Abstract
MicroRNAs (miRNA) are crucial post-transcriptional regulators of gene expression and control cell differentiation and proliferation. However, little is known about their targeting of specific developmental pathways. Hedgehog (Hh) signalling controls cerebellar granule cell progenitor development and a subversion of this pathway leads to neoplastic transformation into medulloblastoma (MB). Using a miRNA high-throughput profile screening, we identify here a downregulated miRNA signature in human MBs with high Hh signalling. Specifically, we identify miR-125b and miR-326 as suppressors of the pathway activator Smoothened together with miR-324-5p, which also targets the downstream transcription factor Gli1. Downregulation of these miRNAs allows high levels of Hh-dependent gene expression leading to tumour cell proliferation. Interestingly, the downregulation of miR-324-5p is genetically determined by MB-associated deletion of chromosome 17p. We also report that whereas miRNA expression is downregulated in cerebellar neuronal progenitors, it increases alongside differentiation, thereby allowing cell maturation and growth inhibition. These findings identify a novel regulatory circuitry of the Hh signalling and suggest that misregulation of specific miRNAs, leading to its aberrant activation, sustain cancer development.
Keywords: cancer, expression profiling, Hedgehog, microRNA, post-transcriptional control
Introduction
Increasing evidence suggests that cancer may be considered as an aberrant morphogenetic process in which molecular signalling pathways regulating cell development are subverted (Beachy et al, 2004; Fogarty et al, 2005). Hedgehog (Hh) signalling pathway has a pivotal function in controlling embryonic patterning and is a master regulator of the development of cerebellar granule cell progenitors (GCPs) which proliferate in response to Purkinje cell-produced Shh (reviewed in Ruiz i Altaba, 2006). Constitutive activation of the Hh signalling alters the development of GCPs that remain overgrown, thus becoming susceptible of malignant transformation into medulloblastoma (MB), the most common paediatric brain malignancy (Ruiz i Altaba et al, 2002, 2006). Signalling from this pathway is triggered by the binding of the Hh extracellular proteins to the Patched receptor (Ptch), thus relieving its inhibitory activity upon Smoothened (Smo) that in turn activates the transcriptional activity of Gli factors. Gli1 is both the downstream effector and target gene of the pathway. Mice carrying a defective Ptch1 receptor or an activated Smo develop MB (Goodrich et al, 1997; Hallahan et al, 2004). Furthermore, a reduced development of MB is observed in Gli1−/−/Ptch1+/− double mutants, supporting the critical function of Gli1 in tumorigenesis (Kimura et al, 2005). A number of genetic and epigenetic abnormalities associated with human MB have been described to affect proteins forming intrinsic components or regulators of the pathway (reviewed in Ferretti et al, 2005; Ruiz i Altaba, 2006). Chromosome 17p, the deletion of which is the most frequent genetic alteration reported in this malignancy, is harbouring some of the genes the deregulation of which contributes to Hh-induced tumorigenesis (Ferretti et al, 2005; Briggs et al, 2008). Although a funneling effect upon the pathway deregulation has been suggested for the various MB-associated abnormalities, they do not account for the whole of Hh signalling subversion observed in this malignancy. More specifically, translational control of Hh pathway components and its deregulation in cancer have not been investigated yet.
MicroRNAs (miRNAs) are a class of small non-coding cellular RNAs that bind to cis-regulatory elements mainly present in the 3′ untranslated regions (3′UTRs) of target mRNAs, resulting in their translational inhibition or degradation (Stefani and Slack, 2008). Recently, miRNAs have been implicated in tumour formation (Calin and Croce, 2006) as well as in the control of neural cell development (Kosik, 2006). These observations suggest the potential involvement of misregulated miRNAs in the malignant transformation of developing neural progenitor cells; however, little is known about miRNA-mediated specific targeting of developmental pathways (i.e. Hh) involved in tumour formation.
In this study, a high-throughput screening of miRNA expression in distinct subsets of human MBs allowed us to identify specific miRNAs involved in the regulation of the Hh signalling: miR-125b, miR-324-5p and miR-326, downregulated in both MB and undifferentiated GCPs, were shown to target the activator components of the pathway, Smo and Gli1, thereby suppressing progenitor and tumour cell growth. Interestingly, the downregulation of miR-324-5p is at least partially due to deletion of chromosome 17p observed in 40% of MBs.
Results
A miRNA signature characterizes MBs with high Hh signalling
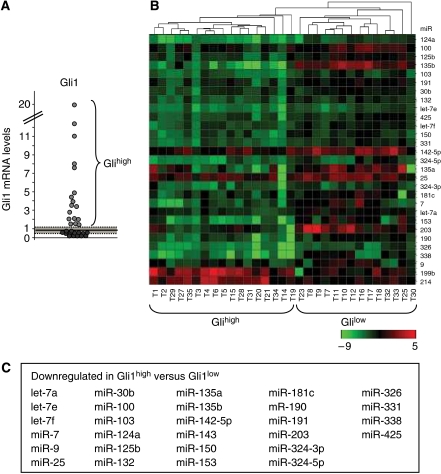
Most MBs have a functional Hh pathway (Dahmane et al, 2001; Berman et al, 2002). As Gli1 is a sensitive read out of the pathway activity, we analysed Gli1 levels in a series of human primary MBs and observed that a subset of them (54%) displays higher levels of Gli1 compared with other tumours or normal cerebellar tissue (Figure 1A). This suggests that they are characterized by an increase of Hh signalling strength due to a hyperactive deregulated pathway. To identify miRNAs able to regulate Hh signalling, we carried out an initial high-throughput expression profile analysis of miRNAs in the two subsets of human primary MBs: a subset with high Hh signalling strength (Gli1high) and a second subset with low normal levels of Gli1 (Gli1low). Gli1high tumours turned out to be characterized by a 34 miRNA signature compared with the Gli1low cluster (Supplementary Figure 1). A further analysis of these 34 differential miRNAs in an enlarged set of MBs (n=31) confirmed the statistical significance for 30 of them. Notably, most miRNAs were downregulated in Gli1high tumours (Figure 1B and C; Supplementary Table 1), suggesting that MB-associated loss of specific miRNAs may be involved in deregulation of the Hh signalling. To elucidate this issue, we selected, by means of bioinformatic analysis, those miRNAs, which may target the 3′UTR sequences of activator components of the pathway. Our analysis suggested that Smo and Gli1 3′UTR, conserved in humans and mice, are potential targets of miR-326, 125b, 103, 203, 338, 324-5p, 135a, 135b and miR-100, 153, 324-5p, 331, respectively.
Figure 1.
Differential expression of miRNAs in Gli1high versus Gli1low expressing MBs. (A) Gli1 mRNA levels in human MBs. Average (continuous line)±2 s.d. (dashed area) levels of Gli1 mRNA in normal adult cerebellum. Gli1high tumours have Gli1 mRNA levels 2 s.d. above the mean value of normal adult cerebellum. (B) Heat map clustering of miRNAs in human MBs. A green–red colour scale depicts normalized miRNA expression levels. (C) List of miRNAs in which the expression is downregulated in Gli1high versus Gli1low MBs.
miR-125b, miR-324-5p and miR-326 target and functionally suppress Smo
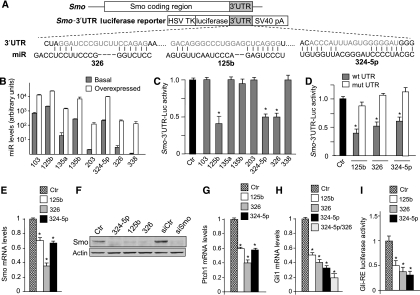
Next, we verified whether these miRNAs were able to regulate directly the Smo and Gli1 mRNAs through binding to their 3′UTRs. When overexpressed in Daoy MB cells (Figure 2B), three out of eight miRNAs (miR-125b, miR-324-5p and miR-326) were able to repress the translation of constructs in which the human Smo-3′UTR was fused to the Renilla luciferase open reading frame (Figure 2A and C; Supplementary Figure 2). As a control, no repression was observed with Smo-3′UTR mutant constructs deleted in the miRNA-binding sites (Figure 2D). Further evidence of Smo repression through miRNA activity was provided by the reduction of Smo mRNA and protein levels following overexpression of miR-125b, miR-326 and miR-324-5p in Daoy cells (Figure 2E and F).
Figure 2.
miRNAs modulate Smo expression and function through binding to the 3′UTR sequence. (A) Schematic representation of 3′UTR sequence from human Smo and the corresponding luciferase reporter vector indicating the potential miRNA-binding sites on the sequence. (B) miRNA levels (evaluated by Q-PCR, relative to RNU6B and RNU66 housekeeping controls) in Daoy cells 24 h after transfection with negative control (basal) or the indicated miRNAs (overexpressed). (C) Levels of luciferase activity in Daoy cells overexpressing the indicated miRNAs and transfected with the Smo wild-type 3′UTR vector. Data are indicated as ratios with respect to pGL4 control vector-transfected cells (ctr). *P<0.05. (D) Levels of luciferase activity in Daoy cells overexpressing the indicated miRNAs and transfected with the Smo wild-type 3′UTR vector (wt) or its mutant derivative lacking the miRNA-binding sites (mut). Data are indicated as ratios with respect to pGL4 control vector-transfected cells (ctr). *P<0.05. (E) Smo mRNA levels in Daoy cells 48 h after transfection with the indicated miRNAs or miRNA mimic negative control (ctr). *P<0.05. (F) Western blot analysis of endogenous Smo protein levels in Daoy cells 48 h after transfection with the indicated miRNAs or mimic negative control (ctr). siRNA against Smo (siSmo) is also shown compared with the silencing control (siCtr). Ptch1 (G) and Gli1 (H) mRNA levels in Daoy cells 48 h after transfection with the indicated miRNAs or negative mimic miRNA as a control (ctr). *P<0.05. miR-325-5p+326 versus either miR-324-5p or miR-326 alone (P<0.05). (I) Luciferase activity from the Gli-responsive reporter transfected in Ptch1−/− MEFs 48 h after transfection with the indicated miRNAs, compared with negative mimic control (ctr). *P<0.05. All data of the above panels (B–I) are average values±s.d. from at least three experiments, each carried out in triplicate.
The use of specific Smo antagonistic drugs or siRNA has demonstrated the role of Smo as an activator of Hh-induced gene expression and activity (Taipale et al, 2000; Chen et al, 2002; Clement et al, 2007). Consistent with these findings, we observed a significant reduction in mRNA levels of endogenous Hh target genes (i.e. Gli1 and Ptch1) in response to miR-125b, miR-326 and miR-324-5p overexpression in MB cells (Figure 2G and H). Notably, a synergistic effect between miRNAs was observed, as indicated by the strong reduction of Gli1 mRNA levels when miR-324-5p and miR-326 were co-overexpressed (Figure 2H). We observed that miRNA regulation of Smo also occurs in mouse as miR-125b-, miR-326- and miR-324-5p-binding sites are conserved on the mouse Smo 3′UTR sequence (Supplementary Figure 3). Accordingly, miR-125b, miR-326 and miR-324-5p overexpression decreases the levels of mouse Smo protein (Supplementary Figure 3).
In the absence of Ptch1-generated inhibitory signals, Smo activates initial Gli transcription factors (e.g. Gli2), which in turn enhance the expression of target genes, including Gli1 (Park et al, 2000; Ikram et al, 2004; Lipinski et al, 2006). Therefore, to finally test the effect of these miRNAs on the Smo-induced transcriptional activation, we used the ex vivo model of Ptch1−/− mouse embryo primary fibroblasts (MEFs) in which the constitutive activation of Gli activity is a consequence of Ptch1 deletion (Goodrich et al, 1997) (Figure 2I). We observed a significant reduction in the activity of a Gli-responsive luciferase reporter in response to the overexpression of miR-125b, miR-326 and miR-324-5p (Figure 2I), which is consistent with the conservation of miRNA-binding sites in the mouse Smo 3′UTR (Supplementary Figure 3). Overall, these findings indicate that miRNA-induced reduction of Smo protein inhibits Hh signalling.
miR-324-5p targets and functionally suppresses Gli1
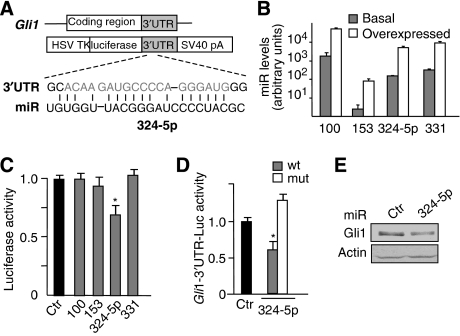
On the basis of possible targeting of the Hh effector Gli1 by miR-100, 153, 324-5p and 331, we hypothesized a further regulation of Hh signalling at a downstream level of the pathway. Indeed, when the Gli1 3′UTR was investigated, one out of four selected miRNAs (miR-324-5p) inhibited the luciferase activity from Gli1-3′UTR-luc construct (Figure 3A–C; Supplementary Figure 4). This effect was abolished on a mutated construct in which the putative miR–mRNA interaction sequence was deleted, confirming that this miRNA was effectively targeting Gli1 mRNA (Figure 3D). As expected, ectopic expression of miR-324-5p caused a reduction of Gli1 protein levels (Figure 3E). Notably, miR-324-5p-binding site in the 3′UTR of Gli1 is also conserved in mouse, and miRNA overexpression decreases the levels of mouse Gli1 protein (Supplementary Figure 3).
Figure 3.
miR-324-5p modulates Gli1 expression and function through binding to the 3′UTR sequence. (A) Schematic representation of 3′UTR sequence from human Gli1 and the corresponding luciferase reporter vector, indicating the potential miRNA-binding sites on the sequence. (B) miRNA levels (evaluated by Q-PCR, relative to RNU6B and RNU66 housekeeping controls) in Daoy cells 24 h after transfection with mimic negative control (basal) or the indicated miRNAs (overexpressed). (C) Levels of luciferase activity in Daoy cells overexpressing the indicated miRNAs and transfected with the Gli1 wild-type 3′UTR vector. Data are indicated as ratios with respect to pGL4 control vector-transfected cells (ctr). *P<0.05. (D) Levels of luciferase activity in Daoy cells overexpressing miR-324-5p and transfected with the Gli1 wild-type 3′UTR vector (wt) or its mutant derivative lacking the miRNA-binding sites (mut). Data are indicated as ratios with respect to pGL4 control vector-transfected cells (ctr). *P<0.05. (E) Western blot analysis of endogenous Gli1 protein levels in Daoy cells overexpressing miR-324-5p or miRNA mimic negative control (ctr). All data in (B–D) are average values±s.d. from at least three experiments, each carried out in triplicate.
Overall, these data indicate that miR-324-5p can elicit a double control on Gli1: an indirect one through Smo repression (Figure 2) and a direct one through interaction with its 3′UTR (Figure 3). The synergistic effect observed between miR-324-5p and miR-326 shown in Figure 2H could be possibly due to this double control.
miR-125b, miR-324-5p and miR-326 inhibit MB cell growth by targeting Hh signalling
Hh signalling is crucial for the proliferation of MB cells (Dahmane et al, 2001; Berman et al, 2002). Therefore, we investigated whether the miRNA-mediated repression of Hh pathway also leads to a decrease in cell proliferation. To this end, we first tested whether these miRNAs are a part of the growth inhibitory pathway triggered by retinoic acid, which is known to promote growth arrest in retinoid-sensitive D283 MB cells (Figure 4A and Hallahan et al, 2003). Notably, the expression of miR-125b, miR-324-5p and miR-326 increased in D283 cells upon retinoic acid treatment (Figure 4A), suggesting that overexpression of these miRNAs may contribute to inhibit tumour cell proliferation. Confirming this hypothesis, we observed that ectopic expression of miR-125b, miR-324-5p and miR-326 was able to significantly inhibit Daoy MB cell proliferation, as evaluated by BrdU uptake (Figure 4B). The miRNA effects on cell proliferation were also confirmed by cell colony formation assays. Indeed, overexpression of miR-125b, miR-324-5p or miR-326 significantly reduced the number of Daoy MB cell colonies (Figure 4C and E). Furthermore, when overexpressed, miR-125b, miR-324-5p or miR-326 was able to inhibit the anchorage-independent growth of the MB cell line, D283, as they significantly reduced the size and the number of colonies grown in soft agar (Figure 4D and F).
Figure 4.
miR-125b, miR-324-5p and miR-326 inhibit MB cell growth. (A) Modulation of BrdU uptake (left) and miRNA levels (right) (means±s.d. from at least three experiments) after retinoic acid (RA) treatment (2.5 μM) of D283 cells for 48 h. *P<0.05. (B) BrdU uptake of Daoy cells 48 h after transfection with the indicated miRNAs. Values shown are relative to mimic negative control miRNA-transfected cells (ctr). *P<0.05 (miRNA-transfected cells versus ctr). (C–F) Colony formation assay on Daoy cells (C, E) and soft agar colony formation in D283 cells (D, F) after transfection with miR-125b or miR-324-5p or miR-326 or miRNA mimic negative control (ctr). (C, D) Representative pictures of the colony growth assay. (E, F) The relative number of colonies formed (the values have been normalized attributing score 100 to the number of colonies grown from control-transfected cells, ctrl). (*P<0.05).
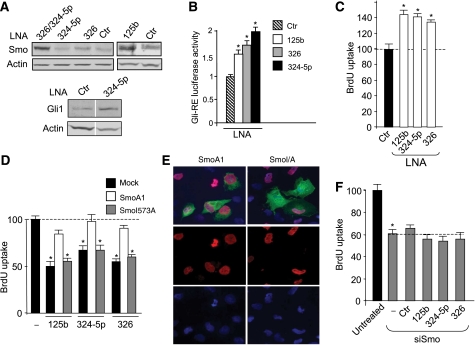
As the expression of miR-125b, miR-324-5p and miR-326 is reduced in human primary MBs together with increased levels of Hh signalling (e.g. high Gli1 expression), we hypothesized that loss of these miRNAs may be responsible for the increased Hh activity and the consequent cell proliferation. In keeping with this view, locked nucleic acid (LNA)-mediated miRNA depletion (Supplementary Figure 5) increased the endogenous levels of Smo and Gli1 proteins (Figure 5A). Depletion of miR-324-5p alone was not sufficient to increase Smo protein levels, likely due to the low miR-324-5p basal expression and the redundancy with other miRNAs that also control Smo (e.g. miR-326 and miR-125b). Interestingly, miR-324-5p cooperates with miR-326, to modulate Smo, as indicated by the enhancing effect of miR-324-5p depletion on the increase in Smo protein levels induced by miR-326 knockdown (Figure 5A). These findings confirm the cooperation between the two miRNAs previously observed (Figure 2H).
Figure 5.
miR-125b, miR-324-5p and miR-326-mediated targeting of Hh signalling control MB cell growth. (A) Western blot analysis of endogenous Smo (upper panel) or Gli1 (bottom panel) protein levels in Daoy cells 48 h after knockdown of miRNA by transfection with LNA against the indicated miRNAs compared with scrambled control LNA (ctr). In bottom panel, Ctr-LNA and 324-5p-LNA lanes showing Gli1 and actin blotting are assembled (see white space between) and come from the same gel after cropping two irrelevant lanes. (B) Luciferase activity from the Gli-responsive reporter transfected in Ptch1−/− MEFs 48 h after miRNA knockdown by transfection with anti-miRNA-LNAs, compared with scrambled control LNA (ctr). *P<0.05. (C) BrdU uptake of Daoy cells 48 h after knockdown of miRNAs by transfection with LNAs against the indicated miRNAs. Values shown are relative to control LNA-transfected cells (ctr). *P<0.05 (LNA-transfected cells versus scrambled LNA). BrdU uptake (D) of Daoy cells 48 h after transfection with the indicated microRNAs in the absence or in the presence of either active SmoA1 or SmoI573A-defective mutant (similarly expressed in 19±2.0 and 21±2.1%, respectively, of transfected cells). Values shown are relative to empty vector-transfected cells (mock). *P<0.05 (miRNA-transfected cells versus mock). (E) The immunofluorescence staining of myc-tagged Smo proteins (green), BrdU (red) and nuclear Hoechst staining (blue). (F) BrdU uptake of Daoy cells 48 h after transfection with Smo siRNA alone (−) or together with the indicated microRNAs or mimic negative control miRNA (ctr). Untreated cells transfected with silencing control siRNA. *P<0.05 (siSmo-transfected versus untreated). All data in (B–F) are average values±s.d. from at least three experiments, each carried out in triplicate.
As expected, LNA-induced miRNA depletion also increased the activity of a luciferase reporter under the control of a Gli-responsive cis element (Figure 5B), further supporting a miRNA-mediated control of Hh signalling. Interestingly, LNA-mediated miRNA depletion also increased the growth rate of MB cells (Figure 5C). The role of Hh signalling by targeting Smo in mediating the miRNA-induced inhibition of cell growth is further supported by the ability of ectopic active Smo cDNA devoid of 3′UTR miRNA-binding sites (SmoA1) to rescue the miRNA effect, whereas a defective Smo mutant (SmoI573A) was ineffective (Figure 5D and E). Consistent with these findings, miR-125b, miR-324-5p and miR-326 were unable to further reduce the proliferation of MB cells once the Hh pathway was inactivated by depletion of Smo protein through Smo siRNA (Figures 2F and 5F), confirming that these miRNAs decrease cell growth through the targeting of the Hh signalling.
miRNAs are downregulated in undifferentiated GCPs and promote their differentiation and inhibition of growth
Retinoic acid is a potent morphogen and induces depletion of Hh-dependent proliferating early GCPs (Yamamoto et al, 1999). These observations together with the retinoic acid-induced upregulation of miR-125b, miR-324-5p and miR-326 (Figure 4A) suggest that these miRNAs may be regulated during development. Confirming this view, the expression of miR-125b and miR-326 dramatically increased in adult compared with 4-day-old mouse cerebella, where undifferentiated and proliferating GCPs are prevailing (Figure 6A). Neonatal cerebellum is mainly constituted by undifferentiated GCPs, which proliferate under the effect of an active Hh signalling (Ruiz i Altaba, 2006). Subsequently, between 7 and 21 days after birth, GCPs progressively exit cell cycle and achieve their terminal differentiation program into mature granule cells. Our findings are consistent with preferential miRNA expression in mature granule cells compared with immature progenitors.
Figure 6.
Role of miR-125b, miR-324-5p and miR-326 in GCP proliferation and differentiation. (A) Expression of the indicated miRNAs during cerebellar development in 4-day (P4) to 60-day (P60)-old mice, measured by real-time Q-PCR. Relative expression levels (means±s.d. from at least three experiments) are indicated, setting P4 expression as 1. *P<0.01. (B) Upregulation of miR-125b, miR-324-5p and miR-326 expression in cultured cerebellar GCPs alongside differentiation, evaluated by Q-PCR. MiR results are expressed as arbitrary units (mean±s.d. from at least three experiments; *P<0.05; **P<0.01). (C) Immunofluorescence with anti-βIII-tubulin antibody (red) and Hoechst staining (blue) (top) in primary cerebellar GCPs cultured for 24 h up to 72 h. Bottom panels show phase-contrast micrographs. (D–F) Primary cerebellar GCPs are cultured in the presence of a mix of miR-125b, miR-324-5p and miR-326 LNAs (LNA) or scrambled LNAs (ctr) for 72 h. Panels show the percentage of BrdU-incorporating cells (D), of cells displaying βIII-tubulin-positive neurite outgrowth (neurite length corresponding to >3 times the size of nuclei) (E) and of NeuN-positive cells (F). *P<0.05. (G) Immunofluorescence staining with anti-βIII-Tub antibody (red) and Hoechst staining (blue) in primary cerebellar GCPs cultured for 72 h. Upper panels show a reduction of cells displaying long βIII-tubulin-positive neurites observed in control cells, after LNA mix or scrambled LNA transfection. Bottom panels show phase-contrast micrographs.
To investigate whether miRNAs were indeed upregulated alongside GCP differentiation, we tested their levels during spontaneous differentiation into mature granule cells in culture. The levels of miR-125b and miR-326 and, to a lesser extent, of miR-324-5p, increased after culturing for 2 and 3 days (Figure 6B), when GCPs reduced their proliferation rate (Supplementary Figure 6E) and significantly differentiated into mature granule cells displaying outgrowth of βIII-tubulin-labelled elongated neurites (Figure 6C). These findings confirm that reduced miRNA levels are a hallmark of poorly differentiated GCPs.
To investigate the functional role of the miRNA increase alongside GCP differentiation, we overexpressed anti-miRNA LNAs to prevent miRNA upregulation and observed an increased cell proliferation (Figure 6D). These findings suggest that in the absence of these miRNAs, GCPs delay growth arrest and differentiation. Indeed, LNA-mediated miRNA depletion also caused a significant reduction in differentiated GCPs that display reduced outgrowth of βIII-tubulin-labelled elongated neurites and decreased expression of the neuronal marker NeuN (Figure 6E–G).
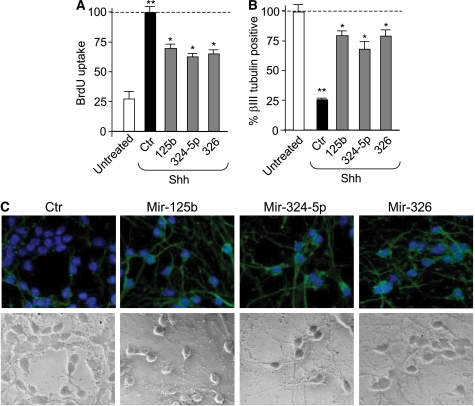
Shh promotes GCP proliferation and limits their differentiation, whereas the addition of the Hh antagonist cyclopamine has opposite effects (Wechsler-Reya and Scott, 1999; Argenti et al, 2005; Di Marcotullio et al, 2006). Consistently with its antidifferentiation function, Shh also prevents the increase in miRNAs levels observed during GCP differentiation in culture (Supplementary Figure 6). Therefore, we investigated whether miR-125b, miR-324-5p and miR-326 were able to antagonize Shh effects. Indeed, overexpression of these miRNAs reduced the Shh-induced proliferation of GCPs (Figure 7A), thus suggesting their active role in the repression of Hh activity upon cell growth. They also antagonized the negative effect triggered by Shh treatment on GCP differentiation, as miRNA overexpression promoted an increase in granule cells displaying outgrowth of elongated neurites expressing βIII-tubulin (Figure 7B and C).
Figure 7.
miR-125b, miR-324-5p and miR-326 antagonize Shh effect of GCP proliferation and differentiation. (A) BrdU uptake of primary GCPs transfected with the indicated miRNAs and cultured in the absence or in the presence of Shh for 48 h. Values shown are relative to mimic negative control miRNA-transfected cells. **P<0.01 (Shh-treated cells versus untreated); *P<0.05 (miRNA-transfected cells versus control, ctr). (B) Percentage of cells displaying βIII-tubulin-positive neurite outgrowth (neurite length corresponding to >3 times the size of nuclei) in primary GCP cultures 72 h after the treatment described in (A). **P<0.01 (Shh-treated cells versus untreated); *P<0.05 (miRNA-transfected cells versus control, ctr). (C) Immunofluorescence staining with anti-βIII-tubulin antibody (green) and Hoechst staining (blue) in primary cerebellar GCPs treated as described in (A), showing cells displaying long βIII-tubulin-positive neurites after miRNA transfection. Bottom panels show phase-contrast micrographs.
Overall, these findings indicate that miR-125b, miR-324-5p and miR-326 are downregulated in poorly differentiated GCPs. They also promote GCP differentiation as well as growth inhibition and antagonize the effects induced by Shh.
miR-324-5p downregulation is caused by chromosome 17p deletion in human MBs
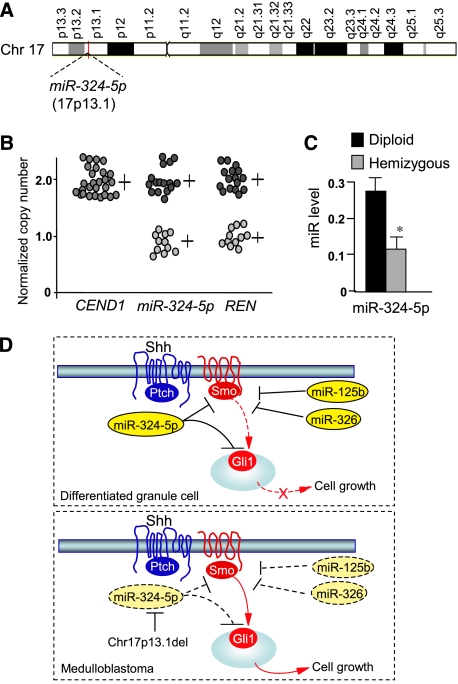
To understand the genetic basis of miRNA downregulation in human cancer, we investigated the miR-324-5p genomic locus that we observed to be mapping to 17p13.1, comprised in the minimal deleted region of chromosome 17p reported in about 40% of MBs (Figure 8A) (Ferretti et al, 2005). Thus, to verify whether chromosome loss resulted in the deletion of the genomic sequence coding for this miRNA, we carried out real-time quantitative PCR (Q-PCR) of the genomic region containing miR-324-5p gene. Allelic deletion of miR-324-5p was observed in 40% of MBs when compared with paired blood genomic DNA from the same patients (Figure 8B), indicating that the downregulation of miR-324-5p expression was at least partially due to genetic mutation in a high percentage of tumours. Indeed, miR-324-5p levels were significantly reduced in 17p hemizygous versus diploid MBs (Figure 8C).
Figure 8.
miR-324-5p downregulation is caused by chromosome 17p deletion in human MBs. (A) Schematic representation of chromosome 17 showing the localization of miR-324-5p locus in 17p13.1. (B) Real-time Q-PCR of miR-324-5p in genomic DNA from 17p wild-type (n, 16) and 17p-deleted human MBs (n, 11). Gene copy numbers of the adjacent hemizygous RENKCTD11 (mapping in 17p13.1) and diploid CEND1 (mapping in 11p) are also indicated, as controls. (C) Loss of 17p correlates with reduced levels of miR-324-5p expression. Q-PCR of miR-324-5p expression in 17p diploid and hemizygous human MBs (mean values±s.d.; *P<0.01). (D) Model illustrating the miRNA control of Hh signalling. In normal differentiated cells, Hh signalling strength is limited by the concerted action of miR-125b, miR-324-5p and miR-326, through inhibition of the activatory components of the pathway, Smo and Gli1. Downregulated expression of miR-324-5p caused by MB-associated deletion of chromosome 17p together with reduced levels of miR-125b and miR-326 determines the overexpression of Smo and of the constitutively active transcription factor Gli1, which result in unrestrained Hh signalling, thereby sustaining cell growth.
Discussion
In this study, we used a high-throughput miRNA expression profiling as a screening method to successfully identify functional miRNAs targeting Hh pathway both in tumours and normal cerebellar cells. We report here the first evidence that miR-125b, miR-326 and miR-324-5p target activator components of the Hh signalling pathway (Smo and Gli1) in mammalian cells. The lowest expression of these miRNAs is observed in Gli1high MBs, which represent about 50% of human MBs. This subset of miRNAs appears to exert an effect in a coordinated way to impair the Hh signalling cascade at the level of the post-receptor transducer Smo and of the downstream transcription factor Gli1. Notably, miR-324-5p targets both Smo and Gli1 and cooperates with miR-326 to further reinforce the inhibitory activity. Therefore, these observations suggest a model (Figure 8D) in which a concerted action by several miRNAs may be required to efficiently impair the oncogenic potential of the Hh pathway. Indeed, experimental evidence indicates that both Smo and Gli1 have a critical function in promoting cerebellar tumorigenesis. Transgenic mice expressing a constitutively active Smo develop MB with a high frequency (Hallahan et al, 2004). Furthermore, Gli1 is involved in tumorigenesis, as indicated by the reduced development of MB in Gli1−/−/Ptch1+/− double mutants (Kimura et al, 2005). Despite the role of Smo and Gli1 in mouse MBs, very rare gain-of-function mutations of Smo and none affecting Gli1 have been described in human MB. Therefore, the loss of function of these miRNAs may represent a major and most frequent mechanism by which these activator components of the oncogenic Hh pathway are unrestrained in human MB.
Our data also suggest that the loss of the miRNA-mediated suppression of Hh signalling in MBs may be caused by a genetic abnormality. Indeed, we observed that miR-324-5p downregulated expression is caused by deletion of miR-324-5p gene in a high percentage of MBs, as a consequence of the loss of chromosome 17p, the most frequent mutation of this tumour. Interestingly, 17p deletion also affects the p53 and HIC1 tumour suppressors as well as the Hh antagonist RENKCTD11. Loss of function of these genes has been reported to cooperate with Hh signalling for MB growth (Wetmore et al, 2001; Di Marcotullio et al, 2004; Ferretti et al, 2005; Briggs et al, 2008). Therefore, these observations suggest that the deletion of chromosome 17p region integrates several genetic defects, including miR-324-5p loss, which coordinately may contribute to Hh-dependent tumorigenesis. This genetic abnormality may either occur early alongside GCP differentiation, thus leading to the loss of a signal withdrawing Hh activity during development, or as a secondary event to maintain high Hh-dependent tumour growth. It remains to be elucidated whether downregulation of miR-125b and miR-326 in MB is caused by either additional genetic abnormalities or other epigenetically determined regulatory mechanisms.
In this regard, the pattern of miRNA expression that we have observed during cerebellar development suggests that their low expression in MB cells is a feature that is reminiscent of the origin of these tumours from immature GCPs. Indeed, the expression of these miRNAs is low in undifferentiated GCPs and increases during their in vitro differentiation into mature granule cells. Furthermore, their expression is required to allow GCPs to reduce the proliferation rate and to differentiate. Notably, they antagonize the opposite effects induced by Shh on GCPs. This behaviour reflects that of other gene products (e.g. RENKCTD11 and Numb) that are upregulated during the transition from undifferentiated GCPs to mature granule cells (Argenti et al, 2005; Di Marcotullio et al, 2006). RENKCTD11 and Numb are also suppressors of Hh signalling and, most interestingly, their downregulated expression is maintained in Hh-dependent MBs (Di Marcotullio et al, 2004, 2006). These observations suggest that, similar to RENKCTD11 and Numb, the miRNAs we have described are also involved in withdrawing Hh signalling at a critical point during GCP development. We therefore suggest that this critical point is characterized by a multifaceted regulation, including a miRNA control mechanism, the misregulation of which during development is responsible for keeping a persistent hyperactive Hh signalling, allowing GCPs to overgrow, a condition of high susceptibility of malignant transformation.
Although we described a miRNA control of agonistic components of the Hh pathway (Smo and Gli1), miRNAs regulating antagonists of this pathway have recently been reported in Drosophila (miR-12 and miR-283 targeting Cos2) and zebrafish (miR-214 targeting suppressor of fused, SUFU) (Flynt et al, 2007; Friggi-Grelin et al, 2008). Drosophila miR-12 and miR-283 target Cos-2, Fu and Smo (Friggi-Grelin et al, 2008). However, both these miRNA sequences and Cos-2 functions (Varjosalo et al, 2006) are not conserved in mammals. SUFU is an inhibitor of Hh signalling (Kogerman et al, 1999; Svärd et al, 2006) and its mutation or loss of function is involved in MB development (Taylor et al, 2002; Lee et al, 2007b). Interestingly, we observed that miR-214 is upregulated in Gli1high tumours (Supplementary Figure 1). These observations suggest that increased expression of miR-214 in human MB may be responsible for the loss of one Hh antagonist. Although zebrafish and human SUFU 3′UTR are not conserved, the miR-214 putative binding site appears to be quite conserved; thus, miR-214 could regulate SUFU in human cells. Conversely, mammalian SUFU has been reported to be targeted by miR-378, although its role in regulating Hh signalling has not been described (Lee et al, 2007a). However, our data did not show a deregulation of miR-378 in human Gli1high MBs (Figure 1).
In conclusion, we described a novel mechanism of regulation of Hh signalling through miRNA-mediated control of Smo and Gli1 and it may have a function in withdrawing Hh activity during cerebellar neuronal progenitor differentiation (Figure 8D). We suggest that abrogation of such a mechanism (e.g. as a consequence of chromosome 17p deletion) may be a relevant event in subverting Hh signalling during neuronal development thereby sustaining brain tumorigenesis. Indeed, this is the first evidence of the involvement of miRNA-mediated control of the Hh pathway in malignancy.
Materials and methods
Human and mouse tissue samples
Thirty-one human primary MB specimens were collected during surgical resection with the approval of Institutional Review Board, as previously described (Ferretti et al, 2006). RNA of normal human cerebellum (nine adult samples from 25- to 70-year-old subjects were purchased from Biocat (Heidelberg, Germany), Ambion (Applied Biosystems, Foster City, CA) and BD Biosciences (San Jose, CA). Human MBs have been divided into two subsets: Glihigh MBs that exhibited Gli1 levels significantly exceeding the average adult cerebella (>2 s.d.) and Glilow subset that presented levels in the range or below this cutoff. Mouse normal cerebella were obtained from CD1 mice (prepared from 4-, 10-, 22- and 60-day-old mice) (Charles River, Calco, Italy).
Cell cultures and treatments
Daoy and D283 MB cells were cultured in MEM medium (Gibco, Carlsbad, CA), supplemented with 10 or 20% fetal bovine serum, respectively, 1% sodium pyruvate, 1% non-essential aminoacid solution, 1% L-glutamine and penicillin/streptomycin. Cerebellar GCPs (prepared from 4-day-old mice) were isolated and cultured as previously described (Di Marcotullio et al, 2004; Argenti et al, 2005). When indicated, GCPs were treated with Shh (3 μg/ml) for up to 72 h (R&D, Minneapolis, MN, USA). Retinoic acid treatment (all-trans-retinoic acid; Sigma-Aldrich, St Louis, MO) (2.5 μM) was performed for 48 h.
RNA isolation and real-time PCR
RNA isolation from tissue samples and cells and cDNA synthesis were performed as previously described (Ferretti et al, 2006; De Smaele et al, 2008) using reagents from Invitrogen (Carlsbad, CA). Quantitative reverse transcription (RT)–PCR analysis of Gli1, Smo, β-actin, GAPDH and HPRT mRNA expression was analysed on cDNAs using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems) employing TaqMan gene expression assay according to the manufacturer's instructions (Applied Biosystems). Each amplification reaction was performed in triplicate, and the average of the three threshold cycles was used to calculate the amount of transcripts in the sample (SDS software, ABI). mRNA quantification was expressed, in arbitrary units, as the ratio of the sample quantity to the calibrator or to the mean values of control samples. All values were normalized to three endogenous controls, GAPDH, β-actin and HPRT.
miRNA expression profiling
Analysis of the expression profiling of 250 miRNAs was carried out on RNA samples according to Applied Biosystems protocols (Foster City, CA). The assay included RT with specific stem-loop primers followed by real-time Q-PCR using the miRNA-specific TaqMan MGB probe and TaqMan universal master mix in an Applied Biosystems 7900HT PCR system. miRNA expression levels were normalized to two internal control small RNAs (RNU6B and RNU66) obtaining similar results. The comparative threshold cycle method was used to calculate the relative miRNA expression. The expression level of each miRNA has been compared with the mean of all adult control samples. Statistical analysis of miRNAs differentially expressed among samples was carried out by non-parametric Mann–Whitney test (P<0.05).
Hierarchical clustering analysis was performed on the miRNA expression levels, representing the values as natural logarithms. miRNA dendrograms were represented using the UPGMA (unweighted average) clustering method (by Euclidean distance) (Spotfire software; Spotfire AB, Göteborg, Sweden).
For northern blot analysis, total RNA from Daoy and MEF cells was fractionated on a 10% polyacrylamide-urea gel and transferred to a nylon membrane. DNA oligonucleotides complementary to the sequences of mature miRNAs and to U2 snRNA (U2R: 5′-GGGTGCACCGTTCCTGGAGGTAC-3′, as loading control) were 32P-labelled and used as probes.
Overexpression and knockdown studies
Synthetic miRNAs or negative control (miRNA mimic negative control code CN-0010000-01) (Dharmacon) were transfected into cells using DharmaFECT-2 transfection reagent (Dharmacon) at 100 nM. SiGlo Green and siGLO Red transfection control reagents (10 nM) (Dharmacon) were used to verify transfection efficiency that ranged between 75 and 85%. miRNA encoding plasmids were cloned as previously described (Laneve et al, 2007) and transfected into Daoy cells by Lipofectamine Plus (Invitrogen), according to the manufacturer's instructions. miRNA knockdown was carried out using miRNA specific (miR-125b, miR-324-5p, miR-326) and scrambled control (mirCURY knockdown probe: code 199002-04) fluorescein-labelled LNA oligonucleotides (Exiqon, Vedbaek, Denmark), transfected into Daoy, GCPs and MEF cells at a final concentration of 40 nM by Hiperfect reagent (Qiagen, Hilden, Germany). Transfection efficiency ranged between 80 and 90%. Myc-tagged-SmoA1, SmoI573A and control empty vectors were kindly provided by J Taipale (Varjosalo et al, 2006) and transfected as described above. DNA sequences encoding for siRNAs designed to target human Smo (siSmo) and silencing control (code AM4611) were obtained from Ambion-Applied Biosystems protocols. Transfection of siRNA duplexes (40 nM) was carried out using Hiperfect reagent (Qiagen) according to the manufacturer's instructions. Cells were subsequently subjected to either western blotting or cell proliferation assays.
Luciferase assays
miRNA target sites in 3′UTR gene regions were identified by bioinformatic analysis using the following online databases: miRanda (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl), Target Scan (http://www.targetscan.org) and MiRBase Sanger (http://microrna.sanger.ac.uk/cgi-bin/targets/v5/search.pl) (only those putative miRNA target sites resulting from at least two databases were considered positive). The entire 3′UTR regions of human Smo and Gli1 were PCR-amplified using the following primers: 5′-atttgcggccgcgcctgcagagcaggacctgggacagg-3′ and 5′-atttgcggccgcagaaaaaccttttattgactgtatttcttctccc-3′; 5′-gctctagaagagtagggaatctcatccatcacagatcg-3′ and 5′-atttgcggccgcctgat gcagttcctttattatcaggaaacagtgt-3′, respectively, and cloned downstream of the stop codon in pRL-TK vector (Promega, Madison, WI). These constructs were used to generate, by inverse PCR, the mutant derivatives lacking miRNA-binding sites. Daoy cells were cultured in 24-well plates and transfected first with 100 nM of synthetic miRNA (100, 103, 125b, 324-5p, 326, 331 and 338) or scramble control and after 24 h with 0.1 μg of specific pRL-TK-3′UTR and 0.3 μg of pGL4 control vector. Cells were harvested and tested 24 h post second transfection with the dual-luciferase assay (Promega). All luciferase activity data are presented as means±s.d. of values from at least three experiments, each performed in triplicate.
Western blot assay
Transfected cells were lysed in a solution containing Tris–HCl pH 7.6, 50 mM deoxycholic acid, sodium salt 1%, NaCl 150 mM, NP40 1%, EDTA 5 mM, NaF 100 mM and protease inhibitors. Lysates were separated on SDS–PAGE and immunoblotted using standard procedures. Anti-Smo N-19 (sc-6366; Santa Cruz Biotechnology, Heidelberg, Germany), anti-Gli1 H-300 (sc-20687; Santa Cruz Biotechnology), anti-actin I19 (sc-1616; Santa Cruz Biotechnology) and HRP-conjugated secondary antisera (Santa Cruz Biotechnology) were used followed by enhanced chemiluminescence (ECL; Amersham).
Gene copy number assay
MicroRNA-324-5p gene copy number was measured by real-time Q-PCR (Livak et al, 1995; Senchenko et al, 2003). DNA from 27 MB specimens and paired blood cells was isolated by overnight treatment with proteinase K at 50°C followed by phenol–chloroform extraction and precipitation with ethanol. Selection of primer and TaqMan probe sequences for miRNA 324-5p was performed using the ABI Primer Express Software (version 1.5). They were as follows: forward 5′-TCCCCTAGGGCATTGGTGTA-3′, reverse 5′-TCTTCCCAGTCGGGTCAGACTAT-3′ and probe 5′-AGCTGGAGACCCACTGA-3′. PCRs were carried out in triplicate in 25 μl (1 × PCR master mix (Applied Biosystems), 200 nM TaqMan probe, forward and reverse primers (900 nm) and 2.5 μl of DNA template (25 ng)). PCR reactions were performed on an ABI Prism 7900 Sequence Detection System (Applied Biosystems) according to the standard thermal profile (2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C). For the relative quantification of miR-324-5p allele copy number, two different methods have been used: the standard curve method and the comparative Ct method, in tumour DNA sample relative to the normal DNA from the same patient (calibrator) and relative to RNaseP as endogenous control. CEND1 and RENKCTD11 have been used as a control of diploid and hemizygous status, respectively (Di Marcotullio et al, 2004).
Cell proliferation and colony assays
Cell proliferation was evaluated by BrdU incorporation (6 or 24 h pulse for Daoy and GCP, respectively). Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and BrdU detection (Roche, Basel, Switzerland) was performed according to the manufacturer's instructions. Nuclei were counterstained with Hoechst reagent. At least 300 nuclei were counted in triplicate, and the number of BrdU-positive nuclei was recorded.
For colony formation assays, 4 × 103 miRNA-transfected Daoy cells were plated in 10-cm diameter dishes and after 2 weeks, cell colonies were counted following staining in 20% methanol and crystal violet.
For anchorage-independent growth in soft agar assay, 6 × 103 miRNA-transfected D283 cells were suspended in culture medium containing 0.3% agarose (Invitrogen) and plated over a layer of 0.7% agarose in the same culture medium. At 21 days after plating, colonies of >50 cells were scored in 10 representative fields in each plate. All assays were performed in triplicate and results shown are the average of three independent experiments.
Immunofluorescence
GCPs were cultured in Lab-Tek chamber slides fixed in 4% paraformaldehyde for 20 min at room temperature, permeabilized with 0.1% Triton X-100 cells, incubated in blocking buffer (PBS with 1% BSA) for 30 min and then with primary antibody overnight in blocking solution at +4°C. Mouse monoclonal antibodies antineuronal-specific nuclear protein (NeuN) (MAB377) and anti-βIII tubulin (MAB1637) were from Chemicon (Temecula, CA) and were used at 1:50 and 1:100, respectively. Samples were then incubated with secondary antibodies for 30 min at room temperature in the blocking solution. Texas red- or FITC-conjugated anti-mouse secondary antibodies were from Molecular Probes (Invitrogen, Eugene, OR). Nuclei were counterstained with Hoechst reagent. Transfected myc-tagged vectors were revealed by immunofluorescence using anti-myc antibody (Upstate).
Supplementary Material
Supplementary Table 1
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Table and Figures Legends
Acknowledgments
We thank MP Scott and L Milenković for the gift of Ptch1−/− MEFs and G Giannini and M Levrero for helpful discussion. This study was supported by Associazione Italiana per la Ricerca sul Cancro, Telethon Grant GGP07118, Ministry of University and Research (PRIN), Ministry of Health, Cenci-Bolognetti Foundation, Rome Oncogenomic Center, Center of Excellence BEMM, 6th Framework Programme of the European Commission, SIROCCO and RIGHT Integrated Projects (LSHG-CT-2006-037900 and LSHB-CT-2004 005276) and the ESF project ‘NuRNASu'.
References
- Argenti B, Gallo R, Di Marcotullio L, Ferretti E, Napolitano M, Canterini S, De Smaele E, Greco A, Fiorenza MT, Maroder M, Screpanti I, Alesse E, Gulino A (2005) Hedgehog antagonist REN(KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors. J Neurosci 25: 8338–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM (2004) Tissue repair and stem cell renewal in carcinogenesis. Nature 432: 324–331 [DOI] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, Beachy PA (2002) Medulloblastoma growth inhibition by hedgehog pathway blockade. Science 297: 1559–1561 [DOI] [PubMed] [Google Scholar]
- Briggs KJ, Corcoran-Schwartz IM, Zhang W, Harcke T, Devereux WL, Baylin SB, Eberhart CG, Watkins DN (2008) Cooperation between the Hic1 and Ptch1 tumor suppressors in medulloblastoma. Genes Dev 22: 770–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Young KE, Maiti T, Beachy PA (2002) Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA 99: 14071–14076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A (2007) HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol 17: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane N, Sánchez P, Gitton Y, Palma V, Sun T, Beyna M, Weiner H, Ruiz i Altaba A (2001) The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development 128: 5201–5212 [DOI] [PubMed] [Google Scholar]
- De Smaele E, Fragomeli C, Ferretti E, Pelloni M, Po A, Canettieri G, Coni S, Di Marcotullio L, Greco A, Moretti M, Di Rocco C, Pazzaglia S, Maroder M, Screpanti I, Giannini G, Gulino A (2008) An integrated approach identifies Nhlh1 and Insm1 as Sonic Hedgehog-regulated genes in developing cerebellum and medulloblastoma. Neoplasia 10: 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, De Smaele E, Argenti B, Mincione C, Zazzeroni F, Gallo R, Masuelli L, Napolitano M, Maroder M, Modesti A, Giangaspero F, Screpanti I, Alesse E, Gulino A (2004) REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci USA 101: 10833–10838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M, Screpanti I, Gulino A (2006) Numb is a suppressor of Hedgehog signaling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol 8: 1415–1423 [DOI] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Di Marcotullio L, Screpanti I, Gulino A (2005) Hedgehog checkpoints in medulloblastoma: the chromosome 17p deletion paradigm. Trends Mol Med 11: 537–545 [DOI] [PubMed] [Google Scholar]
- Ferretti E, Di Marcotullio L, Gessi M, Mattei T, Greco A, Po A, De Smaele E, Giangaspero F, Riccardi R, Di Rocco C, Pazzaglia S, Maroder M, Alimandi M, Screpanti I, Gulino A (2006) Alternative splicing of the ErbB-4 cytoplasmic domain and its regulation by hedgehog signaling identify distinct medulloblastoma subsets. Oncogene 25: 7267–7273 [DOI] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG (2007) Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet 39: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty MP, Kessler JD, Wechsler-Reya RJ (2005) Morphing into cancer: the role of developmental signaling pathways in brain tumor formation. J Neurobiol 64: 458–475 [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F, Lavenant-Staccini L, Therond P (2008) Control of antagonistic components of the hedgehog signaling pathway by microRNAs in Drosophila. Genetics 179: 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Milenković L, Higgins KM, Scott MP (1997) Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277: 1109–1113 [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Chandraratna RA, Ellenbogen RG, Geyer JR, Overland RP, Strand AD, Tapscott SJ, Olson JM (2003) BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat Med 9: 1033–1038 [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, Olson JM (2004) The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res 64: 7794–7800 [DOI] [PubMed] [Google Scholar]
- Ikram MS, Neill GW, Regl G, Eichberger T, Frischauf AM, Aberger F, Quinn A, Philpott M (2004) GLI2 is expressed in normal human epidermis and BCC and induces GLI1 expression by binding to its promoter. J Invest Dermatol 122: 1503–1509 [DOI] [PubMed] [Google Scholar]
- Kimura H, Stephen D, Joyner A, Curran T (2005) Gli1 is important for medulloblastoma formation in Ptc1(+/−) mice. Oncogene 24: 4026–4036 [DOI] [PubMed] [Google Scholar]
- Kogerman P, Grimm T, Kogerman L, Krause D, Undén AB, Sandstedt B, Toftgård R, Zaphiropoulos PG (1999) Mammalian suppressor-of-fused modulates nuclear–cytoplasmic shuttling of Gli-1. Nat Cell Biol 1: 312–319 [DOI] [PubMed] [Google Scholar]
- Kosik KS (2006) The neuronal microRNA system. Nat Rev Neurosci 7: 911–920 [DOI] [PubMed] [Google Scholar]
- Laneve P, Di Marcotullio L, Gioia U, Fiori ME, Ferretti E, Gulino A, Bozzoni I, Caffarelli E (2007) The interplay between microRNAs and the neurotrophin receptor tropomyosin-related kinase C controls proliferation of human neuroblastoma cells. Proc Natl Acad Sci USA 104: 7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Deng Z, Wang CH, Yang BB (2007a) MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA 104: 20350–20355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kawagoe R, Sasai K, Li Y, Russell HR, Curran T, McKinnon PJ (2007b) Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene 26: 6442–6447 [DOI] [PubMed] [Google Scholar]
- Lipinski RJ, Gipp JJ, Zhang J, Doles JD, Bushman W (2006) Unique and complimentary activities of the Gli transcription factors in Hedgehog signaling. Exp Cell Res 312: 1925–1938 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Marmaro J, Todd JA (1995) Towards fully automated genome-wide polymorphism screening. Nat Genet 9: 341–342 [DOI] [PubMed] [Google Scholar]
- Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, Nakashima M, Joyner AL (2000) Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127: 1593–1605 [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A (2006) Hedgehog-Gli Signaling in Human Diseases. Ed Landes Bioscience, New York: Kluwer Academic/Plenum Publishers [Google Scholar]
- Ruiz i Altaba A, Sánchez P, Dahmane N (2002) Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer 2: 361–372 [DOI] [PubMed] [Google Scholar]
- Senchenko V, Liu J, Braga E, Mazurenko N, Loginov W, Seryogin Y, Bazov I, Protopopov A, Kisseljov FL, Kashuba V, Lerman MI, Klein G, Zabarovsky ER (2003) Deletion mapping using quantitative real-time PCR identifies two distinct 3p21.3 regions affected in most cervical carcinomas. Oncogene 22: 2984–2992 [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ (2008) Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Svärd J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergström A, Ericson J, Toftgård R, Teglund S (2006) Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell 10: 187–197 [DOI] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA (2000) Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406: 1005–1009 [DOI] [PubMed] [Google Scholar]
- Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, Hao A, Goldstein AM, Stavrou T, Scherer SW, Dura WT, Wainwright B, Squire JA, Rutka JT, Hogg D (2002) Mutations in SUFU predispose to medulloblastoma. Nat Genet 31: 306–310 [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Li SP, Taipale J (2006) Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell 10: 177–186 [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP (1999) Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22: 103–114 [DOI] [PubMed] [Google Scholar]
- Wetmore C, Eberhart DE, Curran T (2001) Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res 61: 513–516 [PubMed] [Google Scholar]
- Yamamoto M, Ullman D, Dräger UC, McCaffery P (1999) Postnatal effects of retinoic acid on cerebellar development. Neurotoxicol Teratol 21: 141–146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Table and Figures Legends