Abstract
Cotton (Gossypium hirsutum L.) fibers are single highly elongated cells derived from the outer epidermis of ovules. A large number of genes are required for fiber differentiation and development, but so far, little is known about how these genes control and regulate the process of fiber development. Here we examine the role of the cotton-fiber-specific R2R3 MYB gene GhMYB109 in cotton fiber development. Transgenic reporter gene analysis revealed that a 2-kb GhMYB109 promoter was sufficient to confirm its fiber-specific expression. Antisense-mediated suppression of GhMYB109 led to a substantial reduction in fiber length. Consistently, several genes related to cotton fiber growth were found to be significantly reduced in the transgenic cotton. Our results showed that GhMYB109 is required for cotton fiber development and reveal a largely conserved mechanism of the R2R3 MYB transcription factor in cell fate determination in plants.
COTTON (Gossypium hirsutum L.) is an important economic crop that is extensively used in the textile industry. Cotton fibers are single-celled trichomes derived from epidermal cells of the ovule (Basra and Malik 1984). The fiber development usually consists of four overlapping stages: initiation, primary cell-wall formation, secondary cell-wall formation, and maturation. During the initial stage, ∼30% of epidermal cells (fiber initials) on the ovule surface begin to enlarge and elongate rapidly at or just before anthesis. The primary cell-wall formation starts at anthesis and lasts up to 19–20 days post-anthesis (DPA) (Basra and Malik 1984). The quality and productivity of cotton fibers depend mainly on two biological processes: fiber initiation to determine the number of fibers present on each ovule and fiber elongation to control the final length and strength of each fiber (John and Keller 1996). Synthesis of the secondary wall initiates ∼16 DPA, overlapping with the late primary wall formation, and continues for ∼40 DPA, forming a wall (5–10 μm thickness) of almost pure cellulose. Upon maturity, cotton fibers contain ∼90% cellulose. Thus, research of fiber development not only provides a basic understanding of cell differentiation and elongation, but also identifies potential target genes for genetic improvement of cotton fiber production.
Cotton fibers are seed trichomes, which share many similarities with leaf trichomes. Since both the Arabidopsis thaliana trichome and cotton fibers are single-celled structures of epidermal origin, it is likely that Arabidopsis trichomes could serve as a model for elucidating the genetic mechanisms controlling cotton fiber development (Serna and Martin 2006). For the model plant Arabidopsis, trichome development and root epidermal patterning have been studied in depth, and both processes use a common mechanism involving closely related transcription factors and a similar lateral inhibition signaling pathway (Schneider et al. 1997; Schnittger et al. 1999; Larkin et al. 2003). Transcription factors such as the MYB proteins GLABRA1(GL1) or WEREWOLF(WER), the WD40 proteins TRANSPARENT TESTA GLABRA1 (TTG1), and the basic helix-loop-helix proteins GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3) appear to form a transcription factor complex to determine epidermal trichome patterning in Arabidopsis (Glover 2000; Schiefelbein 2003; Hulskamp 2004; Ramsay and Glover 2005; Serna and Martin 2006). This complex is thought to regulate the homeodomain leucine zipper protein GLABRA2 (GL2) and a small family of single-repeat MYB proteins lacking the transcription activation domains TRIPTYCHON (TRY), CAPRICE (CPC), and ENHANCER OF TRY AND CPC1 (ETC1). GL2 encodes a homeobox (HOX) transcription factor that promotes trichome cell differentiation and growth (Rerie et al. 1994; Szymanski et al. 1998; Ohashi et al. 2002). The single-repeat MYB proteins TRY, ETC1, and ETC2 have been shown to negatively regulate trichome formation and act in a partially redundant manner to mediate the lateral inhibition (Schnittger et al. 1999; Schellmann et al. 2002; Kirik et al. 2004a,b). Similar genes and pathways may be involved during seed trichome development in cotton, although cotton fibers are unicellular and never branch.
Compared with the Arabidopsis trichome, little is known about the molecular control of cotton fiber development. Recent studies on cotton fiber development have been focused largely on gene expression profiles during fiber elongation and secondary cell-wall synthesis (Arpat et al. 2004; Shi et al. 2006; Udall et al. 2006; Wu et al. 2006; Yang et al. 2006; Taliercio and Boykin 2007). Previous results suggested that transcription factors could play important roles in cotton fiber development. So far, a dozen genes encoding transcription factors are found to be expressed in developing cotton fiber cells, and some of them show similarity to Arabidopsis trichome regulators in protein sequences. An earlier work isolated six MYB genes (GhMYB1-GhMYB6) from G. hirsutum (Loguerico et al. 1999). Another cotton R2R3 MYB gene, GaMYB2, complements the Arabidopsis gl1, and its ectopic expression induces a single trichome from the epidermis of Arabidopsis seeds (Wang et al. 2004b). GhMYB25, a homolog of AmmIXTA/AmmYBML1 that controls petal conical cell and trichome differentiation in Antirrhinum majus, is predominately expressed in ovules and fiber cell initials (Wu et al. 2006). A recent work has shown that a gene similar to AtCPC that acts as an inhibitor of trichome development in Arabidopsis was identified in fiber initials and appeared to possess the MYB domain but lack the transacting domain, similar to its Arabidopsis counterpart (Taliercio and Boykin 2007). The four putative homologs of TTG1 and GhTTG1–GhTTG4 from G. hirsutum are found to be widely expressed in plant tissues, including ovules and fibers. Two of them were able to complement the Arabidopsis ttg1 mutant (Humphries et al. 2005). Nevertheless, the exact function of these genes in cotton fiber development is not clear. Obviously, cotton fiber cell development is a complex biological process that requires orchestrated changes in gene expression in developmental and physiological pathways (Kim and Triplett 2001; Li et al. 2002; Ji et al. 2003; Arpat et al. 2004; Lee et al. 2006).
Many cotton genes with a fiber-preferential expression have been cloned and characterized. For example, the GhTUB1 gene was preferentially expressed in the elongation stage of fiber development (Li et al. 2002). Fifteen GhACT cDNAs were found to be differentially expressed in various tissues. Specifically, GhACT1 has been found to be predominantly expressed in fiber cells, and its suppression disrupted the actin cytoskeleton and caused reduced fiber elongation, suggesting that GhACT1 plays an important role in fiber elongation but not in fiber initiation (Li et al. 2005). A recent study revealed that the 1-Aminocyclopropane-1-Carboxylic Acid Oxidase1-3 (GhACO1-GhACO3) gene, which is responsible for ethylene production, is expressed at a significantly higher level in rapidly elongating fiber cells, indicating a role of ethylene in cotton fiber cell elongation (Shi et al. 2006). Although several of these genes are involved in fiber development, none of them encodes a transcription factor regulating fiber development.
So far, the molecular control of cotton fiber development remains largely unknown, although cotton is the most important fiber crop for the textile industry. Current understanding of cotton fiber development is limited to computational and expression analyses of high-quality ESTs and the isolation and characterization of fiber-related genes. Therefore, deciphering the molecular control of fiber development will be important for cotton improvement by genetic engineering. In this study, we examined the role of GhMYB109 (Suo et al. 2003), similar to AtGL1/WER, in cotton fiber development using a reverse genetics approach. Our results provide an insight into the molecular mechanism regulating cotton fiber development and reveal a largely conserved mechanism in cell fate determination in plants.
MATERIALS AND METHODS
Plant materials and growth conditions:
Cotton (G. hirsutum cv Coker312 and G. hirsutum L. cv. XZ142) seeds were surface sterilized with 70% ethanol for 30–60 sec and 10% H2O2 for 30–60 min, followed by washing with sterile water. Sterilized seeds were germinated on half-strength MS medium under a 16-hr light/8-hr dark cycle at 28°. Cotyledons and hypocotyls were cut from sterile seedlings as explants for transformation. Tissues for DNA and RNA extraction were derived from cotton plants grown in a greenhouse. Vegetative and reproductive organs and tissues were harvested from the cotton species G. hirsutum L. cv. XZ142 grown under a 30°/21° day/night temperature regime in a greenhouse. Developing ovules were excised from developing flower buds or bolls on various days before or post-anthesis (DPA) relative to the day of anthesis (0 DPA).
Genome Walker PCR and GUS reporter construct:
The unknown regions of the 5′ putative promoter and 3′-end of GhMYB109 were determined using the Universal Genome Walker kit (Clontech, Palo Alto, CA). Briefly, genomic DNA of G. hirsutum L. cv. XZ142 was digested with EcoRV, DraI, PvuII, StuI, and ScaI, respectively. DNA fragments were ligated with a Genome Walker adaptor (5′-GTAATACGACTCACTATAGGGCACGCGTGGTCGACGGCCCGGGCTGGT-3′ and 3′-H2N-CCCGACCA-PO4-5′), which had one blunt end and one end with a 5′ overhang. The primary PCR was performed using the adaptor primer AP1 (5′-GTAATACGACTCACTATAGGGC-3′, forward) and GhMYB109-specific primers GW1 (5′-GAAGTGTGACTGTGTTGTTAAGAACCTG-3′, reverse) for the GhMYB109 promoter. The secondary PCR was performed using primer AP2 (5′-ACTATAGGGCACGCGTGGT-3′, forward) and a nested gene-specific primer GW2 (5′-GAGTAACTTGTCTTCCTCCATTGCCCATAAT-3′, reverse). The 3′-end of GhMYB109 was analyzed in a similar way using primers AP1 and GW3 (5′-GACCATGATTATGAGCTAAGTACACTTGCC-3′, reverse) for primary PCR and AP2 and GW4 (5′-GTACACTTGCCATGATTGACCACTTCCATG-3′, reverse) for secondary PCR. Then a 2-kb putative promoter of GhMYB109 was amplified using two primers (5′-ATAGTCGACTGTGTCAAAGACGACTACTTGAG-3′, forward and 5′-TCTAGAGAGTAACTTGTCTTCCTCCATTGCCCATAAT-3′, reverse).The 2-kb 3′-terminator sequences of GhMYB109 were obtained using two primers (5′-ATGAATTCTATGCTGAGCTTGCCAAGGG-3′, forward and 5′-ATGAGCTCCATCTTAGCTAGAGACTATGTTAT-3′, reverse). The putative promoter region was inserted upstream and the 3′-terminator was inserted downstream of the β-glucuronidase (GUS) reporter gene in pBI101.2 vector (Clontech), giving rise to the GhMYB109∷GUS fuse gene. The construct was completely sequenced to ensure that it did not contain any PCR or cloning errors and used for cotton transformation.
Plasmid constructs:
The coding region of GhMYB109 was subcloned into appropriately digested pBI121 vector (Clontech) in the antisense orientation, downstream of the cauliflower mosaic virus (CaMV) 35S promoter. The primers used were 5′-ATAGAGCTCATGGCCGGGGATACAAAAAGG-3′ (forward) and 5′-TATTCTAGACCCGAATCTAATAACATAGTC-3′ (reverse). The constructs were completely sequenced to ensure that they did not contain any PCR or cloning errors and used for cotton transformation.
Cotton transformation:
Cotton transformation was performed as previously described (Li et al. 2005). The constructs were introduced into Agrobacterium strain AGL-1 used for transformation. Cotyledon and hypocotyl explants from G. hirsutum cv Coker 312 were transformed using Agrobacterium-mediated transformation. Homozygosity of transgenic plants was determined by segregation ratio of kanamycin selection marker and further confirmed by DNA gel blot, real-time PCR, RT–PCR, and histochemical assay.
Histochemical assay of GUS gene expression:
Histochemical assays for GUS activity in transgenic cotton plants were conducted as described previously (Wang et al. 2004a). The samples were cut into 5- to 7-mm-thick sections using a Leica microtome. The sections were examined and photographed under a Leica DMR microscope equipped with dark-field optics.
Scanning electron microscopy:
For examining fiber initiation and elongation, fresh ovules were dissected out and placed on double-sided sticky tape on an aluminum specimen holder and frozen immediately in liquid nitrogen. The frozen sample was viewed with a JSM-5310LV scanning electron microscope (JEOL, Tokyo). Fiber density in the stage of initiation was estimated by counting fiber initials per unit area of 100 ×100 μm using a total of 25 unit areas per ovule from the epidermis of ovules under SEM and statistically analyzed. Eight or nine ovules were used for the transgenic and wild-type plants.
DNA gel blot analysis:
Cotton genomic DNA isolation and Southern blotting analysis were performed as described previously (Suo et al. 2003). Genomic DNA (20 μg) was digested, separated on 0.8% agarose gel, and transferred onto Hybond N+ membrane (Amersham, Buckinghamshire, UK). DNA gel blot analysis of G. hirsutum cv Coker 312 and transgenic cottons was carried out using NPTII and GhMYB109 cDNA as probes.
Real-time PCR:
The expression of the GhMYB genes and other fiber-related genes in cotton tissues was analyzed by real-time quantitative RT–PCR (qRT–PCR). From a pool of three to four plants from each line, the bolls were tagged and harvested at the day of anthesis (0 DPA), 1 DPA, and 3 DPA. Total RNA was extracted from immature ovules or fiber-bearing ovules as previously described and digested with DNase I (TaKaRa, Dalian, China) (Suo et al. 2003). qRT–PCR was performed as previously described in all experiments (Lan et al. 2004). In brief, 2 μg of total RNA was used for cDNA synthesis with a SuperScript III first-strand synthesis kit (Invitrogen). The cDNA samples were diluted to 8 and 2 ng/μl. Triplicate quantitative assays were performed on 1 μl of each cDNA dilution using the SYBR Green Master Mix (Applied Biosystems) with an ABI 7900 sequence detection system according to the manufacturer's protocol (Applied Biosystems). Gene-specific primers (Table 1) were designed by using PRIMEREXPRESS software (Applied Biosystems). The relative quantification method (DDCT) was used to evaluate quantitative variation among replicates examined using a P-value of ≤0.05 and a fold change of expression levels greater than or equal to a twofold change as cutoff. Amplification of 18S rRNA was used as an internal control to normalize all data.
TABLE 1.
Primers used for real-time PCR analysis
| Genes | Primers |
|---|---|
| GhMYB109 | 5′-AAGAAGGTGAAATTCTATACAAAAAGG-3′ (forward) |
| 5′-TCCATGGACATTGACATAATCA-3′ (reverse) | |
| GhMYB102a | 5′-CATGTGGGGGAGAAAGAAGA-3′ (forward) |
| 5′-TGAGGCTGTCAAAACTGCTG-3′ (reverse) | |
| GhMYB111 | 5′-GCAAACCCAACCAGAGTCAT-3′ (forward) |
| 5′-GGTGCTGCAAGTGCAATCT-3′ (reverse) | |
| GhMYB139 | 5′-AAACCTGACCCTGACTTTTTCCT-3′ (forward) |
| 5′-TCGATTTCCGAAACGATTCC-3′ (reverse) | |
| GhMYB149 | 5′-GGGTCCGATTTGAGCGATT-3′ (forward) |
| 5′-GGGCTTGTACACCGTGTGAA-3′ (reverse) | |
| GhACO1 | 5′-CTGACAAATCTCAAGTGTACCCC-3′ (forward) |
| 5′-AAGTTAACTGCAGACTCCACG-3′ (reverse) | |
| GhACO2 | 5′-CCCTAAACCCGACCTAATCA-3′ (forward) |
| 5′-AGGAGTTGAAGCCCACTGAC-3′ (reverse) | |
| GhACT1 | 5′-GGAGACTGGATTGTGGTGCTT-3′ (forward) |
| 5′-CGCGCAAACTGGGACTAACT-3′ (reverse) | |
| GhACT5 | 5′-CTCTGAAGCTCCTCTTGGTTC-3′ (forward) |
| 5′-TATCACAGACGAGGGGTTGA-3′ (reverse) | |
| GhTUB1 | 5′-CGGTACCATGGATAGCGTAA-3′ (forward) |
| 5′-TCCCTTAGCCCAATTGTTTC-3′ (reverse) | |
| 18S rRNA | 5′-CGGCTACCACATCCAAGGAA-3′ (forward) |
| 5′-TGTCACTACCTCCCCGTGTCA-3′ (reverse) |
RESULTS
The GhMYB109 promoter is cotton fiber specific:
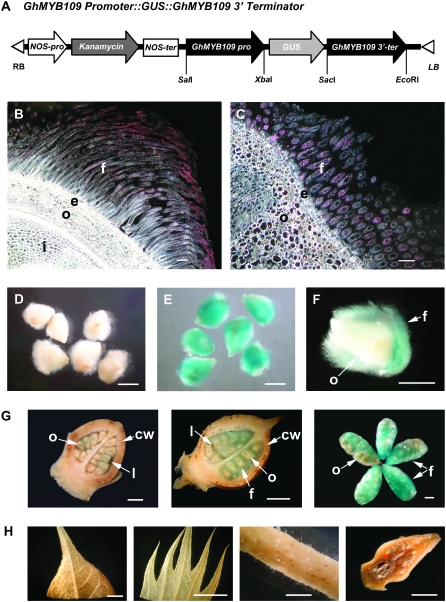
Our previous study showed that the R2R3 MYB transcription factor GhMYB109 was found to be structurally related to AtGL1 and AtWER controlling the trichome initiation in A. thaliana. Our previous study also found that GhMYB109 was specifically expressed in cotton fiber initial cells as well as in elongating fibers (Suo et al. 2003). To better define the expression pattern of GhMYB109 in cotton fibers, a 2-kb putative promoter and 2-kb 3′-terminator sequences of GhMYB109 were inserted downstream of the β-glucuronidase (GUS) reporter gene in the pBI101.2 vector, giving rise to the GhMYB109∷GUS fusion gene (Figure 1A). The GhMYB109∷GUS construct was introduced into the genome of cotton cultivar Coker312 by Agrobacterium tumefaciens-mediated transformation. Twenty progeny from five independent transgenic lines were examined in detail for the GUS expression pattern, using nontransformed wild-type plants as a negative control. In each line, a strong GUS activity was observed only in fibers (Figure 1, B, C, E, F, and G), whereas no or little GUS staining was detected in ovules, petals, sepals, leaves, stems, and flower buds before anthesis (Figure 1H). In comparison, nontransformed plants showed no GUS activity in fibers (Figure 1D) nor in other tissues under the same staining regimen (data not shown). The same pattern of the GhMYB109∷GUS expression was also found in T1 and T2 transgenic plants (data not shown). These results indicated that the 2-kb GhMYB109 putative promoter was sufficient to direct the fiber-specific expression of the GUS reporter gene, confirming that it is a fiber-specific gene.
Figure 1.—
Histochemical localization of GUS activity in the transgenic cotton with the GhMYB109∷GUS fusion gene. (A) A schematic of the GhMYB109 Promoter∷GUS fusion construct used for cotton transformation. (B and C) Dark-field micrographs of 8-μm-thick longitudinal (B) and cross (C) sections of 3-DPA ovules. A high level of GUS activity represented by pink dots was found only in the fiber cells. f, fiber; e, epidermis; o, outer integument of ovule; i, inner integument of ovule. (D–H) Bright field of micrographs and photographs of ovules and other tissues in the transgenic and nontransformed plants.(D–F) GUS staining in ovules at 3 DPA. No GUS staining was detected in the ovules of the nontransformed cotton (D). Strong GUS activity was observed in the fibers of the transgenic plants (E and F). (F) A longitudinal section of a transgenic ovule. (G) GUS staining in each stage of transgenic cotton bolls: 1 DPA, 3 DPA, and 5 DPA (from left to right). The first two panels are longitudinal sections of cotton bolls. cw, carpel wall; l, loculus. (H) GUS staining in other tissues of the transgenic cotton. No GUS activity was detected in leaf, sepal, stem, and flower bud before anthesis (from left to right). Bars, 100 μm in A and B; 1 mm in D–F; 2 mm in G; 1 cm in H.
Generation of antisense GhMYB109 transgenic plants:
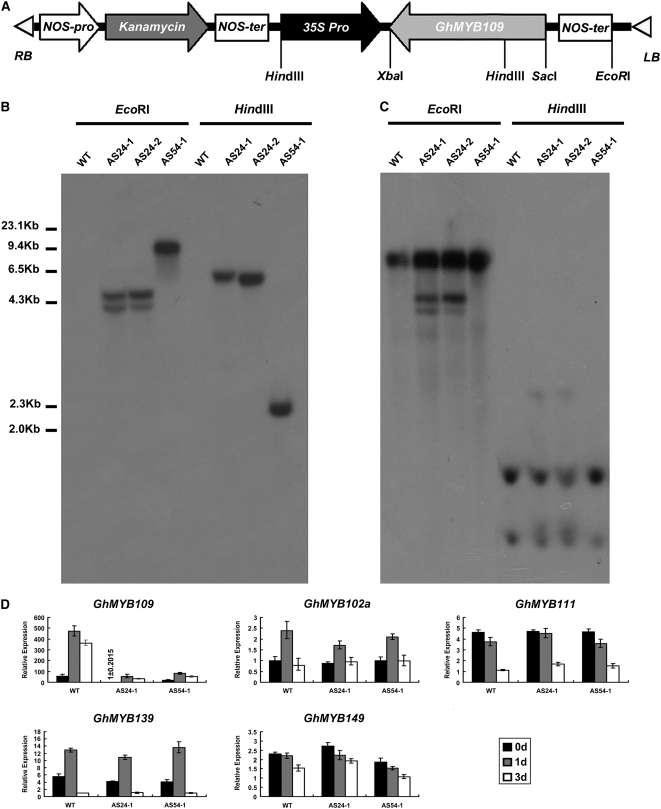
To examine the role of GhMYB109 in fiber development, an antisense GhMYB109 transformation vector driven by the CaMV 35S promoter (Figure 2A) was constructed and introduced into the cotton cultivar Coker312 by A. tumefaciens-mediated transformation. Two independent transgenic T1 lines were subsequently obtained. DNA gel blot analysis using NPTII and GhMYB109 cDNA as probes confirmed that lines AS24-1 and AS24-2 (same transformation event) had two copies and that the other line, AS54-1, had one copy of the antisense GhMYB109 (35S∷GhMYB109AS) transgene (Figure 2, B and C), consistent with the sites of enzymes in genomic DNA and construct.
Figure 2.—
Molecular analysis of the antisense GhMYB109 transgenic cotton. (A) A schematic of the antisense GhMYB109 construct used for cotton transformation. (B and C) DNA gel blot analysis of the transgenic lines. Genomic DNA (20 μg/lane) of two independent transgenic (AS24-1/2 and AS54-1) and wild-type plants was digested with EcoRI and HindIII, respectively, transferred to nylon membrane, and hybridized with 32P-labeled NPTII (B) and 32P-labeled GhMYB109 (C). (D) Quantitative real-time PCR analysis of the transgenic lines. Total RNA was isolated from 0-DPA, 1-DPA, and 3-DPA ovules with their fibers attached from AS24-1, AS54-1, and wild-type plants and subjected to qRT–PCR using GhMYB109-, GhMYB111-, GhMYB139-, GhMYB149-, and GhMYB112a-specific gene primers, respectively, and 18S rRNA as an internal control to normalize all data. The GhMYB109 expression was significantly reduced in the transgenic plants, whereas the expression of the other GhMYB genes was barely affected in the transgenic lines.
To examine the expression of GhMYB109 in the two 35S∷GhMYB109AS transgenic plants, qRT–PCR analysis was performed. Total RNA was extracted from ovules at 0–3 days DPA of AS24-1, AS54-1, and the wild-type plants. The results showed that the level of GhMYB109 mRNAs was reduced significantly (approximately eightfold) in the transgenic plants (Figure 2D). To check if the transgene also affected the expression level of other GhMYB genes, we further analyzed the expression levels of four GhMYBs (GhMYB102a, GhMYB111, GhMYB139, and GhMYB149) (Suo et al. 2003) in ovules and fibers from the transgenic plants by qRT–PCR using the gene-specific primers (Table 1). There was no significant expression reduction of other GhMYB genes (Figure 2D). These results indicated that the expression levels of other MYB genes remained largely unchanged in both the transgenic plants and the wild-type plants, showing that the antisense gene caused a gene-specific significant reduction in GhMYB109 expression.
Fiber development is impaired in the antisense transgenic plants:
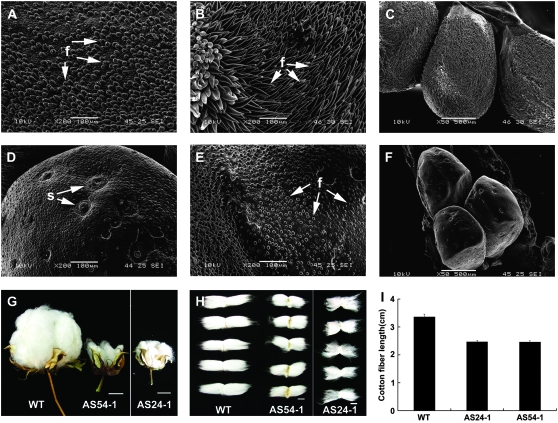
The transgenic plants showed a short-fiber phenotype indicating that the phenotype was a result of the knockdown of GhMYB109 expression. Figure 3 shows the fiber development and seed phenotype of T1 segregants. The impact of GhMYB109 suppression on the cellular development of fiber initials was visualized using scanning electron microscopy. Fiber cells were differentiated and rapidly emerged from the surface of the ovule at 0 DPA in wild-type plants. Figure 3A shows the evenly arranged spherical fiber cells on the surface of wild-type ovules. By contrast, the fiber initials were much slower and smaller in AS54-1 ovules. Many of those cells were shrunken, and some had an abnormal shape and very weak projection above the ovule surface (Figure 3D). Similar shrunken fiber initials also were observed in AS24-1. After initiation on 0 DPA, fiber cells in wild-type plants reached ∼300 μm long at 3 DPA (Figures 3, B and C). This elongation process, however, was inhibited severely in the transgenic plants, and fibers were only <50 μm in length (Figures 3, E and F). In the stage of initiation, there were an estimated 2100 ± 5.58 fiber cells per square millimeter from the ovule epidermis of the wild-type cotton and 1930 ± 5.87 fiber cells in AS54-1 ovules. This result suggested that an incomplete suppression of GhMYB109 had a partial (∼8%) reduction of fiber initials, but it remains unclear if GhMYB109 is directly involved in fiber initiation because of the lack of a null allele. Measurement of the mature fiber length showed that the length of fiber in wild-type cotton reached 3.475 ± 0.19 cm, 2.3 ± 0.12 cm in AS24-1, and 2.315 ± 0.08 cm in AS54-1. Figure 3H shows the fiber length in the transgenic plants reduced ∼33% compared with wild-type plants. Fiber elongation in the transgenic plants was slower than that in wild-type plants (Figure 3I). Most of the bolls of the transgenic plants were smaller than those in the wild type after maturation (Figure 3G), indicating that the GhMYB109 antisense also slightly affected the boll development. The transgenic seeds could be germinated and grown, indicating that suppression of GhMYB109 repressed only the fiber development without affecting embryo development and viability.
Figure 3.—
Comparison of the fiber initiation and length between the antisense transgenic GhMYB109 and wild-type cotton. (A–F) Scanning electron micrographs of the ovule surface of the antisense transgenic GhMYB109 (AS54-1) and wild-type plants. Ovules of the wild-type and transgenic plants are at 0 DPA (A and D), 3 DPA (B and E) and 3 DPA (C and F). The length of fibers in the transgenic plant is much shorter than that in wild-type plant at the same stage. (G) Mature bolls from the transgenic plants AS24-1 and AS54-1 were smaller than that in the wild type. (H) Fibers in the transgenic plants AS24-1 and AS54-1 were much shorter than that in the wild type. (I) Mature fiber lengths of the transgenic antisense GhMYB109 and wild-type cotton seeds. Measurement of the fiber lengths showed that the fiber length in the transgenic plants was reduced ∼33% compared with wild-type plants. f, fiber; s, stoma. Bars: 2 cm in G and 1 cm in H.
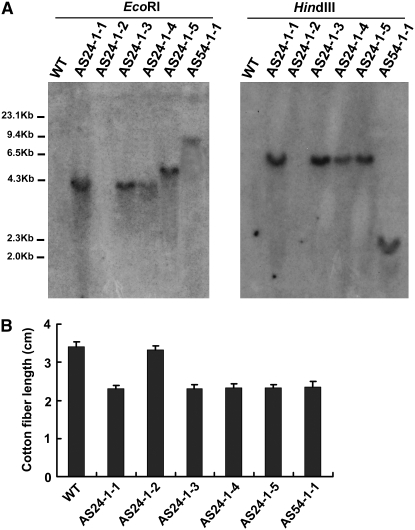
To further examine the effect of the transgene, we analyzed the transgenic plants of the T2 generation (Figure 4). In the line AS54-1, one T2 plant was obtained and had a single copy of the transgene as its parent (Figure 4A). For line AS24-1, among five tested T2 plants, four T2 progeny had the transgene and retained the short-fiber phenotype, and one progeny without the transgene displayed a fiber phenotype similar to wild type (Figure 4B). The results suggested that the antisense gene was effective when it was in both the homozygous and the hemizygous states. Taken together, these results indicated that GhMYB109 plays a direct role in the elongation of cotton fiber cells.
Figure 4.—
Examples of the transgene copy number testing and mature fiber length of the T2 cotton transgenic progeny. (A) Genomic DNA (20 μg/lane) of the wild type and the T2 of the two independent transgenic (AS24-1 and AS54-1) plants was digested with EcoRI (left) and HindIII (right), respectively, transferred to nylon membrane, and hybridized with 32P-labeled NPTII. (Left) WT, wild-type plant; lanes 1–5, five T2 progeny of AS24-1; lane 6, one T2 progeny of AS54-1. Molecular weight markers are indicated in kilobase pairs. (B) Mature fiber lengths of the T2 cotton transformants and wild-type cotton seeds. Measurement of the fiber lengths showed that fiber elongation in the transgenic plants was shorter than that in the wild-type plant. AS24-1-2, one T2 plant of AS24-1 without the transgene copy, displayed a fiber phenotype similar to wild type.
Transcriptional reduction of several fiber-related genes in the transgenic plants:
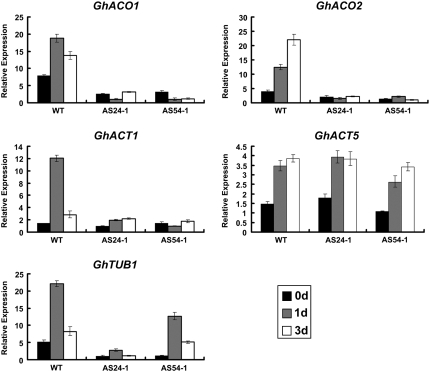
To examine possible targets of GhMYB109 transcript reduction, we selected several known fiber-related genes, GhACO1 and GhACO2 (Shi et al. 2006), GhTUB1 (Li et al. 2002), and GhACT1 and GhACT5 (Li et al. 2005) for a comparative analysis between the transgenic and wild-type cotton using qRT–PCR. Our results revealed that the GhMYB109 suppression led to a substantial reduction of GhACO1, GhACO2, GhTUB1, and GhACT1 expression but had no apparent effect on the expression of GhACT5 (Figure 5), indicating that GhACO and cytoskeleton-encoding genes likely represent potential downstream genes directly or indirectly regulated by GhMYB109.
Figure 5.—
Expression profiling of seven genes important for fiber development in wild-type and transgenic plants. Total RNA samples prepared from 0-DPA, 1-DPA, and 3-DPA ovules with their fibers attached from the two GhMYB109 antisense transgenic and wild-type plants were used for qRT–PCR analysis. 18S rRNA was used as an internal control. The expression of the GhACT5 gene appeared not to be affected in the transgenic lines, whereas GhACO1, GhACO2, GhTUB1, and GhACT1 were expressed at lower levels in the transgenic plant than in wild type.
DISCUSSION
Although the molecular mechanisms controlling cotton fiber initiation and elongation remain largely unknown, we have shown a direct role of the R2R3 MYB transcription factor GhMYB109 in cotton fiber development. This was shown by its role in the knockdown of GhMYB109 expression leading to a substantial reduction in fiber length. This role also is consistent with its fiber-specific expression. To our knowledge, GhMYB109 is the first functional transcriptional factor that has been directly implicated in cotton fiber formation.
Plant MYB genes have been shown to be involved in the regulation of many aspects of plant development, hormone signaling, and metabolism. The MYB family is one of the largest groups of transcription factors in the Arabidopsis genome (Kranz et al. 1998; Stracke et al. 2001). Several MYB transcription factors, such as GhMYB1-6, GaMYB2, and GhMYB25, have been identified in cotton. Although some of them have been characterized with fiber-specific expression, their roles in the cotton fiber development are not yet well defined. The role of GhMYB109 is consistent with its highly conserved R2R3 MYB domain. From previous studies it is clear that many proteins with similar R2R3 MYB factors are involved in the control of development and the determination of cell fate and identity (Schiefelbein 2003; Ramsay and Glover 2005). The role of MYB transcriptional regulators in trichome formation extends beyond Arabidopsis and cotton. The R2R3 MYB-related transcriptional factor MIXTA regulates the formation of conical shape in petal epidermal cells of snapdragon (A. majus) (Noda et al. 1994; Glover et al. 1998; Martin et al. 2002). In Petunia hybrida, conical cell formation in the petals also requires a MYB-related transcription factor named PhMYB1, which is structurally related to MIXTA (Avila et al. 1993; van Houwelingen et al. 1998). The MYB MIXTA LIKE 1 (AmmYBML1) gene from A. majus encodes an R2R3 MYB-related transcriptional regulator identical to that of MIXTA and also promotes trichome and conical cell formation on floral tissues when it is overexpressed under the control of the 35S promoter in tobacco (Glover et al. 1998; Martin et al. 2002; Perez-Rodriguez et al. 2005). In light of these analyses, our study provides a remarkable example of the essential role of the MYB transcription factor in plant growth at the level of a single cell. Because of our findings, we hypothesize that unicellular or multicellular plant hairs develop likely through a similar network of transcription factors (or transcriptional cassette), revealing a functional conservation in cell fate determination in plants.
We have shown that knockdowns of GhMYB109 dramatically reduce cotton fiber elongation, but it remains unclear how the transcription factor controls fiber cell development. In Arabidopsis, AtGL1/AtWER physically interacts with the bHLH proteins AtGL3/AtEGL3 to regulate transcription as part of a multi-protein complex that promotes trichome or root-hair cell fate determination (Schiefelbein 2003; Ramsay and Glover 2005; Serna and Martin 2006). The complex of MYB-bHLH-WD40 appears to regulate the trichome-specific expression of GL2, an activator of downstream trichome-specific differentiation genes, whereas TRY (CPC or ETC1) is a negative regulator that represses trichome differentiation by competing with the MYB factors for binding of the initiation complex (Serna and Martin 2006). It is possible that similar transcription factors in cotton bind to target genes that are involved in the transcriptional regulation of fiber development.
We have found that GhMYB109 suppression induced the expressional reduction of GhACO1, GhACO2 (Shi et al. 2006), GhTUB1 (Li et al. 2002), and GhACT1 (Li et al. 2005) (Figure 5). These results indicate that the MYB-regulated genes are induced prior to the phytohormonal pathway or cytoskeleton-related genes, suggesting that the transcription factor likely regulates these genes for cell fate determination. We hypothesize that the activity of cotton MYB genes is involved in regulating the fiber cell development just at the stage of initiation. When fiber cells begin to enlarge and elongate rapidly at the stage of primary cell-wall formation, the transcription factors activate the transcriptions of the phytohormonal pathway (GhACOs or other related genes), cytoskeleton (GhTUBs and GhACTs), or other fiber-related genes to elaborate and maintain the rapid fiber growth. It is worth examining whether some MYB-binding site elements occur in promoters of GhACOs or cytoskeleton genes. In addition, the cotton homologs related to MIXTA, MYB5, and GL2 are activated during fiber cell initiation (Yang et al. 2006). Wang et al. (2004b) have shown that two cotton transcription factors, GaMYB2/fiber factor 1 (FIF1) and GhHOX3, are able to activate the promoter of a cotton fiber gene, RD22-like1 (RDL1). However, it remains to be seen how these genes are regulated and whether this regulation is directly or indirectly related to cotton fiber development.
In conclusion, the results of this study contribute to an understanding of the developmental mechanism of fiber development and provide direct evidence that GhMYB109 is required for the development of single-celled fibers of cotton. With the demonstration of a fiber-specific promoter from GhMYB109, we will be able to express target gene products in the developing fiber for possible genetic improvement of fiber development.
Acknowledgments
We thank Xiongming Du for cotton seeds of Xuzhou 142. This work was supported by the National High Technology Research and Development Program of China (2003AA222021 and 2006AA10A108).
References
- Arpat, A. B., M. Waugh, J. P. Sullivan, M. Gonzales, D. Frisch et al., 2004. Functional genomics of cell elongation in developing cotton fibers. Plant Mol. Biol. 54 911–929. [DOI] [PubMed] [Google Scholar]
- Avila, J., C. Nieto, L. Canas, M. J. Benito and J. Paz-Ares, 1993. Petunia hybrida genes related to the maize regulatory C1 gene and to animal myb proto-oncogenes. Plant J. 3 553–562. [DOI] [PubMed] [Google Scholar]
- Basra, A. S., and C. P. Malik, 1984. Development of the cotton fiber. Int. Rev. Cytol. 89 65–113. [Google Scholar]
- Glover, B. J., 2000. Differentiation in plant epidermal cells. J. Exp. Bot. 51 497–505. [DOI] [PubMed] [Google Scholar]
- Glover, B. J., M. Perez-Rodriguez and C. Martin, 1998. Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development 125 3497–3508. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., 2004. Plant trichomes: a model for cell differentiation. Nat. Rev. Mol. Cell Biol. 5 471–480. [DOI] [PubMed] [Google Scholar]
- Humphries, J. A., A. R. Walker, J. N. Timmis and S. J. Orford, 2005. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 57 67–81. [DOI] [PubMed] [Google Scholar]
- Ji, S. J., Y. C. Lu, J. X. Feng, G. Wei, J. Li et al., 2003. Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res. 31 2534–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, M. E., and G. Keller, 1996. Metabolic pathway engineering in cotton: biosynthesis of polyhydroxybutyrate in fiber cells. Proc. Natl. Acad. Sci. USA 93 12768–12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. J., and B. A. Triplett, 2001. Cotton fiber growth in planta and in vitro: models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127 1361–1366. [PMC free article] [PubMed] [Google Scholar]
- Kirik, V., M. Simon, M. Huelskamp and J. Schiefelbein, 2004. a The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268 506–513. [DOI] [PubMed] [Google Scholar]
- Kirik, V., M. Simon, K. Wester, J. Schiefelbein and M. Hulskamp, 2004. b ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55 389–398. [DOI] [PubMed] [Google Scholar]
- Kranz, H. D., M. Denekamp, R. Greco, H. Jin, A. Leyva et al., 1998. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16 263–276. [DOI] [PubMed] [Google Scholar]
- Lan, L., W. Chen, Y. Lai, J. Suo, Z. Kong et al., 2004. Monitoring of gene expression profiles and isolation of candidate genes involved in pollination and fertilization in rice (Oryza sativa L.) with a 10K cDNA microarray. Plant Mol. Biol. 54 471–487. [DOI] [PubMed] [Google Scholar]
- Larkin, J. C., M. L. Brown and J. Schiefelbein, 2003. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 54 403–430. [DOI] [PubMed] [Google Scholar]
- Lee, J. J., O. S. Hassan, W. Gao, N. E. Wei, R. J. Kohel et al., 2006. Developmental and gene expression analyses of a cotton naked seed mutant. Planta 223 418–432. [DOI] [PubMed] [Google Scholar]
- Li, X. B., L. Cai, N. H. Cheng and J. W. Liu, 2002. Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol. 130 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. B., X. P. Fan, X. L. Wang, L. Cai and W. C. Yang, 2005. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17 859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loguerico, L. L., J. Q. Zhang and T. A. Wilkins, 1999. Differential regulation of six novel MYB-domain genes defines two distinct expression patterns in allotetraploid cotton (Gossypium hirsutum L.). Mol. Gen. Genet. 261 660–671. [DOI] [PubMed] [Google Scholar]
- Martin, C., K. Bhatt, K. Baumann, H. Jin, S. Zachgo et al., 2002. The mechanics of cell fate determination in petals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, K., B. J. Glover, P. Linstead and C. Martin, 1994. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature 369 661–664. [DOI] [PubMed] [Google Scholar]
- Ohashi, Y., A. Oka, I. Ruberti, G. Morelli and T. Aoyama, 2002. Entopically additive expression of GLABRA2 alters the frequency and spacing of trichome initiation. Plant J. 29 359–369. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez, M., F. W. Jaffe, E. Butelli, B. J. Glover and C. Martin, 2005. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development 132 359–370. [DOI] [PubMed] [Google Scholar]
- Ramsay, N. A., and B. J. Glover, 2005. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10 63–70. [DOI] [PubMed] [Google Scholar]
- Rerie, W. G., K. A. Feldmann and M. D. Marks, 1994. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8 1388–1399. [DOI] [PubMed] [Google Scholar]
- Schellmann, S., A. Schnittger, V. Kirik, T. Wada, K. Okada et al., 2002. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein, J., 2003. Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6 74–78. [DOI] [PubMed] [Google Scholar]
- Schneider, K., B. Wells, L. Dolan and K. Roberts, 1997. Structural and genetic analysis of epidermal cell differentiation in Arabidopsis primary roots. Development 124 1789–1798. [DOI] [PubMed] [Google Scholar]
- Schnittger, A., U. Folkers, B. Schwab, G. Jurgens and M. Hulskamp, 1999. Generation of a spacing pattern: the role of triptychon in trichome patterning in Arabidopsis. Plant Cell 11 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna, L., and C. Martin, 2006. Trichomes: different regulatory networks lead to convergent structures. Trends Plant Sci. 11 274–280. [DOI] [PubMed] [Google Scholar]
- Shi, Y. H., S. W. Zhu, X. Z. Mao, J. X. Feng, Y. M. Qin et al., 2006. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., M. Werber and B. Weisshaar, 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Suo, J., X. Liang, L. Pu, Y. Zhang and Y. Xue, 2003. Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim. Biophys. Acta 1630 25–34. [DOI] [PubMed] [Google Scholar]
- Szymanski, D. B., R. A. Jilk, S. M. Pollock and M. D. Marks, 1998. Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125 1161–1171. [DOI] [PubMed] [Google Scholar]
- Taliercio, E. W., and D. Boykin, 2007. Analysis of gene expression in cotton fiber initials. BMC Plant Biol. 7 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall, J. A., J. M. Swanson, K. Haller, R. A. Rapp, M. E. Sparks et al., 2006. A global assembly of cotton ESTs. Genome Res. 16 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houwelingen, A., E. Souer, K. Spelt, D. Kloos, J. Mol et al., 1998. Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida. Plant J. 13 39–50. [DOI] [PubMed] [Google Scholar]
- Wang, L., L. Dong, Y. Zhang, Y. Zhang, W. Wu et al., 2004. a Genome-wide analysis of S-locus F-box-like genes in Arabidopsis thaliana. Plant Mol. Biol. 56 929–945. [DOI] [PubMed] [Google Scholar]
- Wang, S., J. W. Wang, N. Yu, C. H. Li, B. Luo et al., 2004. b Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., A. C. Machado, R. G. White, D. J. Llewellyn and E. S. Dennis, 2006. Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol. 47 107–127. [DOI] [PubMed] [Google Scholar]
- Yang, S. S., F. Cheung, J. J. Lee, M. Ha, N. E. Wei et al., 2006. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 47 761–775. [DOI] [PMC free article] [PubMed] [Google Scholar]