Abstract
DNA microarray analysis has emerged as a leading technology to enhance our understanding of gene regulation and function in cellular mechanism controls on a genomic scale. This technology has advanced to unravel the genetic machinery of biological rhythms by collecting massive gene-expression data in a time course. Here, we present a statistical model for clustering periodic patterns of gene expression in terms of different transcriptional profiles. The model incorporates biologically meaningful Fourier series approximations of gene periodic expression into a mixture-model-based likelihood function, thus producing results that are likely to be closer to biological relevance, as compared to those from existing models. Also because the structures of the time-dependent means and covariance matrix are modeled, the new approach displays increased statistical power and precision of parameter estimation. The approach was used to reanalyze a real example with 800 periodically expressed transcriptional genes in yeast, leading to the identification of 13 distinct patterns of gene-expression cycles. The model proposed can be useful for characterizing the complex biological effects of gene expression and generate testable hypotheses about the workings of developmental systems in a more precise quantitative way.
ALMOST all of life's phenomena populating the earth can be described in terms of periodic rhythms that result from the planet's rotation and orbit around the sun (Goldbeter 2002). Cell division (Mitchison 2003), circadian rhythms (Crosthwaite 2004; Prolo et al. 2005), morphogenesis of periodic structures such as somites in vertebrates (Dale et al. 2003), and the complex life cycles of some microorganisms (Lakin-Thomas and Brody 2004; Rovery et al. 2005) are all excellent representatives of biological rhythms. From a mechanistic perspective, rhythmic behavior arises in genetic and metabolic networks as a result of nonlinearities associated with various modes of cellular regulation.
The complexity and inherent periodicity of rhythmic processes can be well described by mathematical models. For example, mathematical models of increasing complexity for the genetic regulatory network producing circadian rhythms in the fly Drosophila predict the occurrence of sustained circadian oscillations of the limit cycle type (Leloup et al. 1999). When incorporating the effect of light, the models account for phase shifting of the rhythm by light pulses and for entrainment by light–dark cycles. The models also provide an explanation for the long-term suppression of circadian rhythms by a single pulse of light. Stochastic simulations were developed to test the robustness of circadian oscillations with respect to molecular noise (Gonze et al. 2002, 2003).
The recent development of gene-expression technologies has offered biologists a unique opportunity to more closely study the mechanisms that control a particular biological rhythm. Panda et al. (2002) employed high-density oligonucleotide arrays to trace gene expression in mouse tissue samples taken every 4 hr during two complete circadian cycles. They also identified clusters of circadian-regulated genes among >7000 genes with a cosine wave-fitting algorithm (Harmer et al. 2000). About 650 cycling transcripts were detected to be under circadian regulation specific to either the suprachiasmatic nuclei or the liver. These studies allowed the authors to understand how the major oscillator in the suprachiasmatic nuclei and the liver regulates behavioral and physiological rhythms in the whole organism.
To associate the profile of gene expression with a physiological function of interest, it is crucial to cluster the types of gene expression on the basis of their periodic patterns. Statistical modeling and algorithms play a central role in cataloging dynamic gene-expression profiles. While various computational models have been developed for gene clustering based on static microarray data (Eisen et al. 1998; Ghosh and Chinnaiyan 2002; McLachlan et al. 2002; Ramoni et al. 2002; Fan and Ren 2006), considerable attention has been paid to methodological derivations for detecting temporal patterns of gene expression in a time course based on functional principal component analysis or mixture model analysis (Holter et al. 2001; Qian et al. 2001; Bar-Joseph et al. 2003; Luan and Li 2003; Park et al. 2003; Wakefield et al. 2003; Ernst et al. 2005; Storey et al. 2005; Ma et al. 2006; Ng et al. 2006; Inoue et al. 2007). In particular, Luan and Li (2004) proposed a statistical model for characterizing different types of periodic rhythms of gene expression. These methods show some unique utilization to identify genes with varying expression profiles over time, providing new tools to elucidate a comprehensive picture of the life process.
A common feature of these clustering methods is that they model time-dependent gene-expression profiles using nonparametric approaches, such as cubic splines. Although nonparametric approaches are statistically flexible and computationally fast, they often do not provide rigorous biological interpretations of results even if the gene-expression data analyzed contain some biologically meaningful patterns. This article was motivated by the fundamental idea that key features of many biological processes can be described by parsimonious mathematical equations. For example, Frank (1926) used the Fourier series to approximate periodic and quasi-periodic biological phenomena. Attinger et al. (1966) provided a detailed mathematical formulation of this approximation for a biological rhythmic system through both theoretical and experimental approaches. Fourier series approximation as an analytical tool (Priestley 1981) has been widely applied to study the mechanisms and patterns of biological rhythmicity, including the cyclic organization of preterm and term neonates during the neonatal period (Begum et al. 2006) and pharmacodynamics (Mager and Abernethy 2007).
Fourier approximation has also been used to model periodic gene expression, leading to the detection of periodic signals in various organisms including yeast and human cells (Spellman et al. 1998; Wichert et al. 2004; Kim et al. 2006). Glynn et al. (2006) proposed a Lomb–Scargle periodogram approach based on the fast Fourier transform to model unevenly spaced gene-expression time series and then characterize periodic patterns of gene expression by using a multiple-hypothesis testing procedure with a controlled false discovery rate. Our statistical model presented in this article integrates the Fourier series approximation into a mixture-model framework to mathematically cluster microarray genes on the basis of their distinct patterns of periodic expression. The advantage of this mixture-based model includes its solid statistical foundation of testing the number of gene clusters. Furthermore, through the implementation of the Fourier series approximation, it is possible to test several biologically meaningful characteristics such as sharp peaks in the ordinary periodograms calculated from the Fourier transform of the time series (Durbin 1967). We use a published example for cell cycle-regulated genes in Saccharomyces cerevisiae (Spellman et al. 1998) to validate the new model. The statistical behavior of the model is examined through simulation studies.
METHODS
Mixture model:
Finite mixture models (McLachlan and Peel 2000) have been widely used to model the distributions of a variety of random phenomena. Multivariate normality is generally assumed for multivariate data of a continuous nature. This multivariate normal mixture model is employed to detect different patterns in gene-expression profiles.
Assume that n genes are measured at multiple time points. Our model is able to consider unevenly spaced time intervals and different measurement schedules for each gene. Let 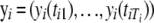 be the Ti-dimensional gene-expression data for gene i. Suppose there are J components in the mixture model. This means that any one of the genes (i) is assumed to arise from one (and only one) of the J possible periodic patterns of expression, the distribution of whose expression data is expressed as the J-component mixture probability density function; i.e.,
be the Ti-dimensional gene-expression data for gene i. Suppose there are J components in the mixture model. This means that any one of the genes (i) is assumed to arise from one (and only one) of the J possible periodic patterns of expression, the distribution of whose expression data is expressed as the J-component mixture probability density function; i.e.,
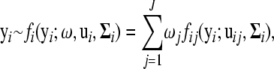 |
(1) |
where 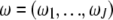 is the mixture proportions that are nonnegative and sum to unity; ui = (ui1,…, uiJ) contains the component- (or pattern-) specific mean vector for gene i; and
is the mixture proportions that are nonnegative and sum to unity; ui = (ui1,…, uiJ) contains the component- (or pattern-) specific mean vector for gene i; and 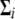 contains residual variances and covariances among the Ti time points for gene i that are common for all gene-expression patterns. The probability density function of the jth gene-expression pattern or cluster,
contains residual variances and covariances among the Ti time points for gene i that are common for all gene-expression patterns. The probability density function of the jth gene-expression pattern or cluster, 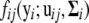 , is assumed to be multivariate normal with mean vector
, is assumed to be multivariate normal with mean vector 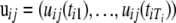 and common covariance matrix
and common covariance matrix 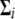 .
.
From the above analysis, different gene-expression patterns can be detected by estimating the number of mixture components, the time-dependent means of each component, and the covariance matrix. By comparing the differences of the mean vector among different components, it is possible to test how response curves differ among different components and, further, to address fundamentally important biological issues regarding the interplay between gene expression and biological rhythms.
Modeling the mean curve:
If the genes studied are periodically regulated, their time-dependent expression can be accurately approximated by a Fourier series approximation (Spellman et al. 1998). Fourier series approximations can assess periodicity, so we can identify the genes whose RNA levels varied periodically within the cell cycle and, further, find the associated amplitudes and phases of these cycles by estimating the mathematical parameters in the Fourier series approximation. A general form of the Fourier signal is given as
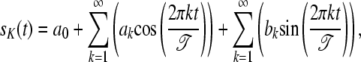 |
(2) |
where a0 is the average value of sK(t) and the other ak and bk coefficients are the amplitude coefficients that determine the times at which the gene achieves peak and trough expression levels, respectively, and 𝒯 is the period of the signal of gene expression.
In practice, the Fourier series of Equation 2 can be approximated by the first K + 1 term. By analyzing the error of the Fourier series approximation, expressed as the difference between the signal and the (K + 1)-term series,
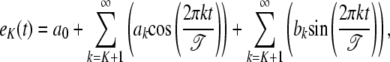 |
it is found that, when a third-order approximation is used (i.e., K = 3), the unused terms (k = 4,…, ∞) from the series together are only ∼3% of the signal. Thus, the time-dependent expression value of a gene can be adequately modeled by a Fourier series approximation of the first three orders. Let 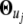 denote the vector containing Fourier parameters of various orders for the genes with pattern j, which is specified as
denote the vector containing Fourier parameters of various orders for the genes with pattern j, which is specified as
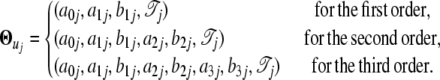 |
(3) |
The mean expression value of gene i when its pattern is j at time point tiτ can be approximated by these Fourier series parameters; i.e.,
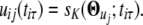 |
Modeling the covariance structure:
A number of approaches can be used to model the covariance structure of serial measurements. A commonly used approach for structuring the covariance is the first-order autoregressive [AR(1)] model (Davidian and Giltinan 1995; Verbeke and Molenberghs 2000). One advantage of using the AR(1) model is that it provides a general expression for calculating the determinant and inverse of the matrix for any number of time points measured. But it assumes variance stationarity and correlation stationarity; i.e., the residual variance at different time points is the same, expressed as σ2, and the correlation between two different time points for gene i, 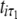 and
and 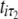 , decreases exponentially in ρ with time lag, expressed as corr(
, decreases exponentially in ρ with time lag, expressed as corr(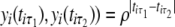 .
.
To remove the heteroscedastic problem of the residual variance, which violates a basic assumption of the simple AR(1) model, two approaches can be used. The first approach is to model the residual variance by a parametric function of time, as proposed by Pletcher and Geyer (1999). But this approach needs to implement additional parameters for characterizing the time-dependent change of the variance. The second approach is to embed Carroll and Ruppert's (1984) transform-both-sides (TBS) model into the growth-incorporated finite mixture model (Wu et al. 2004), which does not need any more parameters. Both empirical analyses with real examples and computer simulations suggest that the TBS-based model can increase the precision of parameter estimation and computational efficiency. Furthermore, the TBS model preserves original biological means of the curve parameters although statistical analyses are based on transformed data. In this study, we used the TBS-based AR(1) model, with the structuring parameters arrayed in 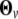 .
.
Computational algorithm:
The EM algorithm and Nelder–Mead simplex algorithm were used to estimate the unknown parameters 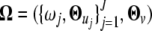 . The observed data
. The observed data 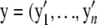 ) are regarded as being incomplete. Let zij be a missing variable, defined as 1 if yi arises from the jth component of the mixture model (i = 1,…, n; j = 1,…, J), and write zi = (zi1,…, ziJ)′. The variables z1,…, zn are independent and obey a multinomial distribution consisting of one category among J possibilities with probabilities (ω1,…, ωJ). Then, we have
) are regarded as being incomplete. Let zij be a missing variable, defined as 1 if yi arises from the jth component of the mixture model (i = 1,…, n; j = 1,…, J), and write zi = (zi1,…, ziJ)′. The variables z1,…, zn are independent and obey a multinomial distribution consisting of one category among J possibilities with probabilities (ω1,…, ωJ). Then, we have
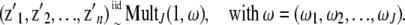 |
The complete data log-likelihood is
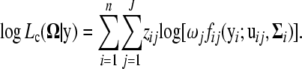 |
In the E step, zij is replaced by the posterior probability (Pij) of the ith gene that belongs to the jth pattern. This is equal to the conditional expectation (E[zij | yi]) of the complete data log-likelihood log Lc(Ω), given the observed data y, and is computed as
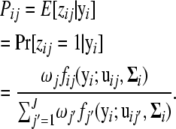 |
(4) |
In the M step, we intend to choose the value of 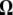 that maximizes
that maximizes 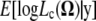 . For the derivation process of the estimates of the unknown parameters in the M step, see the appendix. The E and M steps are iterated repeatedly until the parameter estimates converge to finally obtain the maximum-likelihood estimates (MLEs) of the unknown parameters.
. For the derivation process of the estimates of the unknown parameters in the M step, see the appendix. The E and M steps are iterated repeatedly until the parameter estimates converge to finally obtain the maximum-likelihood estimates (MLEs) of the unknown parameters.
Model selection:
Testing for the number of components in a mixture is an important but very difficult problem that has not been completely resolved. One of the leading selection methods is the Akaike information criterion (AIC) (Akaike 1974). This is designed to be an approximately unbiased estimator of the expected Kullback–Leibler information of a fitted model. The minimum AIC produces a selected model that is close to the best possible choice. Within the context of the mixture model, the number of components, k, is chosen to minimize
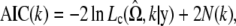 |
where N(k) is the number of independent parameters within 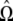 . By the same token, the Bayesian information criterion (BIC) (Schwarz 1978) is also available, expressed as
. By the same token, the Bayesian information criterion (BIC) (Schwarz 1978) is also available, expressed as
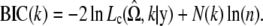 |
The selected model is the one with the smallest BIC. There is no clear consensus on which criterion is best to use, although the empirical work of Fraley and Raftery (1998) seems to favor the BIC. Since information criteria penalize models with additional parameters, the AIC and BIC model order selection criteria are based on parsimony. Note that since the BIC imposes a greater penalty for additional parameters than does the AIC, the BIC always provides a model with a number of parameters not greater than that chosen by the AIC.
Hypothesis testing:
The significance of overall differences in transcriptional expression profile among different groups of microarray genes is tested by formulating the following hypotheses:
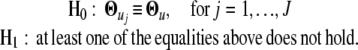 |
(5) |
The log-likelihood ratio (LR) test statistic is then calculated by
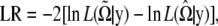 |
(6) |
where the tilde and circumflex stand for the MLEs of the unknown parameters under the null and alternative hypotheses, respectively. The critical threshold for claiming distinguishable expression patterns can be determined on the basis of simulation studies. The null hypothesis means that no different patterns of periodic expression exist among the genes studied, whereas the alternative hypothesis states that at least two different patterns can be identified. Under the null hypothesis, time-dependent expression data for n genes are simulated by assuming that the data follow a multivariate normal distribution with mean vector u and covariance matrix 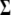 . According to Equation 3, individual elements in u are approximated by the Fourier series function of a particular order. A set of parameters that describes the shape of the Fourier series function can be taken from their estimates obtained from real data analyses. Similarly, the structure of
. According to Equation 3, individual elements in u are approximated by the Fourier series function of a particular order. A set of parameters that describes the shape of the Fourier series function can be taken from their estimates obtained from real data analyses. Similarly, the structure of 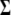 is modeled by AR(1) with the two underlying parameters (ρ and σ2) taken from the estimates.
is modeled by AR(1) with the two underlying parameters (ρ and σ2) taken from the estimates.
The functional clustering model described in the main text is then used to analyze the expression data simulated under the condition of no distinct groups, provide the estimates of the curve parameters and covariance-structuring parameters under the null and alternative hypotheses, and calculate the LR test statistic. This procedure is repeated 1000 times, leading to 1000 LR values. The empirical distribution of the LR test statistic over 1000 replicates is then examined. The 95th percentile of the empirical distribution is then regarded as the critical threshold for claiming the existence of distinct patterns of periodic gene expression.
RESULTS
A worked example:
Spellman et al. (1998) reported the results of 800 periodically expressed transcriptional genes in the genome of yeast (S. cerevisiae). DNA microarrays were used to analyze mRNA levels for six yeast strains in cell cultures that have been synchronized by three independent methods, α-factor arrest, elutriation, and arrest of a cdc15 temperature-sensitive mutant. Each method produces populations of yeast cells synchronized in terms of their phase in the cell cycle.
As described by Spellman et al. (1998), RNA was extracted from each of the samples and from a control sample (unsynchronized cells). cDNAs were labeled with Cy5 fluor (red) for synchronized samples and Cy3 fluor (green) for the controls. Mixed labeled control and experimental cDNAs were hybridized to individual microarrays containing all 6178 yeast genes. The average fluorescence intensity for each fluor within each array spot was recorded. The data of gene expression were measured by normalized fluorescence ratio log2(Cy5/Cy3) at different time points. All the microarray data reported in Spellman et al. (1998) are given at http://cellcycle-www.stanford.edu.
To validate the usefulness of the model proposed in this article, we analyze time-dependent gene-expression data derived from the cdc15 experiment. The data contain 800 genes, 632 of which have complete data during all 24 time points. The remaining 168 genes contain missing values at some time points. All the 800 genes are analyzed simultaneously.
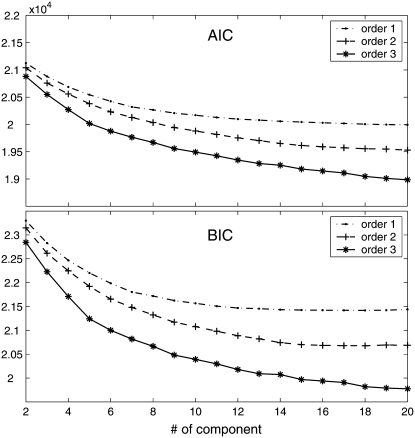
When the Fourier series approximation is used to cluster the periodic patterns of gene expression, two issues should be determined in the following sequence. First, what is the best order for Fourier series function that explains the time-dependent data? Second, what is the optimal number of components for the mixture model that each correspond to a different expression pattern? Model selection criteria, AIC and BIC, were used to determine the best Fourier series order and best number of mixture components for Spellman et al.'s data. The two criteria provide similar results (Figure 1), although our analysis is mostly based on BIC. It seems that a higher order of Fourier series can better fit time-dependent data than a lower order, reflecting the complexity of dynamic changes of gene expression. A lower order of Fourier series tends to detect a smaller number of gene-expression patterns than a higher order. For example, the first order detects 13 components, whereas the second and third orders detect 18 and >20 components, respectively. This makes sense because a higher order of Fourier series function has more power to discern subtle differences in gene-expression profiles. When closely looking at the BIC curves (Figure 1), the first order displays a dramatic decrease when the number of clusters is 6–8, whereas a dramatic decrease for the second and the third order occurs at 12–14 and 18–20 clusters, respectively.
Figure 1.—
Component number-dependent AIC and BIC values of model fitting by a Fourier series function of order 1–3 for 800 genes collected from the yeast genome.
To illustrate different periodic patterns of gene-expression profiles concordant with cell cycles, we used the first-order Fourier series to detect 13 patterns whose profiles (Figure 2) were drawn with the Fourier parameters estimated from the proposed model (Table 1). These 13 patterns differ dramatically in the overall shape of curves as defined by parameter sets 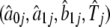 . On the basis of these estimates, a number of hypothesis tests can be made about the developmental patterns of gene expression. Table 1 also provides the estimates of the proportions of mixture components. The 13 patterns occur at different frequencies among the observed genes.
. On the basis of these estimates, a number of hypothesis tests can be made about the developmental patterns of gene expression. Table 1 also provides the estimates of the proportions of mixture components. The 13 patterns occur at different frequencies among the observed genes.
Figure 2.—
Thirteen periodic patterns of gene-expression profiles approximated by a first-order Fourier series function for 800 genes collected from the yeast genome.
TABLE 1.
MLEs of the Fourier parameters for 13 different periodic patterns of gene expression among 800 genes collected from the yeast genome based on the first-order approximation
| Pattern | 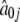 |
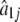 |
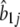 |
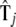 |
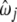 |
|---|---|---|---|---|---|
| 1 | −0.2888 | −0.9178 | 0.7847 | 10.47000 | 0.0151 |
| 2 | −0.0951 | 1.3955 | 0.0150 | 11.05000 | 0.0158 |
| 3 | −0.4384 | 0.0008 | 1.1269 | 9.96970 | 0.0371 |
| 4 | 0.0604 | 0.2396 | −1.2738 | 10.8740 | 0.0142 |
| 5 | −0.0382 | 0.0430 | 0.2508 | 2.0737 | 0.2309 |
| 6 | −0.0339 | 0.6301 | −0.1558 | 10.8620 | 0.0632 |
| 7 | −0.1277 | −0.1248 | 0.3128 | 9.6315 | 0.2011 |
| 8 | 0.1403 | −0.3256 | −0.5849 | 10.6880 | 0.0656 |
| 9 | −0.0177 | −0.5249 | 0.0052 | 10.5780 | 0.0667 |
| 10 | −0.2104 | −0.5480 | 0.0000 | 2.0006 | 0.0099 |
| 11 | 0.4395 | −3.1983 | −1.8160 | 8.2165 | 0.0013 |
| 12 | −0.0124 | −0.1115 | −0.1275 | 2.1848 | 0.1397 |
| 13 | 0.0359 | −0.0277 | −0.2932 | 2.0852 | 0.1394 |
The AR(1) parameters used to model the covariance structure are estimated as 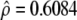 and
and 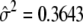 when 13 gene-expression patterns are fitted to the data.
when 13 gene-expression patterns are fitted to the data.
Computer simulation:
We performed Monte Carlo simulation studies to investigate statistical properties of the functional clustering model proposed. A total of 1000 genes were simulated whose time-dependent expression was measured at 24 equally spaced time points. All the genes were sorted into three distinct patterns with varying proportions. The simulated gene-expression profiles follow an arbitrary form of periodic function. As has been mathematically clear, a periodic function can be approximated by a Fourier series function. The Fourier parameters used for our simulation were assigned by values that are within their spaces according to Spellman et al.'s (1998) data. The time-dependent covariance matrix of gene expression was structured by the AR(1) model. Different residual variances were used in the simulation to examine the effect of residual errors on parameter estimation.
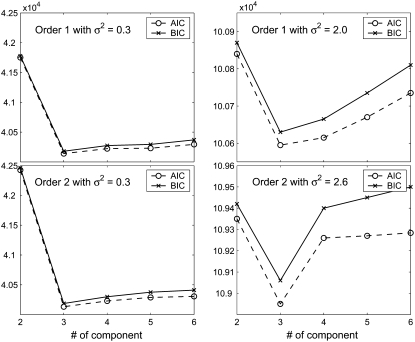
The optimal number of components for the simulated data was determined by calculating AIC and BIC values. As shown in Figure 3, the model can correctly estimate the number of components. On the basis of results from 1000 simulation replicates, the model can provide reasonably accurate and precise estimates of all Fourier parameters (Tables 2 and 3). The precision of parameter estimation depends on the proportion of a gene-expression pattern being better for a more frequent than for a less frequent pattern. As expected, increasing residual variance will reduce the estimation precision of parameters (Table 2 vs. Table 3). To show the robustness of the model, an additional simulation study based on a second-order Fourier series approximation was performed. The results suggest that all parameters can be reasonably estimated even if the number of parameters being estimated is increased (Tables 4 and 5).
Figure 3.—
Component number-dependent AIC and BIC values of model fitting by a first- and a second-order Fourier series function for 1000 simulated genes under different residual variances.
TABLE 2.
MLEs of the first-order Fourier parameters for three periodic patterns of gene expression among 1000 simulated genes with a given set of values (a0, a1, b1, T) under the residual variance of σ2 = 0.3
| Pattern
|
|||
|---|---|---|---|
| 1 | 2 | 3 | |
| Proportion | |||
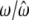
|
0.5850/0.5851 (0.0002) | 0.1000/0.1001 (0.0002) | 0.3150/0.3148 (0.0005) |
| Mean vector | |||
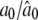
|
0.3000/0.30571 (0.0218) | 0.0300/0.0511 (0.0378) | 0.0600/0.0696 (0.0171) |
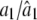
|
1.0000/1.0041 (0.0081) | 1.0000/1.0007 (0.0081) | 0.9000/0.9061 (0.0121) |
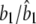
|
0.2000/0.1910 (0.0118) | 0.0200/0.0084 (0.0141) | 0.0100/0.0160 (0.0150) |
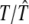
|
6.0000/6.0034 (0.0043) | 10.0000/10.0150 (0.0188) | 16.0000/15.9730 (0.0392) |
| Covariance | |||
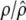
|
0.6000/0.5888 (0.0044) | ||
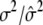
|
0.3000/0.3013 (0.0036) | ||
The averages of parameter estimates are calculated from 100 simulations and the mean square errors of the estimates are given in parentheses.
TABLE 3.
MLEs of the first-order Fourier parameters for three periodic patterns of gene expression among 1000 simulated genes with a given set of values (a0, a1, b1, T) under the residual variance of σ2 = 2.0
| Pattern
|
|||
|---|---|---|---|
| 1 | 2 | 3 | |
| Proportion | |||
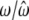
|
0.5850/0.5839 (0.0070) | 0.1000/0.1007 (0.0093) | 0.3150/0.3154 (0.0112) |
| Mean vector | |||
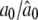
|
0.3000/0.2983 (0.0385) | 0.0300/0.0480 (0.0606) | 0.0600/0.0535 (0.0519) |
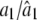
|
1.0000/1.0016 (0.0148) | 1.0000/0.9956 (0.0631) | 0.9000/0.9035 (0.0383) |
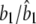
|
0.2000/0.2022 (0.0290) | 0.0200/0.0296 (0.0435) | 0.0100/0.0157 (0.0199) |
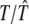
|
6.0000/5.9997 (0.0088) | 10.0000/10.0030 (0.0575) | 16.0000/15.9930 (0.1212) |
| Covariance | |||
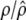
|
0.6000/0.5993 (0.0041) | ||
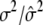
|
2.0000/1.9945 (0.0250) | ||
The averages of parameter estimates are calculated from 100 simulations and the mean square errors of the estimates are given in parentheses.
TABLE 4.
MLEs of the second-order Fourier parameters for three periodic patterns of gene expression among 1000 simulated genes with a given set of values (a0, a1, b1, T) under the residual variance of σ2 = 0.3
| Pattern
|
|||
|---|---|---|---|
| 1 | 2 | 3 | |
| Proportion | |||
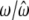
|
0.5850/0.5850 (0.0004) | 0.1000/0.1000 (0.0003) | 0.3150/0.3150 (0.0003) |
| Mean vector | |||
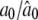
|
0.3000/0.2972 (0.0166) | 0.0300/0.0340 (0.0291) | 0.0600/0.0592 (0.0212) |
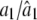
|
1.0000/1.0002 (0.0056) | 1.0000/1.0018 (0.0169) | 0.9000/0.8994 (0.0109) |
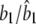
|
0.2000/0.1998 (0.0103) | 0.0200/0.0211 (0.0214) | 0.0100/0.0150 (0.0147) |
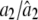
|
0.2000/0.2000 (0.0030) | 0.2000/0.1997 (0.0112) | 0.4000/0.3985 (0.0081) |
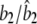
|
0.0400/0.0398 (0.0051) | 0.0100/0.0121 (0.0103) | 0.0400/0.0441 (0.0155) |
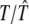
|
6.0000/6.0002 (0.0030) | 10.0000/10.0010 (0.0164) | 16.0000/15.9880 (0.0397) |
| Covariance | |||
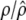
|
0.6000/0.5998 (0.0044) | ||
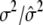
|
0.3000/0.2995 (0.0039) | ||
The averages of parameter estimates are calculated from 100 simulations and the mean square errors of the estimates are given in parentheses.
TABLE 5.
MLEs of the second-order Fourier parameters for three periodic patterns of gene expression among 1000 simulated genes with a given set of values (a0, a1, b1, T) under the residual variance of σ2 = 2.6
| Pattern
|
|||
|---|---|---|---|
| 1 | 2 | 3 | |
| Proportion | |||
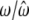
|
0.5850/0.5836 (0.0088) | 0.1000/0.1010 (0.0088) | 0.3150/0.3154 (0.0109) |
| Mean vector | |||
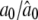
|
0.3000/0.3028 (0.0452) | 0.0300/0.0627 (0.0874) | 0.0600/0.0619 (0.0638) |
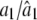
|
1.0000/1.0042 (0.0174) | 1.0000/0.9910 (0.0753) | 0.9000/0.9014 (0.0416) |
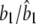
|
0.2000/0.2004 (0.0302) | 0.0200/0.0388 (0.0569) | 0.0100/0.0232 (0.0367) |
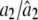
|
0.2000/0.2008 (0.0079) | 0.2000/0.1987 (0.0425) | 0.4000/0.3979 (0.0256) |
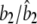
|
0.0400/0.0409 (0.0163) | 0.0100/0.0231 (0.0325) | 0.0400/0.0478 (0.0416) |
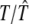
|
6.0000/6.0006 (0.0092) | 10.0000/9.9890 (0.0522) | 16.0000/15.9770 (0.1049) |
| Covariance | |||
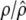
|
0.6000/0.5994 (0.0040) | ||
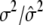
|
2.6000/2.5935 (0.0323) | ||
The averages of parameter estimates are calculated from 100 simulations and the mean square errors of the estimates are given in parentheses.
Ng et al. (2006) proposed a random-effect mixture model for clustering gene-expression profiles through the incorporation of covariate information. When the covariate is time, Ng et al.'s model functions as ours does. However, these two models are different in three aspects. First, our model allows gene expression measured at unevenly spaced time intervals and gene-specific differences in measurement schedule, although these issues can be incorporated into Ng et al.'s model through extensive modifications. Second, we derived a closed form for the estimation of all the Fourier parameters for each gene cluster within the EM framework, whereas Ng et al. estimated the period of the expression cycle for the mean Fourier curve of all genes by using the least-squares estimation approach. Third, our model is able and flexible enough to consider Fourier series approximation of any arbitrary order. We conducted an additional simulation experiment to compare the results from our and Ng et al.'s models. The data from 1000 time-dependent gene expressions were simulated by assuming that these follow a multivariate normal distribution with gene cluster-specific mean vectors each fitted by a group of Fourier parameters and covariance matrix structured by the AR(1) model. Both our and Ng et al.'s models can correctly estimate the number of gene clusters, i.e., three in this simulated example (Table 6). While our model is able to provide reasonably accurate estimates of the Fourier parameters, Ng et al.'s model was quite biased for many parameter estimates. This comparative analysis suggests that our model will perform better than Ng et al.'s model when gene-expression profiles follow Fourier series approximations.
TABLE 6.
Comparisons between the results from our and Ng et al.'s (2006) models for clustering time series gene-expression profiles that are approximated by the first-order Fourier series
| Cluster | Model | Proportion | a0 | a1 | b1 | T |
|---|---|---|---|---|---|---|
| 1 | True | 0.585 | 0.300 | 1.000 | 0.200 | 6.000 |
| Ours | 0.584 | 0.298 | 1.002 | 0.202 | 5.999 | |
| Ng | 0.421 | 1.012 | −0.273 | −0.591 | 14.770 | |
| 2 | True | 0.100 | 0.030 | 1.000 | 0.020 | 10.000 |
| Ours | 0.101 | 0.048 | 0.996 | 0.030 | 10.003 | |
| Ng | 0.262 | 0.078 | 0.857 | −0.120 | 14.770 | |
| 3 | True | 0.315 | 0.060 | 0.900 | 0.010 | 16.000 |
| Ours | 0.315 | 0.054 | 0.904 | 0.016 | 15.993 | |
| Ng | 0.316 | 0.041 | 0.548 | 0.018 | 14.770 |
DISCUSSION
The use of microarray gene-expression technologies to understand developmental questions has received considerable attention in recent years (Panda et al. 2002). Statistical approaches for analyzing time-dependent gene expression data have been proposed (Holter et al. 2001; Qian et al. 2001; Bar-Joseph et al. 2003; Luan and Li 2003; Park et al. 2003; Wakefield et al. 2003; Ernst et al. 2005; Storey et al. 2005; Ma et al. 2006; Ng et al. 2006; Inoue et al. 2007), but most of them are limited in both biological and statistical aspects. First, these approaches mostly based on a clustering analysis were not implemented with biological principles of gene expression that are related to a life process. For this reason, the results obtained from these approaches may not be biologically relevant and, thus, may be less useful for deciphering the developmental machinery of gene expression. The model proposed in this article integrates mathematical aspects of periodic gene expression into the analytical framework, thereby allowing for the interplay between gene expression and development.
Second, many existing approaches to clustering genes of a similar expression pattern on the basis of a similarity measure have not considered the autocorrelation of time series data and, therefore, fail to remove systematical measurement errors. Although some authors implemented time-dependent correlations into their models (e.g., Luan and Li 2003; Ng et al. 2006), biological meanings of gene expression were not well considered. In the model proposed, the statistical principle of functional data analysis has been embedded into the model by structuring the time-dependent covariance matrix. This, on the one hand, de-noises repeated measurement errors and increases the effectiveness of the model and, on the other hand, enhances the model's power due to a reduced number of parameters being estimated. As an illustration, we used a simple AR(1) model to approximate the covariance structure. Other models, such as a structured antedependence model (Zimmerman and Núñez-Antón 2001), can also be incorporated (see Jaffrézic et al. 2003; Zhao et al. 2005), allowing the choice of an optimal model for structuring the covariance matrix. Finally, the mixture model-based approach allows for the estimation of the frequencies of various patterns of gene expression and the calculation of the posterior probability of each gene that belongs to a particular pattern.
The mixture-based approach incorporated by Fourier series approximation is a promising technique for detecting periodic gene-expression patterns. A major advantage of this approach lies in its remarkable flexibility to ask and address fundamental biological questions at the interplay between gene-expression and developmental patterns. Several important hypotheses can be made from this approach, including those about the differences of gene expression in Fourier curve shapes, curve features, and duration of gene expression based on individual Fourier parameters that describe biological characteristics of periodic cycles. For example, the peak-to-trough ratio, am/bm, reflects the amplitude of expression profile and can be tested for its differences among the gene groups detected. If the mean curve is modeled with the Fourier series of order one, i.e.,
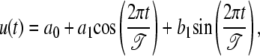 |
the hypothesis can be expressed as
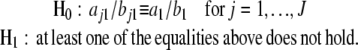 |
The slope of the gene expression profile may change with time, which suggests the occurrence of gene expression × time interaction effects during a time course. The differentiation of u(t) with respect to time t represents a slope of gene expression. If the slopes at a particular time point t* are different between the curves of different gene groups, this means that significant gene expression × time interaction occurs between this time point and next. The test for gene expression × time interaction can be formulated with the hypotheses
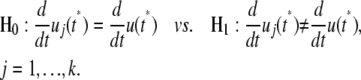 |
The effect of gene expression × time interaction can be examined during a given time course.
The new approach was used to analyze a real data set for periodic gene expression. The results from this approach suggest that it would be useful for the identification of gene clusters in terms of their periodic expression patterns. Through simulation studies, this approach has proved to provide reasonable accuracy and precision of parameter estimation and can be directly used to analyze a real data set of periodic gene expression. This approach can be modified or extended in the following areas. First, the clustering and estimation of different gene-expression profiles depends on the precise estimation of covariance functions. Fan et al. (2007b) proposed a semiparametric approach for modeling the covariance structure, which has been shown to be particularly powerful for functional data collected at irregular and subject-specific time points. The incorporation of Fan et al.'s approach into our functional clustering model is expected to improve its power for gene clustering. Second, when repeated measurement includes a high number of time points, the structuring of the covariance matrix may be quickly problematic. A handful of statistical models for dimension reduction proposed by J. Fan and his group (Fan et al. 2007a; Fan and Lv 2008) can be incorporated into our model, in a hope to increase the tractability of high-dimensional data. With these and other modifications, the approach for gene clustering presented in this article could be useful for addressing some development-relevant questions in genetic control of complex biological processes. The computer code for the approach proposed in this article is available at statgen.ufl.edu.
Acknowledgments
We thank the two anonymous referees for their constructive comments on the manuscript, which led to significant improvement of its presentation. Part of this work was conducted when R. Wu spent his sabbatical leave at Princeton University. The preparation of this manuscript was partially supported by National Science Foundation grant no. 0540745 to R. Wu and the Brain Korea 21 project in 2007.
APPENDIX
In what follows, we derive the log-likelihood equations for estimating the unknown vector 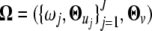 . The log-likelihood of parameters
. The log-likelihood of parameters 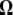 constructed on the basis of the mixture model is expressed as
constructed on the basis of the mixture model is expressed as
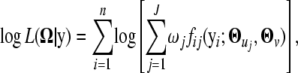 |
and the posterior probability with which the ith gene belongs to the jth pattern is defined by Equation 4. Since 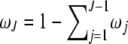 , we have
, we have
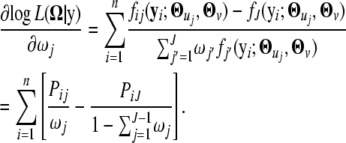 |
By setting it equal to zero, we have
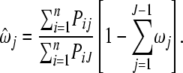 |
(A1) |
By plugging in 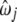 into the right side of Equation A1, we have
into the right side of Equation A1, we have
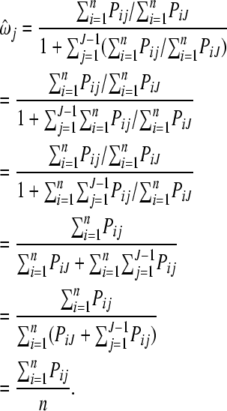 |
(A2) |
Assuming that the model is implemented by a first-order Fourier series approximation, unknown Fourier parameters are specified as 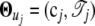 , where cj = (a0j, a1j, b1j). We have
, where cj = (a0j, a1j, b1j). We have
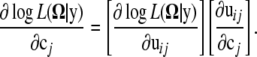 |
Note that
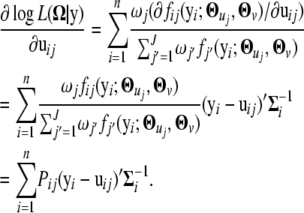 |
Let
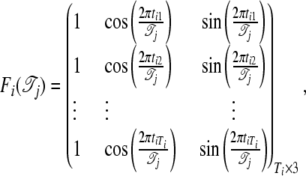 |
and then we have
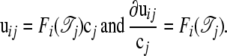 |
To solve cj, we conduct the differentiation as
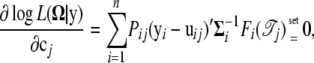 |
which leads to
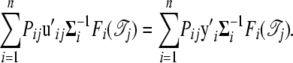 |
We further have
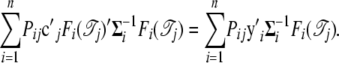 |
If 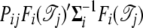 is invertable, we have
is invertable, we have
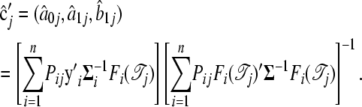 |
(A3) |
Note that 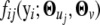 can be written as
can be written as
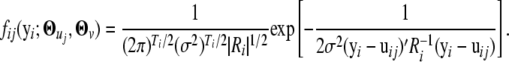 |
If we write 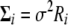 , where
, where
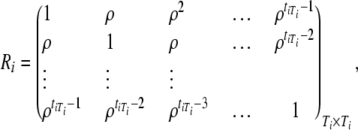 |
we have
 |
and
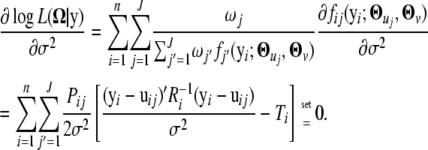 |
By solving it for σ2, we have
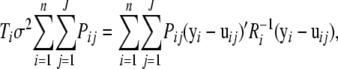 |
so
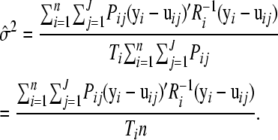 |
(A4) |
For the AR(1) model, we have
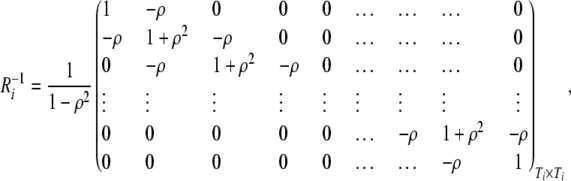 |
and
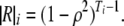 |
Let
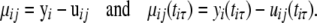 |
Then, we have
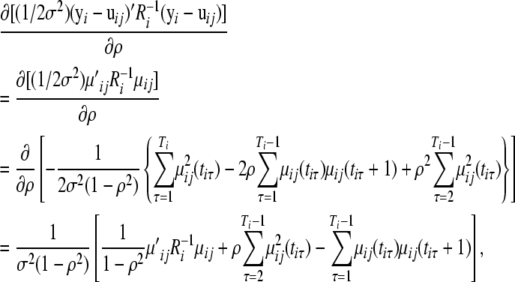 |
and
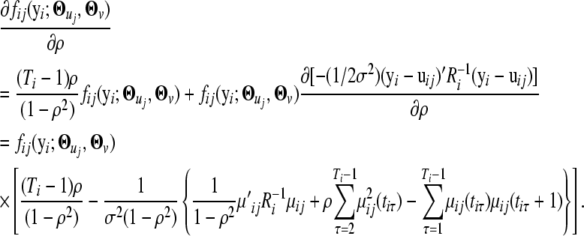 |
Therefore, we have
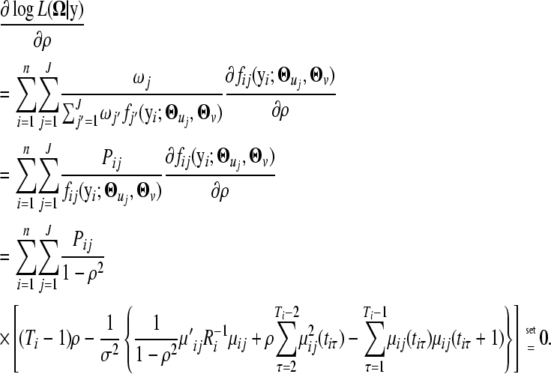 |
By solving the above log-likelihood equation, the MLE of ρ can be obtained as
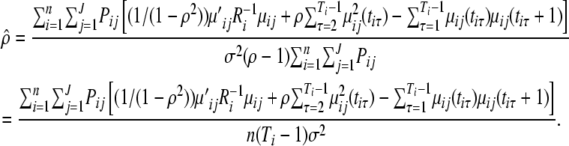 |
(A5) |
The conditional expectation estimated with Equation 4 in the E step is used to solve the unknown parameters with log-likelihood Equations A2–A5 in the M step. But since it is impossible to derive a closed form for 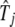 , we implement the simplex algorithm to estimate this parameter in the M step. The E and M steps are repeated until the estimates are stable.
, we implement the simplex algorithm to estimate this parameter in the M step. The E and M steps are repeated until the estimates are stable.
References
- Akaike, H., 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19 716–723. [Google Scholar]
- Attinger, E. O., A. Anne and D. A. McDonald, 1966. Use of Fourier series for the analysis of biological systems. Biophys. J. 6 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph, Z., G. K. Gerber, D. K. Gifford, T. S. Jaakkola and I. Simon, 2003. Continuous representations of time-series gene expression data. J. Comput. Biol. 10 341–356. [DOI] [PubMed] [Google Scholar]
- Begum, E. A., B. Motoki, O. Makoto, Y. Hatsumi, K. Masatoshi et al., 2006. Emergence of physiological rhythmicity in term and preterm neonates in a neonatal intensive care unit. J. Circadian Rhythms 4 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, R. J., and D. Ruppert, 1984. Power-transformations when fitting theoretical models to data. J. Am. Stat. Assoc. 79 321–328. [Google Scholar]
- Crosthwaite, S. K., 2004. Circadian clocks and natural antisense. RNA FEBS Lett. 567 49–54. [DOI] [PubMed] [Google Scholar]
- Dale, J. K., M. Maroto, M. L. Dequeant, P. Malapert, M. McGrew et al., 2003. Periodic notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature 421 275–278. [DOI] [PubMed] [Google Scholar]
- Davidian, M., and D. M. Giltinan, 1995. Nonlinear Models for Repeated Measurement Data. Chapman & Hall, London.
- Durbin, J., 1967. Tests of serial independence based on the cumulated periodogram. Bull. Int. Stat. Inst. 42 1039–1049. [Google Scholar]
- Eisen, M. B., P. T. Spellman, P. O. Brown and D. Botstein, 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, J., G. J. Nau and Z. Bar-Joseph, 2005. Clsutering short time series gene expression data. Bioinformatics 21 i159–i168. [DOI] [PubMed] [Google Scholar]
- Fan, J., and J. Lv, 2008. Sure independence screening for ultra-high dimensional feature space (with discussion). J. R. Stat. Soc. Ser. B 70 849–911. [DOI] [PMC free article] [PubMed]
- Fan, J., and Y. Ren, 2006. Statistical analysis of DNA microarray data in cancer research. Clin. Cancer Res. 12 4469–4473. [DOI] [PubMed] [Google Scholar]
- Fan, J., Y. Fan and J. Lv, 2007. a High dimensional covariance matrix estimation using a factor model. J. Econom (in press).
- Fan, J., T. Huang and R. Z. Li, 2007. b Analysis of longitudinal data with semiparametric estimation of covariance function. J. Am. Stat. Assoc. 35 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley, C., and A. E. Raftery, 1998. How many clusters? Which clustering method? Answers via model-based cluster analysis. Technical Report 329. Department of Statistics, University of Washington, Seattle.
- Frank, O., 1926. Die theorie der pulswellen. Zool. Biol. 85 91–130. [Google Scholar]
- Ghosh, D., and A. M. Chinnaiyan, 2002. Mixture modeling of gene expression data from microarray experiments. Bioinformatics 18 275–286. [DOI] [PubMed] [Google Scholar]
- Glynn, E. F., J. Chen and A. R. Mushegian, 2006. Detecting periodic patterns in unevenly spaced gene expression time series using Lomb-Scargle periodograms. Bioinformatics 22 310–316. [DOI] [PubMed] [Google Scholar]
- Goldbeter, A., 2002. Computational approaches to cellular rhythms. Nature 420 238–245. [DOI] [PubMed] [Google Scholar]
- Gonze, D., J. Halloy and A. Goldbeter, 2002. Stochastic versus deterministic models for circadian rhythms. J. Biol. Phys. 28 637–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonze, D., J. Halloy, J. C. Leloup and A. Goldbeter, 2003. Stochastic models for circadian rhythms: influence of molecular noise on periodic and chaotic behavior. C. R. Biol. 326 189–203. [DOI] [PubMed] [Google Scholar]
- Harmer, S. L., J. B. Hogenesch, M. Straume, H.-S. Chang, B. Han et al., 2000. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113. [DOI] [PubMed] [Google Scholar]
- Holter, N. S., A. Maritan and M. Cieplak, 2001. Dynamic modeling of gene expression data. Proc. Natl. Acad. Sci. USA 98 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, L. Y., M. Neira, C. Nelson, M. Gleave and R. Etzioni, 2007. Cluster-based network model for time-course gene expression data. Biostatistics 8 507–525. [DOI] [PubMed] [Google Scholar]
- Jaffrézic, F., R. Thompson and W. G. Hill, 2003. Structured antedependence models for genetic analysis of multivariate repeated measures in quantitative traits. Genet. Res. 82 55–65. [DOI] [PubMed] [Google Scholar]
- Kim, B.-R., R. C. Littell and R. L. Wu, 2006. Clustering the periodic pattern of gene expression using Fourier series approximations. Curr. Genomics 7 197–203. [Google Scholar]
- Lakin-Thomas, P. L., and S. Brody, 2004. Circadian rhythms in microorganisms: new complexities. Annu. Rev. Microbiol. 58 489–519. [DOI] [PubMed] [Google Scholar]
- Leloup, J.-C., D. Gonze and A. Goldbeter, 1999. Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J. Biol. Rhythms 14 433–448. [DOI] [PubMed] [Google Scholar]
- Luan, Y., and H. Li, 2003. Clustering of time-course gene expression data using a mixed-effects model with B-splines. Bioinformatics 19 474–482. [DOI] [PubMed] [Google Scholar]
- Luan, Y., and H. Li, 2004. Model-based methods for identifying periodically expressed genes based on time course microarray gene expression data. Bioinformatics 20 332–339. [DOI] [PubMed] [Google Scholar]
- Ma, P., C. I. Castillo-Davis, W. Zhong and J. S. Liu, 2006. A data-driven clustering method for time course gene expression data. Nucleic Acids Res. 34 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager, D. E., and D. R. Abernethy, 2007. Use of wavelet and fast Fourier transforms in pharmacodynamics. J. Pharmacol. Exp. Ther. 321 423–430. [DOI] [PubMed]
- McLachlan, G., and D. Peel, 2000. Finite Mixture Models. John Wiley & Sons, New York.
- McLachlan, G. J., R. W. Bean and D. Peel, 2002. A mixture model-based approach to the clustering of microarray expression data. Bioinformatics 18 414–422. [DOI] [PubMed] [Google Scholar]
- Mitchison, J. M., 2003. Growth during the cell cycle. Int. Rev. Cytol. 226 165–258. [DOI] [PubMed] [Google Scholar]
- Ng, S. K., G. J. McLachlan, K. Wang, L. B.-T. Jones and S.-W. Ng, 2006. A mixture model with random-effects components for clustering correlated gene-expression profiles. Bioinformatics 22 1745–1752. [DOI] [PubMed] [Google Scholar]
- Panda, S., T. K. Sato, A. M. Castrucci, M. D. Rollag, W. J. DeGrip et al., 2002. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science 298 2213–2216. [DOI] [PubMed] [Google Scholar]
- Park, T., S. G. Yi, S. Lee, S. Y. Lee, D. H. Yoo et al., 2003. Statistical tests for identifying differentially expressed genes in time-course microarray experiments. Bioinformatics 19 694–703. [DOI] [PubMed] [Google Scholar]
- Pletcher, S. D., and C. J. Geyer, 1999. The genetic analysis of age-dependent traits: modeling a character process. Genetics 153 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestley, M. B., 1981. Spectral Analysis and Time Series. Academic Press, San Diego.
- Prolo, L. M., J. S. Takahashi and E. D. Herzog, 2005. Circadian rhythm generation and entrainment in astrocytes. J. Neurosci. 25 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, J., B. Stenger, C. A. Wilson, J. Lin, R. Jansen et al., 2001. Partslist: a web-based system for dynamically ranking protein folds based on disparate attributes, including whole-genome expression and interaction information. Nucleic Acids Res. 29 1750–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramoni, M. F., P. Sebastiani and I. S. Kohane, 2002. Cluster analysis of gene expression dynamics. Proc. Natl. Acad. Sci. USA 99 9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovery, C., M. V. La, S. Robineau, K. Matsumoto, P. Renesto et al., 2005. Preliminary transcriptional analsysis of spoT gene family and of membrane proteins in Rickettsia conorii and Rickettsia felis. Ann. NY Acad. Sci. 1063 79–82. [DOI] [PubMed] [Google Scholar]
- Schwarz, G., 1978. Estimating the dimension of a model. Ann. Stat. 6 461–464. [Google Scholar]
- Spellman, P. T., S. Sherlock, M. Q. Zhang, V. R. Iyer, K. Aners et al., 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J. D., W. Xiao, J. T. Leek, R. G. Tompkins and R. W. Davis, 2005. Significant analysis of time course microarray experiments. Proc. Natl. Acad. Sci. USA 102 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke, G., and G. Molenberghs, 2000. Linear Mixed Models for Longitudinal Data. Springer-Verlag, New York.
- Wakefield, J., C. Zhou and S. G. Self, 2003. Modelling gene expression data over time: curve clustering with informative prior distributions, pp. 721–732 in Bayesian Statistics 7, Proceedings of the Seventh Valencia International Meeting, edited by J. M. Bernardo, M. J. Bayarri, J. O. Berger, A. P. Dawid, D. Heckerman, A. F. M. Smith and M. West. Oxford University Press, London/New York/Oxford.
- Wichert, S., K. Fokianos and K. Strimmer, 2004. Identifying periodically expressed transcripts in microarray time series data. Bioinformatics 20 5–20. [DOI] [PubMed] [Google Scholar]
- Wu, R. L., C.-X. Ma, M. Lin, Z. H. Wang and G. Casella, 2004. Functional mapping of quantitative trait loci underlying growth trajectories using a transform-both-sides logistic model. Biometrics 60 729–738. [DOI] [PubMed] [Google Scholar]
- Zhao, W., Y. Q. Chen, G. Casella, J. M. Cheverud and R. L. Wu, 2005. A nonstationary model for functional mapping of complex traits. Bioinformatics 21 2469–2477. [DOI] [PubMed] [Google Scholar]
- Zimmerman, D. L., and V. Núñez-Antón, 2001. Parametric modeling of growth curve data: an overview (with discussion). Test 10 1–73. [Google Scholar]