Abstract
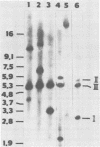
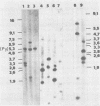
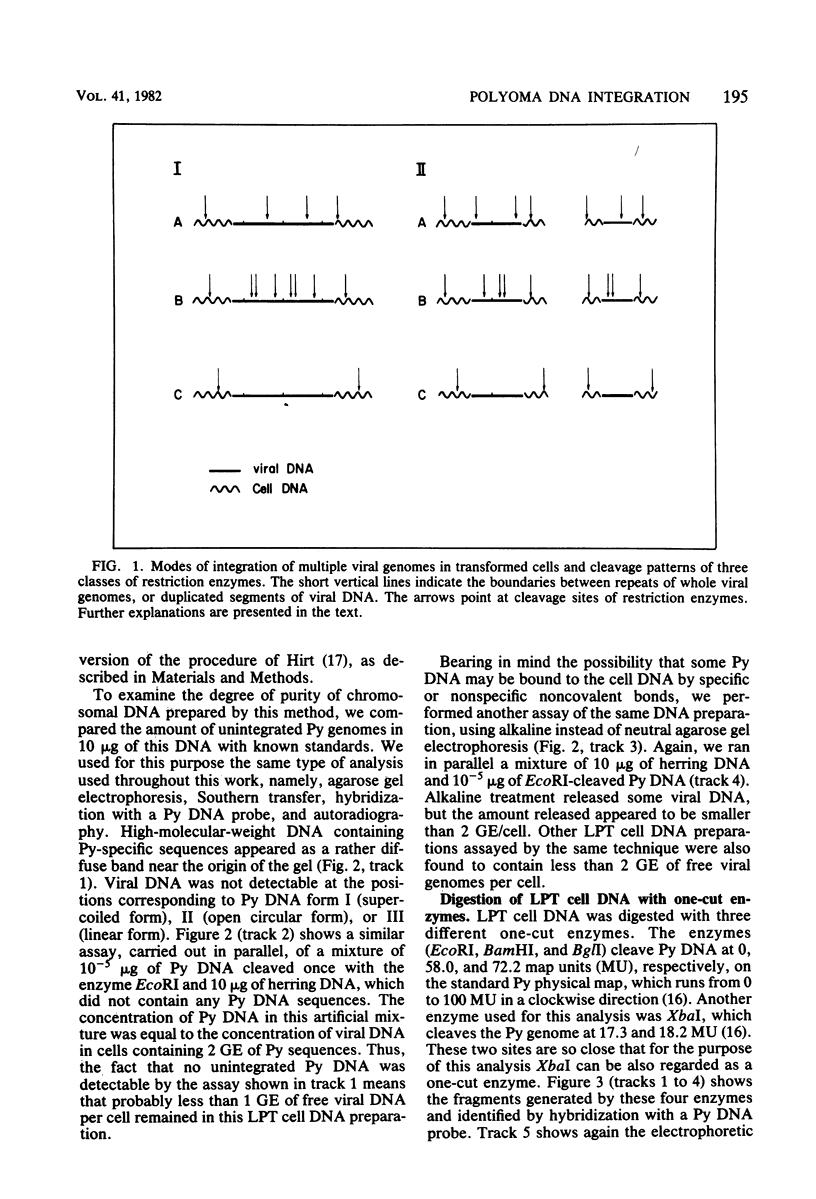
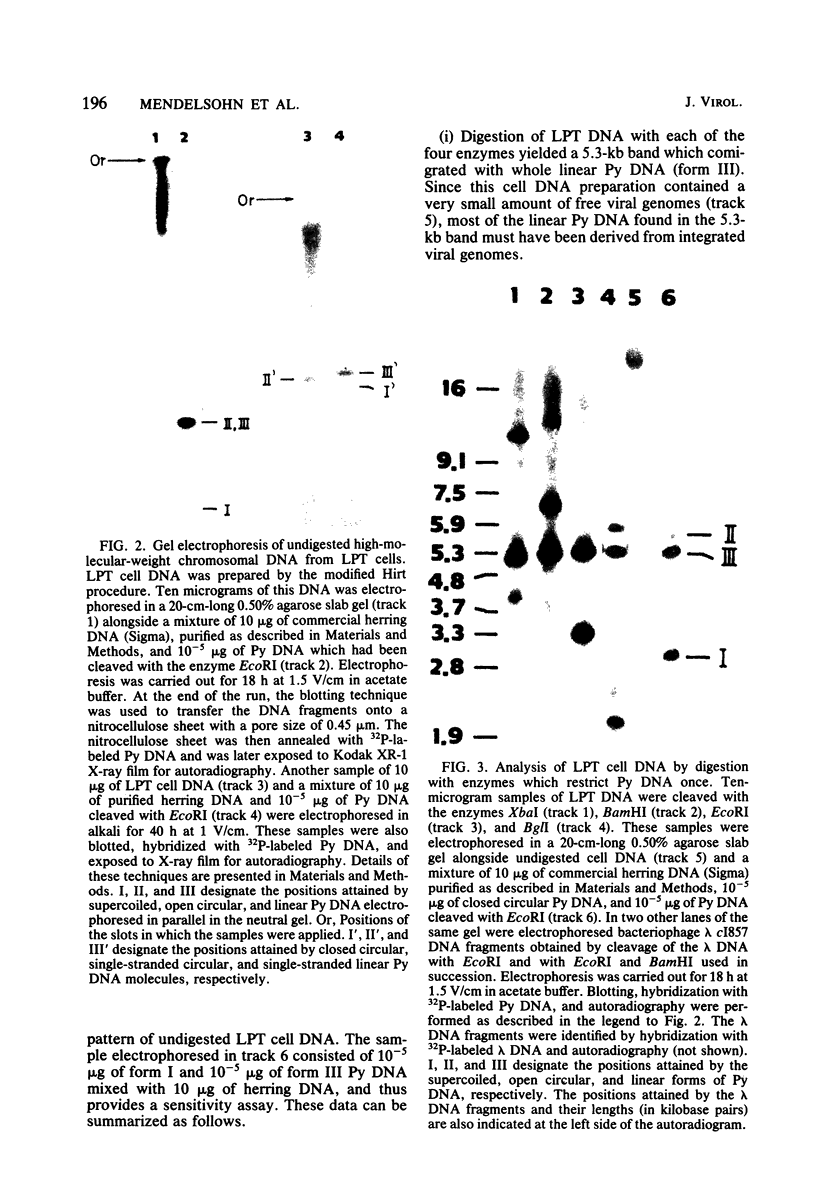
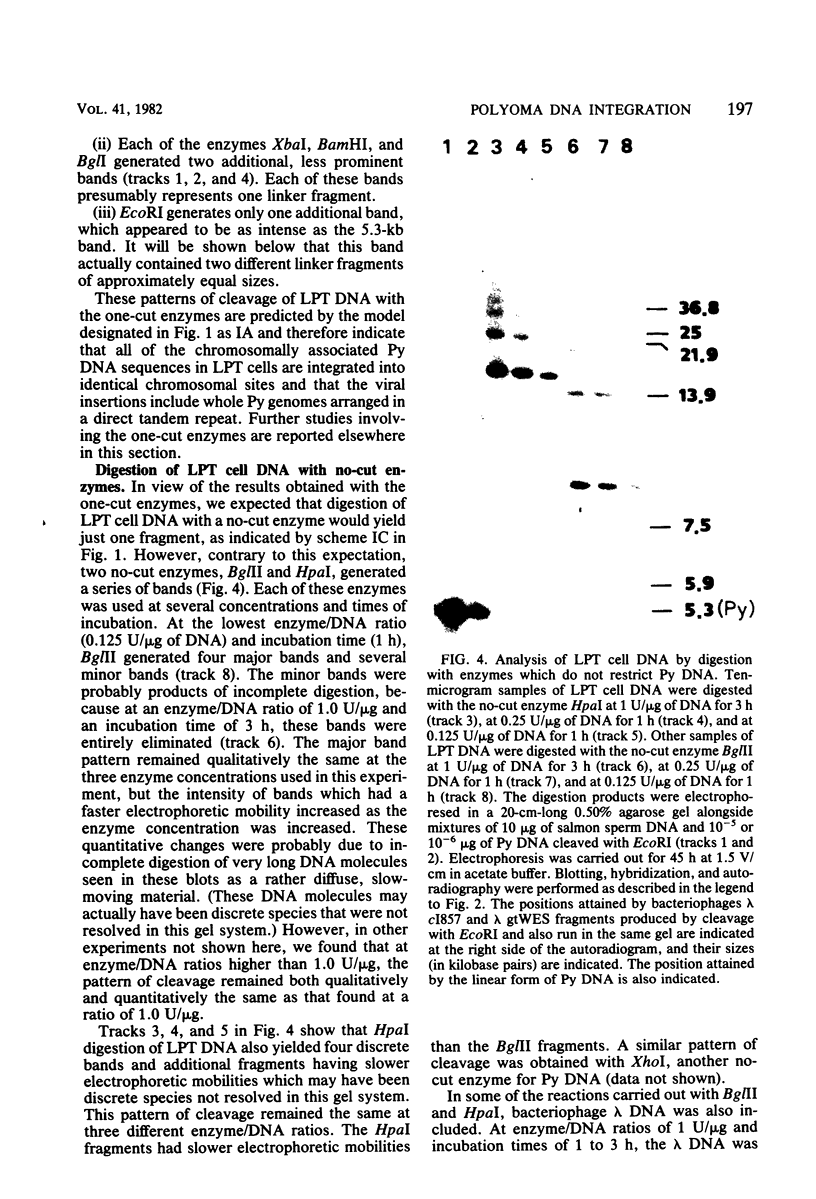
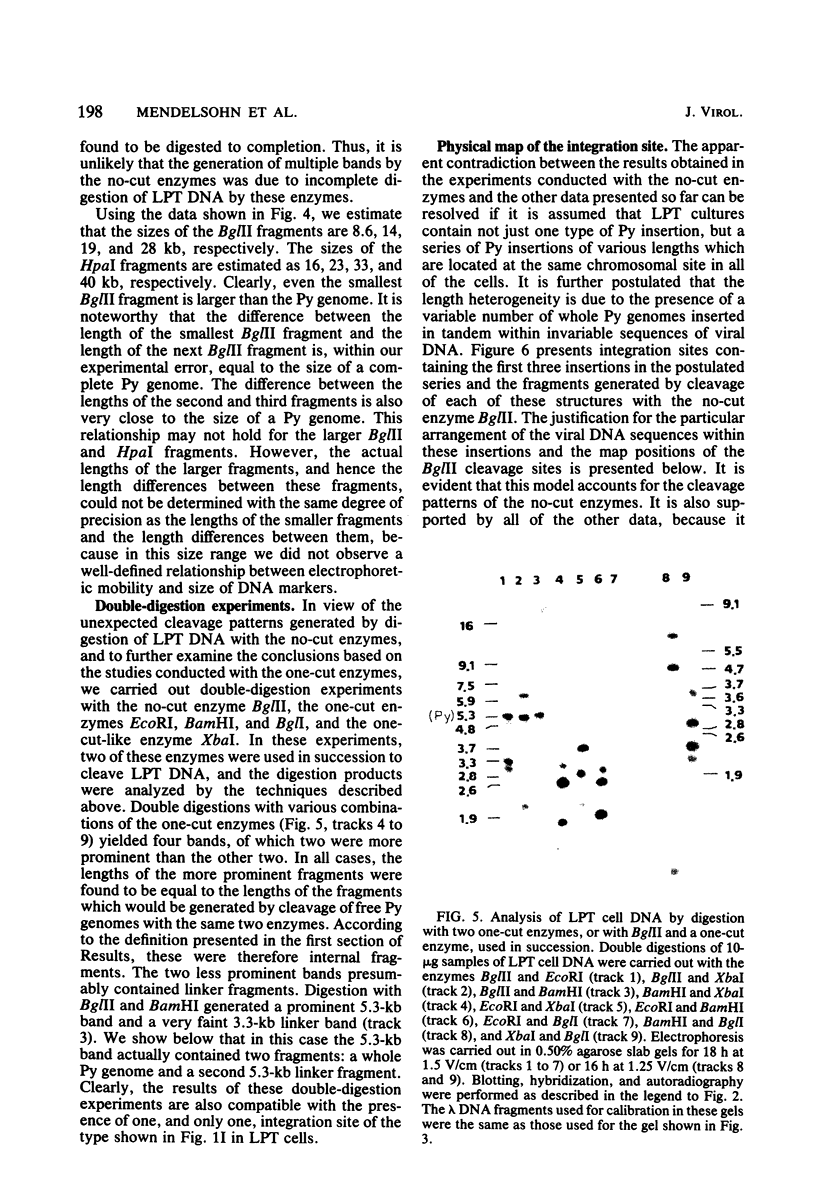
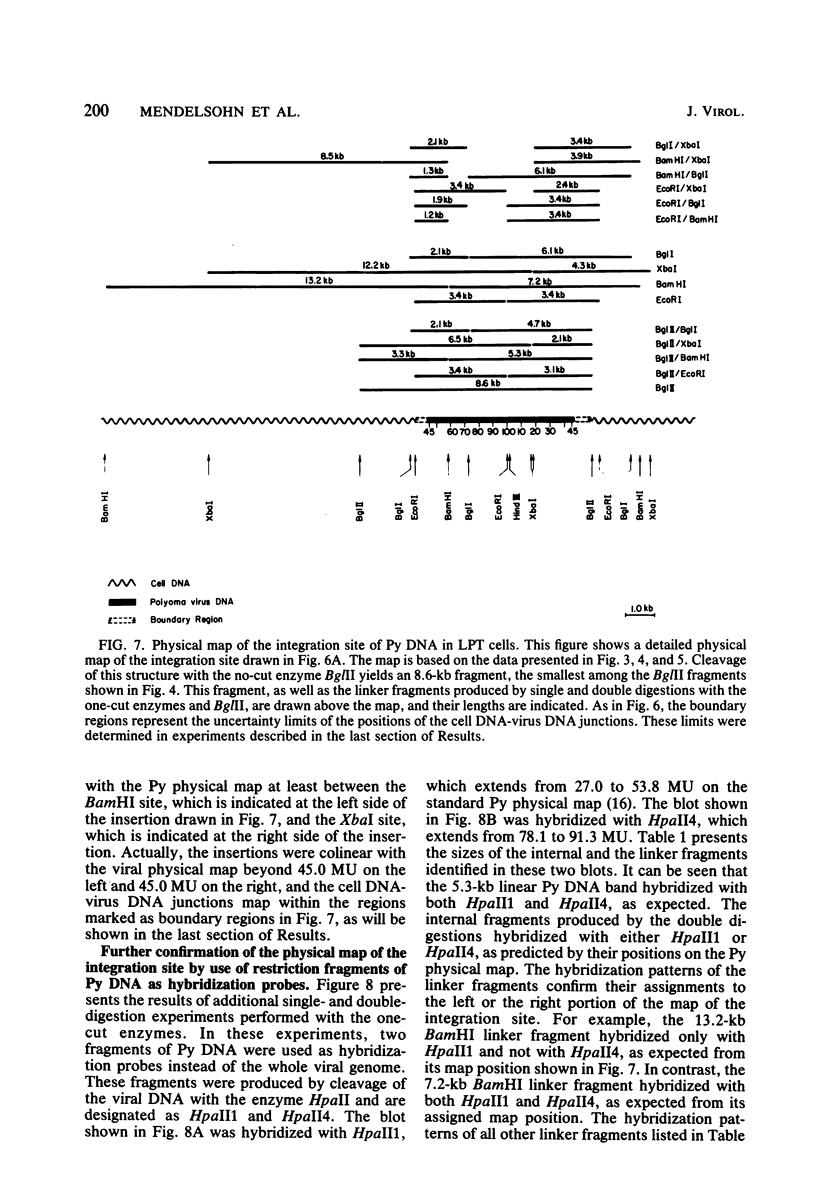
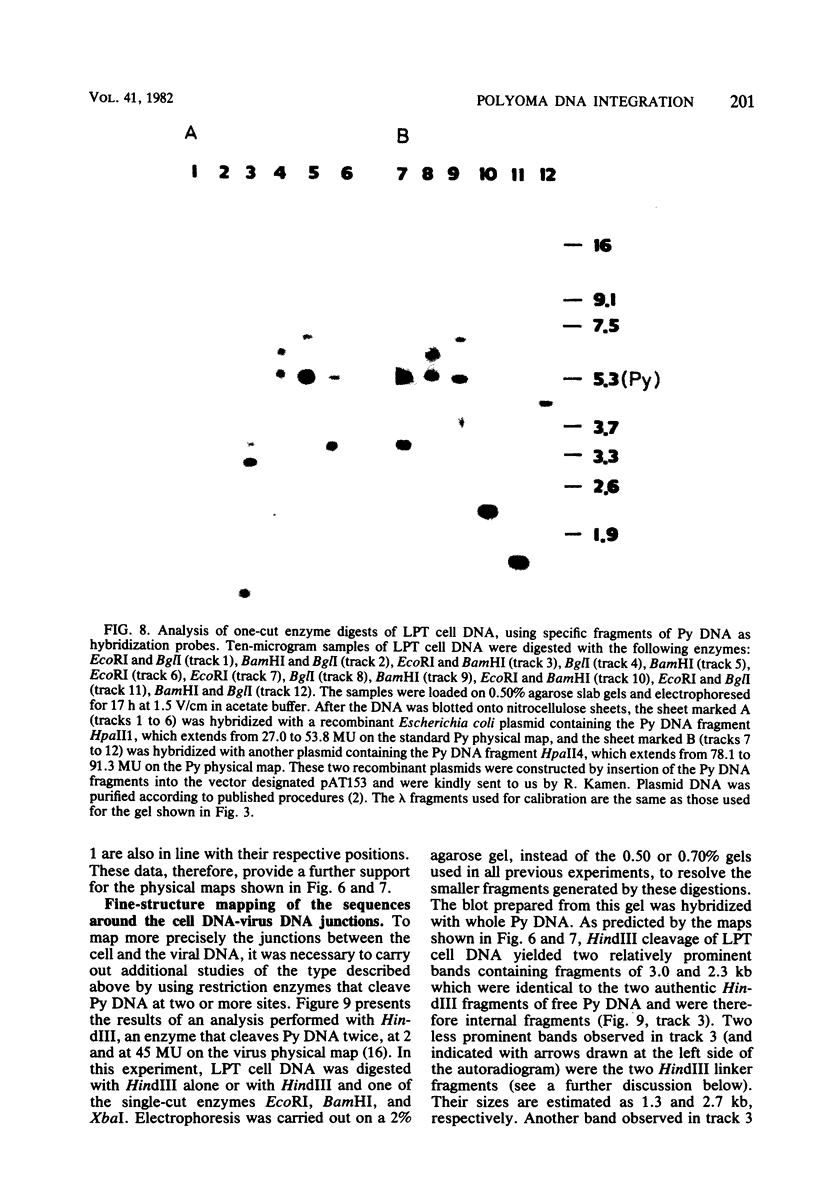
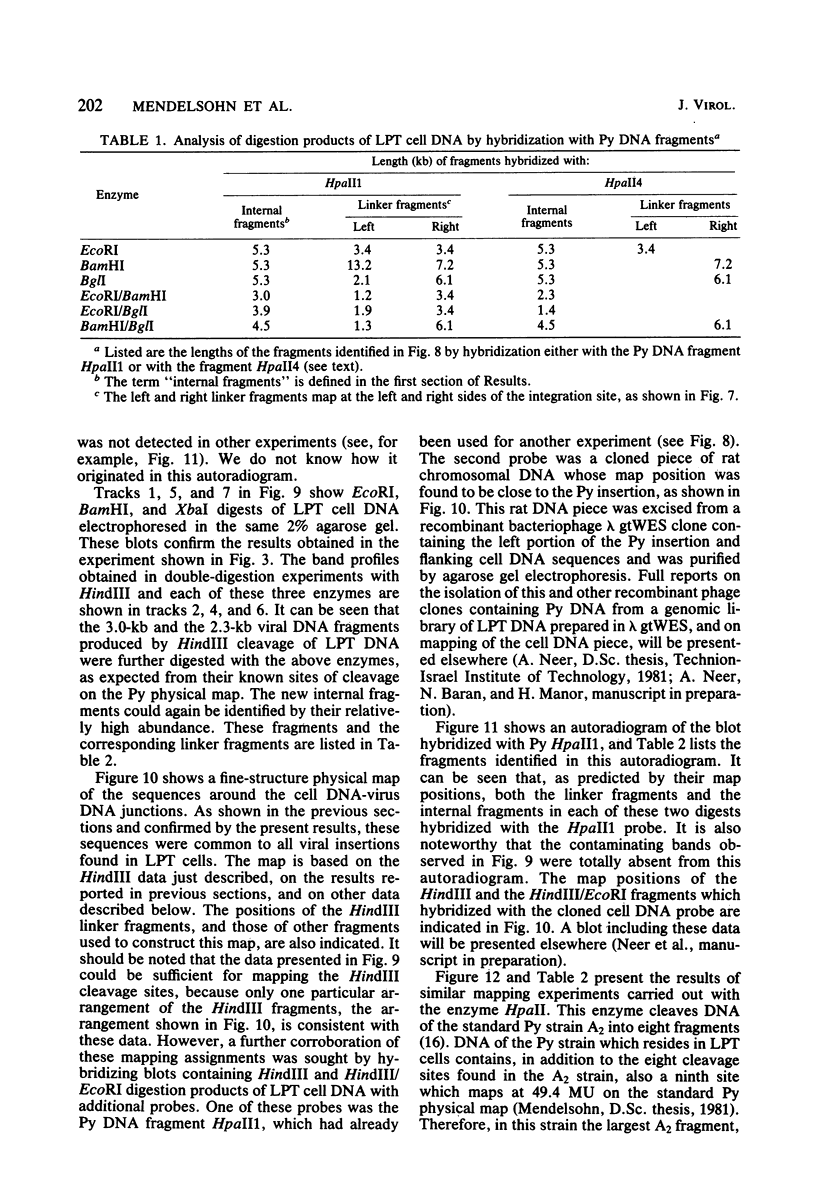
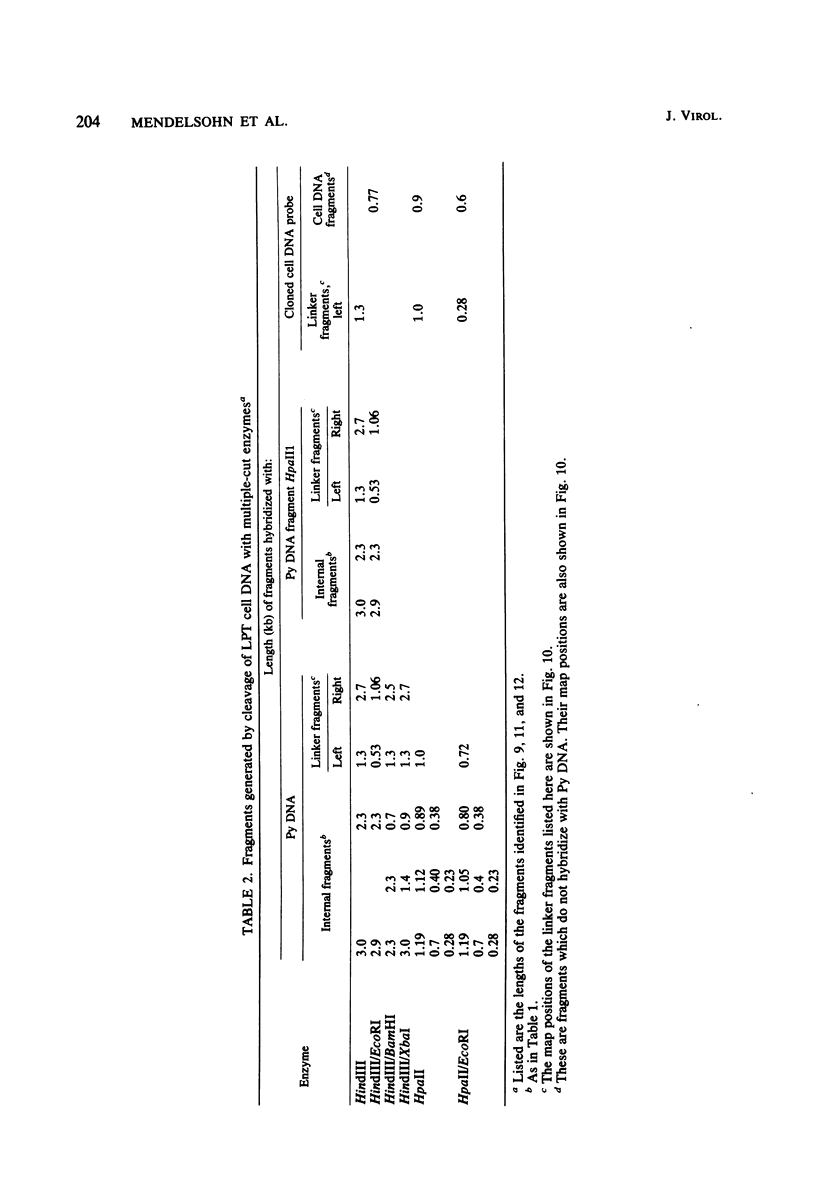
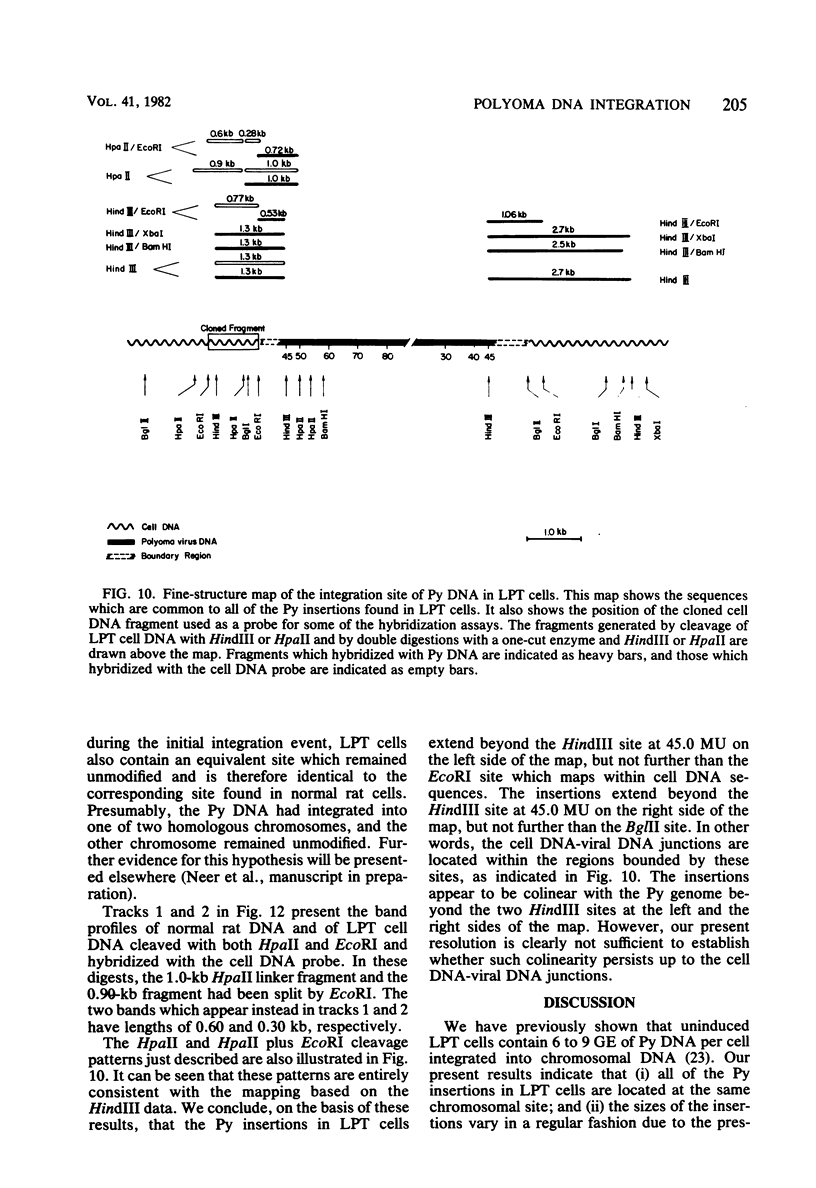
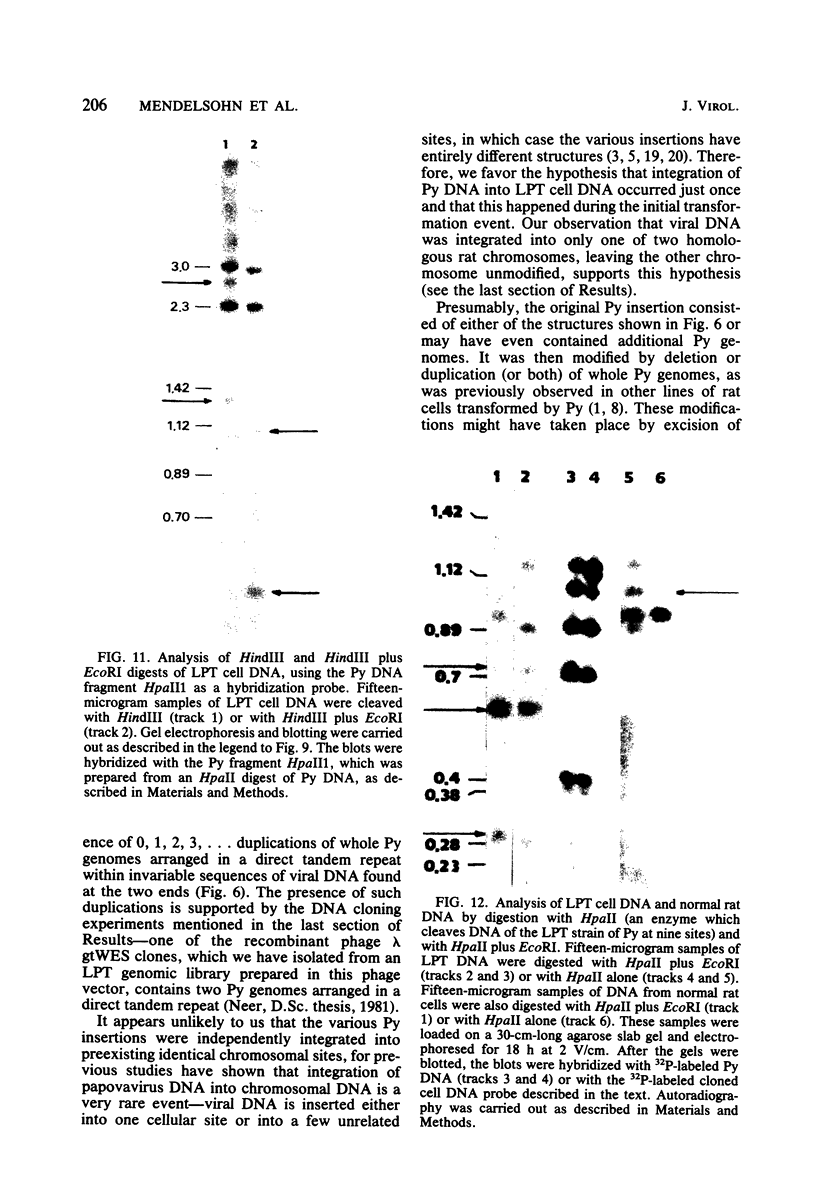
The structure of the polyoma virus (Py) integration site in the inducible LPT line of Py-transformed rat cells was determined by biochemical methods of gene mapping. LPT cell DNA was digested with various restriction enzymes. The digestion products were electrophoresed in agarose gels and transferred onto nitrocellulose sheets by Southern blotting. Fragments containing viral or cell DNA sequences, or both, were identified by hybridization with Py DNA or with a cloned flanking cell DNA probe. Cleavage of LPT DNA with enzymes that restrict the Py genome once generated linear Py DNA molecules and two fragments containing both cell and viral DNA sequences. Cleavage of LPT DNA with enzymes which do not restrict Py DNA generated series of fragments whose lengths were found to differ by increments of a whole Py genome; the smallest fragment in each series was found to be longer than the viral genome. These data indicate that LPT cultures contain Py insertions of various lengths integrated into the same chromosomal site in all the cells. The length heterogeneity of the viral insertions is due to the presence of 0, 1, 2, 3. . . Py genomes arranged in a direct tandem repeat within invariable sequences of viral DNA. Double-digestion experiments were also carried out with the above enzymes and with enzymes that cleave the Py genome at multiple sites. The data obtained in these experiments were used to construct a physical map of the integration site. This map showed that the early region of the virus remained intact even in the smallest insertion (which contains no whole duplicated genomes), whereas the late region was partially duplicated and split during integration. The smallest insertion is colinear with the Py physical map over a region including the entire Py genome and at least a part of the duplicated segment. This structure could give rise to nondefective circular viral DNA molecules by single homologous recombination events. Similar recombination events may occur at a higher frequency in the longer insertions, which include longer regions of homology, and may yield many more free viral genomes. The presence of these insertions in LPT cells could thus be one of the factors which account for the high inducibility of the LPT line.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basilico C., Gattoni S., Zouzias D., Valle G. D. Loss of integrated viral DNA sequences in polyomatransformed cells is associated with an active viral A function. Cell. 1979 Jul;17(3):645–659. doi: 10.1016/0092-8674(79)90272-1. [DOI] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Stringer J., Mitchison T., Sambrook J. Integration and excision of SV40 DNA from the chromosome of a transformed cell. Cell. 1980 May;20(1):143–152. doi: 10.1016/0092-8674(80)90242-1. [DOI] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. Studies on simian virus 40 excision from cellular chromosomes. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):709–719. doi: 10.1101/sqb.1979.043.01.079. [DOI] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Campo M. S., Cameron I. R., Rogers M. E. Tandem integration of complete and defective SV40 genomes in mouse-human somatic cell hybrids. Cell. 1978 Dec;15(4):1411–1426. doi: 10.1016/0092-8674(78)90065-x. [DOI] [PubMed] [Google Scholar]
- Colantuoni V., Dailey L., Basilico C. Amplification of integrated viral DNA sequences in polyoma virus-transformed cells. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3850–3854. doi: 10.1073/pnas.77.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DULBECCO R., VOGT M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J Exp Med. 1954 Feb;99(2):167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Valle G., Fenton R. G., Basilico C. Polyoma large T antigen regulates the integration of viral DNA sequences into the genome of transformed cells. Cell. 1981 Feb;23(2):347–355. doi: 10.1016/0092-8674(81)90130-6. [DOI] [PubMed] [Google Scholar]
- Fogel M. Induction of virus synthesis in polyoma-transformed cells by DNA antimetabolites and by irradiation after pretreatment with 5-bromodeoxyuridine. Virology. 1972 Jul;49(1):12–22. doi: 10.1016/s0042-6822(72)80003-5. [DOI] [PubMed] [Google Scholar]
- Fogel M., Sachs L. Induction of virus synthesis in polyoma transformed cells by ultraviolet light and mitomycin C. Virology. 1970 Jan;40(1):174–177. doi: 10.1016/0042-6822(70)90391-0. [DOI] [PubMed] [Google Scholar]
- Fogel M., Sachs L. The activation of virus synthesis in polyoma-transformed cells. Virology. 1969 Mar;37(3):327–334. doi: 10.1016/0042-6822(69)90216-5. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Ketner G., Kelly T. J., Jr Integrated simian virus 40 sequences in transformed cell DNA: analysis using restriction endonucleases. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1102–1106. doi: 10.1073/pnas.73.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Lev Z., Kamen R., Manor H. Topography of polyoma virus-specific giant nuclear RNA molecules containing poly(A) sequences. Virology. 1979 Mar;93(2):445–457. doi: 10.1016/0042-6822(79)90248-4. [DOI] [PubMed] [Google Scholar]
- Manor H., Fogel M., Sachs L. Integration of viral into chromosomal deoxyribonucleic acid in an inducible line of polyoma-transformed cells. Virology. 1973 May;53(1):174–185. doi: 10.1016/0042-6822(73)90476-5. [DOI] [PubMed] [Google Scholar]
- Manor H., Kamen R. Polyoma virus-specific RNA synthesis in an inducible line of polyoma virus-transformed rat cells. J Virol. 1978 Mar;25(3):719–729. doi: 10.1128/jvi.25.3.719-729.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor H., Neer A. Effects of cycloheximide on virus RNA replication in an inducible line of polyoma-transformed rat cells. Cell. 1975 Jul;5(3):311–318. doi: 10.1016/0092-8674(75)90106-3. [DOI] [PubMed] [Google Scholar]
- Mougneau E., Birg F., Rassoulzadegan M., Cuzin F. Integration sites and sequence arrangement of SV40 DNA in a homogeneous series of transformed rat fibroblast lines. Cell. 1980 Dec;22(3):917–927. doi: 10.1016/0092-8674(80)90569-3. [DOI] [PubMed] [Google Scholar]
- Neer A., Baran N., Manor H. In situ hybridization analysis of polyoma DNA replication in an inducible line of polyoma-transformed cells. Cell. 1977 May;11(1):65–71. doi: 10.1016/0092-8674(77)90317-8. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tartof K. D. Unequal mitotic sister chromatin exchange as the mechanism of ribosomal RNA gene magnification. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1272–1276. doi: 10.1073/pnas.71.4.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINOCOUR E., SACHS L. Cell-virus interactions with the polyoma virus. 1. Studies on the lytic interaction in the mouse embryo system. Virology. 1960 Aug;11:699–721. doi: 10.1016/0042-6822(60)90115-x. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Lis J., Wu R. Elution of DNA from agarose gels after electrophoresis. Methods Enzymol. 1979;68:176–182. doi: 10.1016/0076-6879(79)68012-6. [DOI] [PubMed] [Google Scholar]