Abstract
Rifamycin antibacterial agents inhibit bacterial RNA polymerase (RNAP) by binding to a site adjacent to the RNAP active center and preventing synthesis of RNA products >2–3 nt in length. Recently, Artsimovitch et al. [(2005) Cell 122:351–363] proposed that rifamycins function by allosteric modulation of binding of Mg2+ to the RNAP active center and presented three lines of biochemical evidence consistent with this proposal. Here, we show that rifamycins do not affect the affinity of binding of Mg2+ to the RNAP active center, and we reassess the three lines of biochemical evidence, obtaining results not supportive of the proposal. We conclude that rifamycins do not function by allosteric modulation of binding of Mg2+ to the RNAP active center.
Keywords: rifampicin, rifapentine, rifabutin, RNA polymerase inhibitors, antibacterial agents
The rifamycins—notably rifampicin, rifapentine, and rifabutin—are potent, broad-spectrum antibacterial agents and are the lynchpin of current antituberculosis therapy (1) [supporting information (SI) Fig. S1]. The activity of rifamycins stems from their high-affinity binding to, and inhibition of, bacterial RNA polymerase (RNAP) (2).
The molecular mechanism of inhibition of RNAP by rifamycins has been investigated for four decades. Rifamycins have no or only small effects on RNAP–promoter interaction and RNAP–NTP interaction and generally have no or only small effects on formation of the RNA first phosphodiester bond (3, 4). The predominant effect of rifamycins is to block formation of the RNA second phosphodiester bond or third phosphodiester bond (when transcription is initiated with an NTP, or with an NDP or NMP, respectively) (4). RNAP that has synthesized a sufficiently long RNA product to enter into the transcription–elongation phase is resistant to rifamycins (5). These properties led to the proposal that rifamycins inhibit RNAP through a simple steric-occlusion mechanism, in which the rifamycin binds adjacent to the RNAP active center, along the path of the RNA product, and physically prevents synthesis or retention of RNA products >2–3 nt in length (4).
The crystal structure of Thermus aquaticus RNAP in complex with rifampicin showed that rifampicin binds to a site adjacent to the RNAP active center, along the path of the RNA product, in a position to physically prevent synthesis or retention of RNA products >2–3 nt in length—in complete agreement with the prediction of the steric-occlusion mechanism (6) (Figs. S2 and S3A). The structure accounts for biochemical results defining the mechanism of rifamycins and genetic results defining amino acid substitutions in RNAP that confer rifamycin resistance, and provides a basis for structure-based design of improved RNAP inhibitors (6).
Recently, Artsimovitch et al. (7) proposed a new mechanism for inhibition of RNAP by rifamycins—a mechanism that is proposed to operate in addition to, or instead of, the steric-occlusion mechanism. Artsimovitch et al. propose that an essential component of inhibition of RNAP by rifamycins is allosteric modulation of binding of Mg2+ to the RNAP active center. Specifically, Artsimovitch et al. propose that rifamycins induce an allosteric signal that is transmitted, over a distance of ≈19 Å, from the rifamycin binding site to the RNAP active center and that decreases binding of Mg2+ to the RNAP active center, resulting in decreased RNAP activity (Fig. S3B). Artsimovitch et al. determined two crystal structures of Thermus thermophilus RNAP in complex with rifamycins and observed that, in these crystal structures, in contrast to in most other crystal structures of RNAP, the RNAP active center did not contain bound Mg2+ (7). Artsimovitch et al. inferred a causal connection between the presence of rifamycins and the absence of Mg2+ in the two crystal structures, proposed that rifamycins induce an allosteric signal transmitted from the rifamycin binding site to the RNAP active center that decreases the affinity of binding of Mg2+ to the RNAP active center, and proposed that allosteric modulation of the affinity of binding of Mg2+ to the RNAP active center is essential for inhibition of RNAP by rifamycins. Artsimovitch et al. presented three lines of biochemical evidence consistent with their allosteric model:
High Mg2+ concentrations confer resistance to transcription inhibition by rifamycins.
The classic rifamycin-resistant mutants β-D516N and β-D516V, which substitute a residue located on the proposed allosteric signaling pathway, confer resistance to rifamycins but do not correspondingly reduce affinity of RNAP for rifamycins.
The designed rifamycin-resistant mutant β-L1235A, which substitutes a residue located on the proposed allosteric signaling pathway, confers resistance to rifamycins but does not correspondingly reduce affinity of RNAP for rifamycins.
In this work, we directly tested the principal premise of the model of Artsimovitch et al. (7): i.e., the premise that rifamycins decrease the affinity of binding of Mg2+ to the RNAP active center. We find that rifamycins do not affect the affinity of binding of Mg2+ to the RNAP active center. In addition, we reanalyzed the three lines of biochemical evidence presented by Artsimovitch et al. We find that the three lines of biochemical evidence are incorrect. We conclude that rifamycins do not function by allosteric modulation of binding of Mg2+ to the RNAP active center.
Results and Discussion
Absence of Effects of Rifamycins on Metal-Ion Binding.
Artsimovitch et al. (7) determined two crystal structures of T. thermophilus RNAP in complex with rifamycins and observed that the RNAP-active-center Mg2+ was absent in both crystal structures. We suggest that this observation, by itself, does not constitute sufficient basis to infer a causal relationship between the presence of a rifamycin and the absence of Mg2+ in the two crystal structures.
Artsimovitch et al. did not directly test the assertion that rifamycins decrease the affinity of binding of metal ions to RNAP active center in solution. In this work, we directly tested the effect of rifamycins on metal-ion binding in the RNAP active center.
It has been shown that Fe2+ competes with Mg2+ for binding to the RNAP active center (8) and that Fe2+ bound at the RNAP active center can be used to generate hydroxyl radicals that cause localized cleavage of the RNAP β′ and β subunits (8, 9). At appropriate concentrations of Fe2+, the Fe2+-mediated cleavage of the RNAP β′ and β subunits involves a single Fe2+ binding site corresponding to the Mg2+ binding site at the RNAP active center (8, 9). Mapping of Fe2+-mediated cleavage sites has been used to demonstrate that the three aspartic acid (D) residues of the β′ NADFDGD motif coordinate Mg2+ at the RNAP active center (8), and to identify additional residues of the RNAP β′ and β subunits located near the RNAP active center (9)—findings that were confirmed when the three-dimensional structure of RNAP was determined (10).
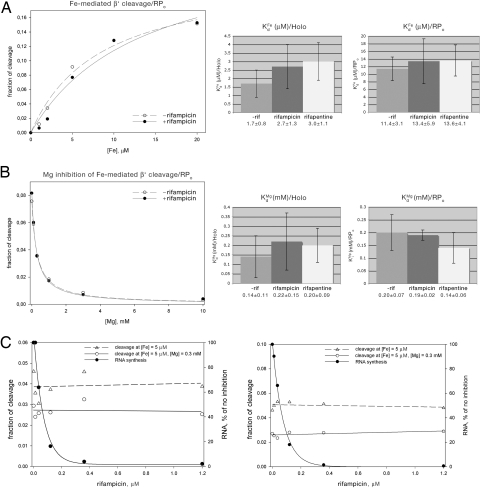
To assess the equilibrium dissociation constant for Fe2+, KdFe, and to assess effects of rifamycins on KdFe, we quantified Fe2+-mediated cleavage as a function of Fe2+ concentration. Fig. 1A and Fig. S4A present measurements of Fe2+-mediated cleavage as a function of Fe2+ concentration in the absence of rifamycins and in the presence of saturating concentrations of rifamycins. Data are presented both for experiments performed with RNAP holoenzyme (holo) and with RNAP–promoter open complexes (RPo). The data shown are for cleavage of the RNAP β′ subunit within the NADFDGD motif, directly at the Fe2+ binding site corresponding to the Mg2+ binding site at the RNAP active center (i.e., cleavage resulting in cleavage product VI of ref. 9). Data for cleavage of the RNAP β′ and β subunits at other sites in and adjacent to the RNAP active center are equivalent (data not shown). The results indicate that the value of the equilibrium dissociation constant for Fe2+, KdFe, in the absence of rifamycins is ≈1.7 μM for RNAP holo and ≈11 μM for RPo, consistent with previous measurements (9). The results further indicate that, within experimental error, values of KdFe in the presence of rifamycins are indistinguishable from values of KdFe in the absence of rifamycins. We conclude that rifamycins do not affect the affinity of binding of Fe2+ to the RNAP active center in solution.
Fig. 1.
Absence of effects of rifamycins on metal-ion binding. (A) Absence of effects of rifamycins on Fe2+ binding (as inferred from measurement of Fe2+-mediated cleavage within the RNAP β′ subunit NADFDGD motif as a function of Fe2+ concentration). (A Left) Representative data (data for RPo at 0 and 1.2 μM rifampicin). (A Center and Right) Summary. (B) Absence of effects of rifamycins on Mg2+ binding (as inferred from measurement of Fe2+-mediated cleavage within the RNAP β′ subunit NADFDGD motif as a function of competing Mg2+ concentration). (B Left) Representative data (RPo at 0 and 1.2 μM rifampicin). (B Center and Right) Summary. (C) Absence of effects of rifamycins on Fe2+ binding and Mg2+ binding. Data for Fe2+ binding and Mg2+ binding to RNAP. (C Left) RNAP holo. (C Right) RPo.
To assess the equilibrium dissociation constant for Mg2+, KdMg, and to assess effects of rifamycins on KdMg, we performed competition experiments, quantifying Fe2+-mediated cleavage as a function of competing Mg2+ concentration. Fig. 1B and Fig. S4B present measurements of Fe2+-mediated cleavage as a function of competing Mg2+ concentration in the absence of rifamycins and in the presence of saturating concentrations of rifamycins. The results indicate that the value of the equilibrium dissociation constant for Mg2+, KdMg, in the absence of rifamycins is ≈140 μM for RNAP holo and ≈200 μM for RPo. The results further indicate that, within experimental error, values of KdMg in the presence of rifamycins are indistinguishable from values of KdMg in the absence of rifamycins. We conclude that rifamycins do not affect the affinity of binding of Mg2+ to the RNAP active center in solution.
To verify that the rifamycin concentrations used in the above experiments in fact were saturating rifamycin concentrations, we performed measurements of transcription, Fe2+ binding (as inferred from Fe2+-mediated cleavage at 5 μM Fe2+), and Mg2+ binding (as inferred from Fe2+-mediated cleavage at 5 μM Fe2+ and 300 μM Mg2+), as a function of rifamycin concentration (Fig. 1C). The results indicate that transcription is essentially completely inhibited at a rifamycin concentration of 0.3 μM and is completely inhibited at a rifamycin concentration of 1.2 μM, the concentration used in the above experiments. The results further indicate that, in contrast, Fe2+ binding and Mg2+ binding are unaltered at any rifamycin concentration tested. We conclude that rifamycins have no effect on the affinities of binding of Fe2+ and Mg2+ to the RNAP active center in solution.
Absence of Effects of Mg2+ on Rifamycin Function.
Artsimovitch et al. (7) reported that high concentrations of Mg2+ (2.5–10 mM) interfere with transcription inhibition by rifamycins. Artsimovitch et al. briefly incubated rifampicin with preformed RPo at low or high Mg2+, added nucleotides to initiate transcription, and observed higher transcription inhibition by rifampicin at low Mg2+ than at high Mg2+. We repeated experiments using the experimental design of Artsimovitch et al. and obtained results qualitatively consistent with the reported results: i.e., a decrease in transcription inhibition by rifampicin at high Mg2+ concentrations (Fig. S5A). However, consideration of the experimental design used by Artsimovitch et al. reveals two issues of potential concern. First, the incubation time used by Artsimovitch et al. for rifampicin–RNAP interaction in RPo is <10% of the incubation time required to reach saturation of rifampicin–RNAP interaction in RPo [1 min vs. >10 min (Fig. S5B; see also ref. 11)]. Second, with this short incubation time used, small effects of Mg2+ on the on-rate for rifampicin–RNAP interaction translate into large effects on the fractional occupancies of RNAP with rifampicin (Fig. S5B; see also ref. 12). As a result of these two issues, the experimental design used by Artsimovitch et al. does not yield complete saturation of RNAP with rifampicin at either low or high Mg2+ and yields a different, higher, extent of saturation at low Mg2+ than at high Mg2+. {Artsimovitch et al. performed a control experiment to document that RNAP was saturated with rifampicin in the transcription experiments (figure 5B of ref. 7). However, the control experiment was performed using an incubation time of 15 min [which is sufficient to reach, or nearly reach, saturation (see Fig. S5B)], and not using the short incubation time used in the transcription experiments [1 min, which is not sufficient to reach, or nearly reach, saturation (see Fig. S5B)].}
We reassessed whether high concentrations of Mg2+ interfere with transcription inhibition by rifamycin using experimental conditions that yield saturation of rifampicin–RNAP interaction (see Fig. S5B). The results show that, under these conditions, there is no decrease in transcription inhibition by rifampicin at high Mg2+ concentrations (Fig. 2A) and there is no decrease in fractional occupancy of RNAP by rifampicin at high Mg2+ concentrations (Fig. 2B). We conclude that high concentrations of Mg2+ do not interfere with transcription inhibition by rifampicin.
Fig. 2.
Absence of effects of Mg2+ on rifamycin function. (A) Absence of effects of Mg2+ on transcription inhibition by rifampicin. Data are from experiments with preincubation of 0.5 μM rifampicin with RNAP holo for 5 min (see Fig. S5B). Data are reported as (Yo/Y)100%, where Y is the yield of run-off transcript at the specified Mg2+ concentration, and Yo is the yield of run-off transcript at the lowest tested Mg2+ concentration (0.4 mM). (B) Absence of effects of Mg2+ on rifampicin–RNAP interaction. Data are from experiments with preequilibration of 0.5 μM rifampicin with RNAP holo for 5 min (see Fig. S5B). Data are reported as (θ/θo)100%, where θ is the fractional occupancy of RNAP by rifampicin at the specified Mg2+ concentration, and θo is the fractional occupancy of RNAP by rifampicin at the lowest tested Mg2+ concentration (0.4 mM).
Artsimovitch et al. (7) also reported that high concentrations of Mg2+ interfere with inhibition of bacterial growth by rifampicin. In view of our finding that, under conditions that result in saturation of rifampicin–RNAP interaction, high concentrations of Mg2+ do not interfere with transcription inhibition by rifampicin in vitro, we infer that this observation is unlikely to reflect interactions of Mg2+ and rifampicin with RNAP and, instead, is likely to reflect other effects of Mg2+. We point out that it is well established that Mg+2 concentrations affect rates of bacterial growth and affect multiple biochemical processes in bacterial cells. It also is possible that Mg2+ concentrations affect stability of rifampicin in culture media, uptake of rifampicin into bacterial cells, degradation of rifampicin by bacterial cells, or other rifampicin-dependent, but RNAP-independent, processes.
Absence of Putative Allosteric Effects of the Classic Mutants β-D516N and β-D516V.
Artsimovitch et al. (7) reported that the classic rifamycin-resistant substitutions β-D516N and β-D516V (13–15), which involve RNAP β subunit residue 516, a residue that is part of the rifamycin binding site and is located on the proposed rifamycin-mediated allosteric signaling pathway (Fig. S3B), reduce transcription inhibition by rifampicin but do not correspondingly reduce rifampicin–RNAP interaction. Artsimovitch et al. concluded that the rifampicin-resistance properties of these substitutions cannot be fully accounted for by effects on rifampicin–RNAP interaction and, instead, must involve effects on rifampicin-mediated allosteric signaling.
However, consideration of the experimental design used by Artsimovitch et al. (7) reveals three issues of potential concern. First, the experiments assessing transcription inhibition and rifampicin–RNAP interaction were performed under different, potentially quantitatively incommensurate, conditions (in vivo vs. in vitro). Second, the experiments assessing transcription inhibition and rifampicin–RNAP interaction were performed at only one rifampicin concentration and thus do not permit determination of the rifampicin concentration dependence of transcription inhibition and rifampicin–RNAP interaction. Third, the experiments assessing rifampicin–RNAP interaction were performed by using RNAP concentrations that were ≥10- to ≥100-fold higher than the equilibrium dissociation constant, Kd,RNAP, for rifampicin–RNAP interaction for wild-type RNAP [RNAP concentration = 25 nM at start of filtration step in experiments; RNAP concentration = 250 nM at end of filtration step in experiments; Kd,RNAP = 0.3–1 nM (see refs. 11 and 12)] and thus do not permit, even in principle, detection of up to 10- to 100-fold differences between affinity of rifampicin–RNAP interactions for wild-type RNAP vs. for mutant RNAP. As a result of these issues, the experimental design used by Artsimovitch et al. does not permit quantitative comparison of transcription inhibition and rifampicin–RNAP interaction, and does not permit determination of absolute or even relative affinities for rifampicin–RNAP interaction.
We reassessed the issue of whether the classic rifamycin-resistant substitutions β-D516N and β-D516V reduce transcription inhibition by rifampicin but do not correspondingly reduce rifampicin–RNAP interaction. We used an experimental design that employed equivalent experimental conditions for analysis of transcription inhibition and rifampicin–RNAP interaction, multiple rifampicin concentrations for analysis of transcription inhibition and rifampicin–RNAP interaction, and kinetic measurements, which enable determination of absolute and relative affinities even when concentrations of binding partners are high relative to equilibrium dissociation constants, for analysis of rifampicin–RNAP interaction. The results show that, with this experimental design, observed effects of the β-D516N and β-D516V substitutions on transcription inhibition can be accounted for by observed effects on rifampicin–RNAP interaction (Table 1 and Fig. S6).
Table 1.
Absence of putative allosteric effects of classic mutants β-D516N and β-D516V
| RNAP derivative | ISAT,* % | ISAT,X/ISAT,RNAP | IC50,† nM | IC50X/IC50RNAP | kon, M−1·s−1 | koff, s−1 | Kd, nM | Kd,X/Kd,RNAP |
|---|---|---|---|---|---|---|---|---|
| Transcription inhibition by rifampicin | ||||||||
| RNAP | 100 | [1] | 2 | [1] | ||||
| [Asn516]β-RNAP | 100 | 1 | 400 | 200 | ||||
| [Val516]β-RNAP | 100 | 1 | 4,000 | 2,000 | ||||
| Rifampicin-RNAP interaction | ||||||||
| RNAP | 3.6 × 105 | 1.5 × 10−4 | 0.41 | [1] | ||||
| [Asn516]β-RNAP | 4.6 × 105 | 1.3 × 10−2 | 28‡ | 68‡ | ||||
| 69§ | 170§ | |||||||
| [Val516]β-RNAP | ≤7.1 × 105 | 1.7 × 10−1 | ≥240‡ | ≥590‡ | ||||
| 850§ | 2100§ |
*ISAT is the percent inhibition of transcription at a saturating concentration of rifampicin.
†IC50 is the concentration of rifampicin (unbound rifampicin only) resulting in 50% inhibition of transcription.
‡Data for [Asn516]β-RNAP and [Val516]β-RNAP obtained from association and dissociation kinetics.
§Data for [Asn516]β-RNAP and [Val516]β-RNAP obtained from equilibrium binding assays.
Results of experiments assessing transcription inhibition in vitro are presented in Table 1 (left columns). The results show that the β-D516N and β-D516V substitutions affect the rifampicin concentration dependence for transcription inhibition, increasing the rifampicin concentration dependence for transcription inhibition by factors of ≈200 and ≈2,000, respectively (IC50X/IC50RNAP = ≈200 and ≈2,000; values consistent with values in ref. 13), but do not affect transcription inhibition at saturating rifampicin (ISAT,X/ISAT,RNAP = 1 and 1). Results of experiments assessing rifampicin–RNAP interaction in vitro are presented in Table 1 (right columns) and Fig. S6. The results show, contrary to Artsimovitch et al., that the β-D516N and β-D516V substitutions profoundly affect rifampicin–RNAP interaction, increasing the rifampicin concentration dependence for rifampicin–RNAP interaction by factors of ≈70–200 and ≈600–2,000, respectively (Kd,X/Kd,RNAP = ≈70–200 and ≈600–2,000). Within error, the quantitative effects of the substitutions on the rifampicin concentration dependence for transcription inhibition [Table 1 (left columns)] can be accounted for by the quantitative effects of the substitutions on rifampicin–RNAP interaction [Table 1 (right columns)], without invoking putative additional effects on rifampicin-mediated allosteric signaling subsequent to rifampicin–RNAP interaction. The finding that the substitutions affect the rifampicin concentration dependence for transcription inhibition but do not affect transcription inhibition at saturating rifampicin further indicates that effects of the substitutions on transcription inhibition are accounted for by effects on rifampicin–RNAP interaction (i.e., rifampicin-concentration-dependent, rifampicin-saturable effects), without invoking putative additional effects on rifampicin-mediated allosteric signaling subsequent to rifampicin–RNAP interaction (i.e., rifampicin-concentration-independent, rifampicin-nonsaturable effects). Indeed, the finding that the substitutions do not affect transcription inhibition at saturating rifampicin unequivocally rules out putative additional effects on rifampicin-mediated allosteric signaling subsequent to rifampicin–RNAP interaction.
We conclude that the classic rifamycin-resistant mutants β-D516N and β-D516V confer resistance through effects on rifampicin interaction and not through putative additional effects on rifampicin-mediated allosteric signaling.
Absence of Putative Allosteric Effects of Designed Mutant β-L1235A.
Artsimovitch et al. (7) reported that the β-L1235A substitution, which was designed to substitute RNAP β residue 1235, a residue that is not part of the rifamycin binding site but is located on the proposed rifamycin-mediated allosteric signaling pathway (Fig. S3B), reduces transcription inhibition by rifampicin, in vivo and in vitro, but does not correspondingly reduce rifampicin–RNAP interaction. Artsimovitch et al. concluded that the rifampicin-resistance properties of this substitution cannot be fully accounted for by effects on rifampicin–RNAP interaction and, instead, must involve effects on rifampicin-mediated allosteric signaling.
The report that substitution of RNAP β residue 1235 confers significant rifampicin-resistance was surprising. To our knowledge, substitutions of this residue that confer rifampicin-resistance have not been reported elsewhere, neither among reports of sequenced rifampicin-resistant mutants of E. coli RNAP isolated after spontaneous, random, and saturation mutagenesis [≫500 independent isolates (refs. 1 and 16 and references therein)], nor among reports of sequenced rifampicin-resistant clinical isolates of Mycobacterium tuberculosis RNAP and Staphylococcus aureus RNAP [≫500 independent isolates (refs. 17 and 18 and references therein)].
We have been unable to substantiate the report of Artsimovitch et al. that substitution of RNAP β residue 1235 confers significant rifampicin-resistance.
Results of experiments assessing effects of the β-L1235A substitution in vivo are presented in Table 2(left columns) and in Tables S1 and S2. The results show that the substituted protein—produced using the same plasmid used by Artsimovitch et al. (pIA594; provided by I. Artsimovitch, Ohio State University, Columbus)—does not result in significant rifampicin resistance in vivo. Observed minimal inhibitory concentrations of rifampicin for cells producing the substituted protein are equal to, or within a factor of two of, those for cells producing the wild-type protein. Equivalent results are obtained by using E. coli strain DH5α (the strain used by Artsimovitch et al.) and E. coli strain D21f2/tolC—a strain with cell-envelope defects resulting in increased uptake and retention of small molecules and increased sensitivity to small-molecule antibacterial agents, including rifamycins (19) (Q.J. and R.H.E., unpublished data) [Table 2 (left columns)]. Equivalent results also are obtained in assays performed in liquid medium and on solid medium [Table 2 (left columns)], in assays performed at 37°C and at 32°C (a temperature that could facilitate detection of resistance mediated by an unstable, thermosensitive RNAP derivative) [Table 2 (left columns) and Table S1], and in assays performed using four different rifamycins: rifampicin, rifapentine, rifabutin, and rifamycin SV [Table 2 (left columns) and Table S2]. We conclude that substitution of RNAP β residue 1235 does not confer significant rifamycin resistance in vivo.
Table 2.
Absence of putative allosteric effects of designed mutant β-L1235A
| RNAP derivative | Liquid medium |
Solid medium |
ISAT,† % | ISAT,X/ISAT,RNAP | IC50,‡ nM | IC50,X/IC50,RNAP | kon, M−1·s−1 | koff, s−1 | Kd, nM | Kd,X/Kd,RNAP | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC,* μg/ml | MIC ratio | MIC, μg/ml | MIC ratio | |||||||||
| Growth inhibition by rifampicin (E. coli strain DH5α) | ||||||||||||
| RNAP | 6.25 | [1] | 0.4 | [1] | ||||||||
| [Ala1235]β-RNAP | 12.5 | 2 | 0.3 | 0.8 | ||||||||
| Growth inhibition by rifampicin (E. coli strain D21f2/tolC) | ||||||||||||
| RNAP | 0.195 | [1] | 0.1 | [1] | ||||||||
| [Ala1235]β-RNAP | 0.391 | 2 | 0.2 | 2 | ||||||||
| Transcription inhibition by rifampicin | ||||||||||||
| RNAP | 100 | [1] | 2 | [1] | ||||||||
| [Ala1235]β-RNAP | 100 | 1 | 2 | 1 | ||||||||
| Rifampicin–RNAP interaction | ||||||||||||
| RNAP | 3.6 × 105 | 1.5 × 10−4 | 0.41 | [1] | ||||||||
| [Ala1235]β-RNAP | 3.6 × 105 | 2.4 × 10−4 | 0.68 | 1.7 | ||||||||
*MIC is the concentration of rifampicin resulting in 90% inhibition of growth.
†ISAT is the percent inhibition of transcription at a saturating concentration of rifampicin.
‡IC50 is the concentration of rifampicin (unbound rifampicin only) resulting in 50% inhibition of transcription.
Results of experiments assessing the effects of the β-L1235A substitution in vitro are presented in Table 2 (center and right columns), Table S1, and Fig. S7. The results show that the substituted protein has no significant effect on the rifampicin concentration dependence of transcription inhibition (Kd,X/Kd,RNAP = 1) and has no significant effect on transcription inhibition at saturating rifampicin (ISAT,X/ISAT,RNAP = 1) [Table 2 (center columns)]. The results further show that the substituted protein has no significant effect on the rifampicin concentration dependence for rifampicin–RNAP interaction (Kd,X/Kd,RNAP = 1.7) [Table 2 (right columns)]. Equivalent results are obtained in assays performed at 37°C with protein produced from cells grown at 37°C, and in assays performed at 32°C with protein produced from cells cultured at 32°C [Table 1 (center and right columns) and Table S1]. Equivalent results are obtained in assays performed with two independent preparations of the substituted protein produced using the rpoBL1235A expression plasmid pIA594 (provided by I. Artsimovitch), and in assays with two independent preparations of the substituted protein produced using rpoABL1235ACZ expression plasmid pEcA(H10-PPX)BL1235ACZ (constructed in this work) [Table 1 (center and right columns)] (V.M. and R.H.E., unpublished data). We conclude that substitution of RNAP β residue 1235 does not confer significant rifampicin resistance in vitro.
We sequenced the entire rpoB genes of the plasmids used to produce the substituted protein in vivo and in vitro: pIA594 (provided by I. Artsimovitch), pIA597 (provided by I. Artsimovitch), and pEcA(H10-PPX)BL1235ACZ (constructed in this work). The plasmids all contain the rpoBL1235A mutation, and all are free of additional rpoB mutations.
Conclusions
Rifamycins are among the most potent and broad-spectrum antibiotics against bacterial pathogens and remain a key component of antituberculosis therapy (1). Bacteria develop resistance to rifamycins with relatively high frequency, however, limiting the utility of rifamycin therapy (1). A detailed understanding of the mechanism of inhibition of RNAP by rifamycins and of effects of rifamycin-resistant mutants is essential to guide further research.
Structure–function studies of rifamycin–RNAP complexes have led to two mechanistic models for inhibition of RNAP by rifamycins: a model in which rifamycins sterically prevent extension and retention of RNA products >2–3 nt (“steric-occlusion model”) (4, 6) (Fig. S3A) and a model in which, instead or in addition, rifamycins allosterically decrease the affinity of binding of Mg2+ to the RNAP active center (“allosteric model”) (7) (Fig. S3B). The two models make different predictions regarding effects of rifamycin-resistant mutants and have different implications for structure-based design of improved, next-generation rifamycins (6, 7). For example, the steric-occlusion model predicts that rifamycin-resistant mutants of RNAP involve amino acid substitutions that decrease the affinity of binding of rifamycins to RNAP, whereas the allosteric model predicts that rifamycin-resistant mutants of RNAP can involve amino acid substitutions that do not decrease the affinity of binding of rifamycins to RNAP but, instead, disrupt allosteric signaling.
The allosteric model was proposed based on a structural observation, i.e., the absence of the RNAP-active-center Mg2+ in two crystal structures of T. thermophilus RNAP in complex with rifamycins, and three sets of biochemical observations (7). Overall, Artsimovitch et al. made four testable assertions:
Rifamycins decrease the affinity of binding of Mg2+ to the RNAP active center.
High Mg2+ concentrations confer resistance to transcription inhibition by rifamycins.
The classic rifamycin-resistant mutants β-D516N and β-D516V, which substitute a residue located on the proposed allosteric signaling pathway, confer resistance to rifamycins but do not correspondingly reduce affinity of RNAP for rifamycins.
The designed rifamycin-resistant mutant β-L1235A, which substitutes a residue located on the proposed allosteric signaling pathway, confers resistance to rifamycins but does not correspondingly reduce affinity of RNAP for rifamycins.
Here, we have directly tested these four assertions. We find that all four assertions are incorrect. We further note that the allosteric model, at least in its simplest form, is inconsistent with the fact that most rifamycins, in most contexts, have no effect on the formation of the first phosphodiester bond (see ref. 4). We conclude that there is no basis for the proposal that allosteric modulation of the affinity of binding of Mg2+ to the RNAP active center is essential for inhibition of RNAP by rifamycins.
Methods
Full details of the methods used are presented in SI Materials and Methods.
Fe2+-Mediated Cleavage Experiments.
Fe2+-mediated cleavage experiments using HMPK-tagged RNAP with 32P incorporated at the C terminus of the β′ subunit were performed as described in ref. 9.
Transcription-Inhibition Assays.
Transcription-inhibition assays were performed as described in refs. 6 and 20.
Growth-Inhibition Assays.
Growth-inhibition assays were performed essentially as described in ref. 21.
Rifampicin–RNAP Interaction Assays.
Rifamycin–RNAP interaction was detected by monitoring quenching of fluorescence emission of the fluorescent probe fluorescein incorporated site-specifically into RNAP (serving as fluorescence resonance energy transfer donor) by the naphthyl group of rifamycin [serving as fluorescence resonance energy transfer acceptor (22)]. Data shown are for experiments using holo derivatives and RPo derivatives having fluorescein incorporated site-specifically at residue 517 of σ70 (methods as in ref. 22). Parallel experiments were performed by using RNAP holo derivatives having fluorescein incorporated site-specifically at residue 36 of σ70, at residue 59 of σ70, or at residue 459 of σ70; there was no detectable effect of the labeling-site position on association kinetics, dissociation kinetics, or equilibrium dissociation constants (V.M. and R.H.E., unpublished data).
Supplementary Material
Acknowledgments.
We thank I. Artsimovitch and R. Landick for plasmids and I. Artsimovitch, G. Höfle, V. Nikiforov, V. Svetlov, and D. Vassylyev for discussion. This work was supported by a Howard Hughes Medical Institute Investigatorship (to R.H.E.) and National Institutes of Health Grants GM30717 (to A.M.), GM61898 (to S.A.D.), GM41376 (to R.H.E.), and AI072766 (to R.H.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802822105/DCSupplemental.
References
- 1.Floss HG, Yu TW. Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev. 2005;105:621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 2.Hartmann G, Honikel KO, Knusel F, Nuesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta. 1967;145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 3.Hinkle DC, Mangel WF, Chamberlin MJ. Studies of the binding of Escherichia coli RNA polymerase to DNA. IV. The effect of rifampicin on binding and on RNA chain initiation. J Mol Biol. 1972;70:209–220. doi: 10.1016/0022-2836(72)90534-7. [DOI] [PubMed] [Google Scholar]
- 4.McClure WR, Cech CL. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem. 1978;253:8949–8956. [PubMed] [Google Scholar]
- 5.Sippel AE, Hartmann GR. Rifampicin resistance of RNA polymerase in the binary complex with DNA. Eur J Biochem. 1970;16:152–157. doi: 10.1111/j.1432-1033.1970.tb01066.x. [DOI] [PubMed] [Google Scholar]
- 6.Campbell EA, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 7.Artsimovitch I, et al. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122:351–363. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Zaychikov E, et al. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 9.Mustaev A, et al. Modular organization of the catalytic center of RNA polymerase. Proc Natl Acad Sci USA. 1997;94:6641–6645. doi: 10.1073/pnas.94.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang G, et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 11.Wehrli W, Handschin J, Wunderli W. In: RNA Polymerase. Chamberlin MJ, Losick R, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1976. pp. 397–412. [Google Scholar]
- 12.Barh W, Stender W, Scheit K-H, Jovin T. In: RNA Polymerase. Chamberlin MJ, Losick R, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1976. pp. 369–396. [Google Scholar]
- 13.Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 14.Lisitsyn NA, Gur'ev SO, Sverdlov ED, Moiseeva EP, Nikiforov VG. Nucleotide substitutions in the rpoB gene leading to rifampicin resistance of E. coli RNA polymerase. Bioorg Khim. 1984;10:127–128. [PubMed] [Google Scholar]
- 15.Ovchinnikov YA, et al. Primary structure of Escherichia coli RNA polymerase nucleotide substitution in the b subunit gene of the rifampicin resistant rpoB255 mutant. Mol Gen Genet. 1981;184:536–538. doi: 10.1007/BF00352535. [DOI] [PubMed] [Google Scholar]
- 16.Garibyan L, et al. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair. 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 17.Musser J. Antimicrobial agent resistance in mycobacteria: Molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Neill A, Huovinen T, Fishwick C, Chopra I. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob Agents Chemother. 2006;50:298–309. doi: 10.1128/AAC.50.1.298-309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fralick J, Burns-Keliher L. Additive effect of tolC and rfa mutations on the hydrophobic barrier of the outer membrane of Escherichia coli K-12. J Bacteriol. 1994;176:6404–6406. doi: 10.1128/jb.176.20.6404-6406.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuske S, et al. Inhibition of bacterial RNA polymerase by streptolydigin: Stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight J, Mekler V, Mukhopadhyay J, Ebright RH, Levy R. Distance-restrained docking of rifampicin and rifamycin SV to RNA polymerase using systematic FRET measurements: Developing benchmarks of model quality and reliability. Biophys J. 2005;88:925–938. doi: 10.1529/biophysj.104.050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.