MoDcl2 mRNA was ∼13.5 times higher than that of MoDcl1 mRNA in vegetative mycelia of M. oryzae.
TABLE 1.
Absolute quantitative RT–PCR (qRT–PCR) analysis of MoDcl1 and MoDcl2 in the overexpressing transformants and wild type of Magnaporthe oryzae
| mRNA copies (per 25 ng total RNA)
|
Relative fold to wild type
|
|||
|---|---|---|---|---|
| Strain | MoDcl1 | MoDcl2 | MoDcl1 | MoDcl2 |
| Wild type (Br48) | 689.1 ± 97.8a | 9,267.7 ± 935.8 | 1 | 1 |
| tMoDcl1-OE11 | 248,853.2 ± 15,017.6 | NE | 361.2 | |
| tMoDcl1-OE12 | 154,310.5 ± 8,766.6 | NE | 223.9 | |
| tMoDcl1-OE13 | 224,692.2 ± 3,004.1 | NE | 326.1 | |
| tMoDcl2-OE21 | NE | 24,372.4 ± 1,662.2 | 2.6 | |
| tMoDcl2-OE22 | NE | 252,251.5 ± 19,721.7 | 27.2 | |
| tMoDcl2-OE23 | NE | 13,400.5 ± 1,140.8 | 1.4 | |
Quantitative RT–PCR (qRT–PCR) analysis was performed with real-time PCR (Applied Biosystems 7500 or 7300 real-time PCR system) and SYBR Green fluorescence detection (SYBR GreenER two-step qRT–PCR kit for universal; Applied Biosystems, Foster City, CA). Total RNA was extracted from fungal mycelia grown in liquid media as described previously (Kadotani et al. 2004), and reverse transcribed into cDNAs using a SuperScript III RT–PCR system (Invitrogen). Sets of specific primers for MoDcl1 (MDL1-Fw: CCCGTGGATACTGTGGTAATGG, MDL1-Rv: GCAATGTCGATTGGAATATCAAGAC) and MoDcl2 (MDL2-Fw: CGCTCTACTTCCAGTCCCTTTC, MDL2-Rv: TGGTCACGTCTGGATCAAAGC) as well as for two internal controls, actin (Mo-actin-F: GCGGTTACACCTTCTCTACCAC, Mo-actin-R: AGTCTGGATCTCCTGCTCAAAG), and β-tubulin genes (Mo-tub-F: CGAGACCTTCTGCATTGACAAC, Mo-tub-R: GGCCGAAACCAGGTAGTTCA), were used in the analysis. The copy numbers of the MoDcl1 and MoDcl2 transcripts relative to the total RNA were determined by real-time RT–PCR with reference to standard plasmids carrying MoDcl1 and MoDcl2 cDNAs, respectively. NE, not examined.
Standard deviation.
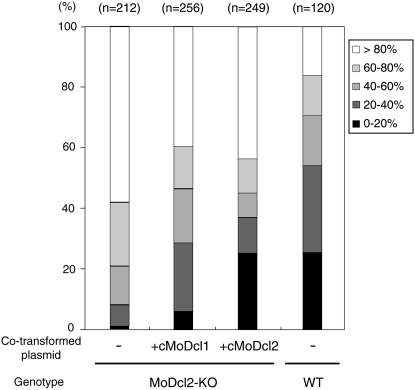
To examine whether different levels of transcriptional activity could play a role in the functional diversification of MoDcl1 and MoDcl2 proteins in RNA silencing pathways in M. oryzae, we performed overexpression (OE) experiments. The full-length MoDcl1 and MoDcl2 cDNAs were introduced into the OE vector, pGT, which carries multiple cloning sites between the Aspergillus nidulans gpdA (glyceraldehyde-3-phosphate dehydrogenase) promoter (∼2 kb in length) and A. nidulans trpC terminator (∼0.5 kb). A GFP-expressing MoDcl2 knockout (KO) mutant, which was previously constructed by a conventional gene replacement method, was transformed with the MoDcl1 or MoDcl2 OE vectors together with a GFP-silencing plasmid, pEGFP-SA-neo (Kadotani et al. 2004). As a control, pEGFP-SA-neo was introduced into a GFP-expressing wild-type strain. The resulting transformants were classified into five categories on the basis of the relative GFP fluorescence levels to their parent strains (Figure 1). The results revealed that the RNAi phenotype of the parent strain (MoDcl2 KO mutant) was completely restored to the wild-type levels in the MoDcl2-OE lines (tMoDcl2-OE). Surprisingly, GFP silencing was also observed in the MoDcl1-OE lines (tMoDcl1-OE), although the average level of silencing was significantly lower than those in the tMoDcl2-OE and wild-type strains (P < 0.01 by t-test).
Figure 1.—
Overexpression of MoDcl1 cDNA partially restored hairpin RNA-induced gene silencing in the MoDcl2 KO mutant. The overexpression (OE) vectors of cMoDcl1 and cMoDcl2 were introduced into the GFP-expressing MoDcl2 knockout mutant by the cotransformation method with the GFP silencing construct, pEGFP-SA-neo carrying the geneticine resistant gene (Kadotani et al. 2004). As a control, pEGFP-SA-neo was introduced into a GFP-expressing strain in a wild-type background. The resulting transformants were classified into five categories (0–20%, 21–40%, 41–60%, 61–80%, and >80%) on the basis of relative fluorescence to the parent strain, and the number of the transformants in each category was plotted in the graph. The full-length cDNAs of the MoDcl1 and MoDcl2 genes were obtained by reverse transcription with oligo dT primers and SuperScript III (Invitrogen, San Diego) and subsequent 5′-RACE.
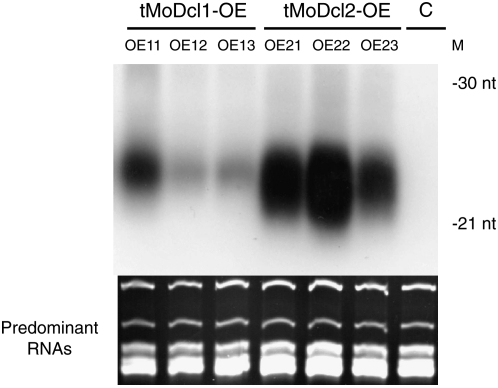
To address whether MoDcl1 protein is capable of producing siRNAs, three strongly silenced transformants (categorized in the 0–20% class in Figure 1) were picked up from the tMoDcl1-OE lines as well as from the tMoDcl2-OE lines and subjected to Northern blot analysis. Consistent with the phenotypic observations in Figure 1, lower levels of GFP siRNA accumulation were, in general, detected in the tMoDcl1-OE lines compared to the tMoDcl2-OE lines (Figure 2). Next we performed qRT–PCR to examine the transcriptional levels of MoDcl1 and MoDcl2 mRNA in the OE lines. The results showed that MoDcl1 and MoDcl2 mRNAs were overexpressed, on average, by 303.7- and 10.4-fold, respectively, relative to the wild-type strain (Table 1). The absolute expression levels of the target genes differed significantly among the OE lines but were at comparable levels in lines strongly expressing either MoDcl1 or MoDcl2 (e.g., tMoDcl1-OE13 and tMoDcl2-OE22 in Table 1). These results indicated the following. First, the overexpressed MoDcl1 protein was capable of generating siRNAs that functioned in the RNAi pathway in M. oryzae. Second, the MoDcl1 protein appeared to be less efficient in Dicer activity since siRNA accumulation was lower in the tMoDcl1-OE lines than in the tMoDcl2-OE lines, despite the relatively higher gene expression levels of the MoDcl1 gene in the transformants.
Figure 2.—
Northern blot analysis of GFP siRNAs in the MoDcl1 and MoDcl2 overexpressing transformants. Three MoDcl1 and MoDcl2 overexpressing transformants each were subjected to siRNA analysis. Low-molecular-weight RNA was extracted and hybridized with a GFP probe as described previously (Kadotani et al. 2004). DNA oligonucleotides (30 and 21 mer) were used as molecular size markers. Equal loading of total RNA was estimated by ethidium bromide staining of predominant RNAs. M, oligonucleotide markers; C, parent strain (GFP-expressing MoDcl2 knockout mutant); tMoDcl1-OE11–13, transformants with the cMoDcl1 OE vector; tMoDcl2-OE21–23, transformants with the cMoDcl2 OE vector.
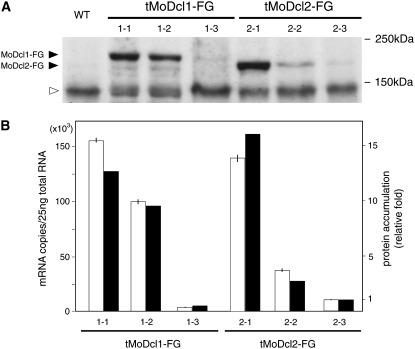
To examine the possibility that the lower Dicer activities in the tMoDcl1-OE lines resulted from lower protein stability, the levels of protein accumulation in the cells were examined by Western blotting. For an accurate comparison using a single antibody, MoDcl1 and MoDcl2 proteins were N-terminally-tagged with 3xFLAG. The FLAG-tagged MoDcl1 (MoDcl1-FG) and MoDcl2 (MoDcl2-FG) proteins were capable of producing siRNAs and inducing GFP silencing with slightly lower efficiencies than the wild-type proteins (data not shown). Three M. oryzae transformants each with the FLAG-tagged MoDcl1 or MoDcl2 gene were subjected to the analysis. The FLAG-tagged MoDcl1 and MoDcl2 proteins were detected at the expected sizes of 191 and 169 kDa, respectively. The levels of protein accumulation were mostly consistent with the mRNA levels assessed by qRT–PCR (Figure 3B), indicating that the MoDcl1 and MoDcl2 proteins have overall similar stabilities in M. oryzae cells. These results support the idea that MoDcl1 protein was less efficient than MoDcl2 protein in producing functional siRNAs in the RNAi pathway.
Figure 3.—
Western blot (A) and qRT–PCR (B) analyses of M. oryzae transformants overexpressing 3xFLAG-tagged MoDcl1 and MoDcl2. (A) By introducing synthesized oligonucleotides (ACCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATCATGATGGT) into the overexpression vectors using a PCR-based cloning strategy, we constructed N-terminally 3xFLAG-tagged MoDcl1 (MoDcl1-FG) and MoDcl2 (MoDcl2-FG). Mycelia of tMoDcl1-FG and tMoDcl2-FG were homogenized in buffer composed of 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 1% Nonidet P-40. The homogenates were centrifuged (10,000 × g, 10 min) and the supernatants were collected. Equal amounts of homogenate protein (75 μg) were applied to 10% SDS–polyacrylamide gel electrophoresis (PAGE), electroblotted onto a Immobilon-P PVDF membrane (Millipore, Bedford, MA), and probed with an anti-FLAG M2 monoclonal antibody (Sigma, St. Louis). Proteins reacting with the antibody were visualized with ECL plus Western blotting detection reagents (GE Healthcare, Piscataway, NJ). The wild-type (WT) strain, Br48, was used as a control. (B) The transformants used in the Western blot analysis were subjected to qRT–PCR as described in the legend of Table 1. On the basis of qRT–PCR, the copy numbers of the transcripts per total RNA quantity were determined with reference to the standard plasmids (open bars). To estimate protein accumulation levels in A, the X-ray film was scanned and densitometrically analyzed with the 1D Quantifier software (Phoretix). Accumulation levels of MoDcl1-FG and MoDcl2-FG protein were normalized with the levels of a nonspecific protein signal (open triangle in A). Relative fold of protein accumulation (solid bars) was calculated with reference to the data of a dilution series (threefold) of MoDcl1-FG protein (data not shown).
Overall, the results of this study lead to two main conclusions. First, MoDcl1 and MoDcl2 are functionally diversified at the protein level, even though both proteins are fundamentally capable of producing siRNAs that guide RNAi. The levels of siRNA accumulation and consequent RNAi efficiencies were significantly lower in the tMoDcl1-OE lines than in the tMoDcl2-OE lines, even when both were overexpressed at similar levels. These findings may suggest that the MoDcl1 and MoDcl2 proteins have roles in different RNA silencing pathways in M. oryzae. The possible differences in the size distributions of the siRNAs produced by MoDcl1 and MoDcl2 proteins (Figure 2) may support this assumption since siRNAs of different sizes have been shown to have roles in discrete RNA silencing pathways in other eukaryotes (Hamilton et al. 2002). The intercellular localizations and/or interacting proteins of the DCL proteins may be involved in protein specialization as suggested in higher eukaryotes (Hiraguri et al. 2005).
Second, transcriptional control seems to play a role in the functional diversification between MoDcl1 and MoDcl2 proteins in M. oryzae. Despite the capability of MoDcl1 protein to produce siRNAs when overexpressed, no detectable RNAi or siRNA accumulation either triggered by hairpin-RNA-expressing transgenes or endogenous repetitive sequences was observed under normal conditions in the vegetative mycelia of the MoDcl2 KO mutant, in which the MoDcl1 gene should remain intact (Kadotani et al. 2004; Murata et al. 2007). This is likely due to transcriptional control that suppresses MoDcl1 expression in the mycelia. To support this hypothesis, we found that the expression levels of MoDcl1 mRNA were upregulated by 10- to 15-fold during the sexual stage (data not shown) where the MoDcl1 protein may function as a Dicer in the MSUD pathway even though it is, so far, not clear whether or not M. oryzae possesses this pathway. In addition, upregulation of MoDcl2 orthologs has been reported in response to RNAi induction in other fungi (Choudhary et al. 2007; Zhang et al. 2008), suggesting that transcriptional control is an important factor for their function.
Accumulating evidence suggests that orthologs of MoDcl2 such as N. crassa Dcl-2, and Cryphonectria parasitica Dcl2, are the major DCL proteins responsible for the RNAi pathway in ascomycete fungi (Catalanotto et al. 2004; Kadotani et al. 2004; Alexander et al. 2008; Zhang et al. 2008). However, the extent of redundancy of the DCL proteins in the RNAi pathway seems to differ among fungal species. In N. crassa, two DCL proteins, Dcl-1/Sms-3 and Dcl-2, were shown to be redundantly involved in the RNAi pathway (Catalanotto et al. 2004). In contrast, only the MoDcl2 gene in M. oryzae was responsible for RNAi under normal conditions (Kadotani et al. 2004). Our results suggest that this apparent discrepancy between N. crassa and M. oryzae may be due to differences in the transcriptional regulation of the DCL proteins in these fungal species.
Fungal DCL proteins could represent a model for studying the functional diversification of paralogous proteins during evolution. At the time of gene duplication, the two resulting gene products should be identical and therefore completely redundant. During evolution, natural selection pressure sustains genetic sequence diversity, leading to protein specialization as well as spatiotemporally specific patterns of gene expression when it occurs in the promoter region. The fungal DCL proteins may be at slightly different stages of functional diversification as paralogous proteins, or different selection pressure may be exerted on the DCL proteins among fungal species. In either case, protein specialization and transcriptional control are important factors for functional diversification of paralogous proteins, as clearly shown in this study.
Acknowledgments
This work was supported in part by funds from Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- Alexander, W. G., N. B. Raju, H. Xiao, T. M. Hammond, T. D. Perdue et al., 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 45 719–727. [DOI] [PubMed] [Google Scholar]
- Aramayo, R., and R. L. Metzenberg, 1996. Meiotic transvection in fungi. Cell 86 103–113. [DOI] [PubMed] [Google Scholar]
- Billy, E., V. Brondani, H. Zhang, U. Muller and W. Filipowicz, 2001. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 98 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C., M. Pallotta, P. ReFalo, M. S. Sachs, L. Vayssie et al., 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, S., H. C. Lee, M. Maiti, Q. He, P. Cheng et al., 2007. A double-stranded-RNA response program important for RNA interference efficiency. Mol. Cell. Biol. 27 3995–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 2000. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. 10 638–643. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422 859–868. [DOI] [PubMed] [Google Scholar]
- Hamilton, A. J., O. Voinnet, L. Chappell and D. C. Baulcombe, 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri, A., R. Itoh, N. Kondo, Y. Nomura, D. Aizawa et al., 2005. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 57 173–188. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl et al., 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293 834–838. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz, L., and W. Filipowicz, 2008. Role of Dicer in posttranscriptional RNA silencing, pp. 77–97 in RNA Interference, edited by P. J. Paddison and P. K. Vogt. Springer, Heidelberg, Germany. [DOI] [PubMed]
- Kadotani, N., H. Nakayashiki, Y. Tosa and S. Mayama, 2004. One of the two dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related siRNA accumulation. J. Biol. Chem. 279 44467–44474. [DOI] [PubMed] [Google Scholar]
- Lee, D. W., R. J. Pratt, M. McLaughlin and R. Aramayo, 2003. An argonaute-like protein is required for meiotic silencing. Genetics 164 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti, M., H. C. Lee and Y. Liu, 2007. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 21 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, T., N. Kadotani, M. Yamaguchi, Y. Tosa, S. Mayama et al., 2007. siRNA-dependent and -independent post-transcriptional cosuppression of the LTR-retrotransposon MAGGY in the phytopathogenic fungus Magnaporthe oryzae. Nucleic Acids Res. 35 5987–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, N., and G. Macino, 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6 3343–3353. [DOI] [PubMed] [Google Scholar]
- Shiu, P. K. T., N. B. Raju, D. Zickler and R. L. Metzenberg, 2001. Meiotic silencing by unpaired DNA. Cell 107 905–916. [DOI] [PubMed] [Google Scholar]
- Zhang, X., G. C. Segers, Q. Sun, F. Deng and D. L. Nuss, 2008. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J. Virol. 82 2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]