Abstract
Forkhead box class O (FoxO) transcription factors are key regulators of growth, metabolism, life span, and stress resistance. FoxOs integrate signals from different pathways and guide the cellular response to varying energy and stress conditions. FoxOs are modulated by several signaling pathways, e.g., the insulin-TOR signaling pathway and the stress induced JNK signaling pathway. Here, we report a genome wide RNAi screen of kinases and phosphatases aiming to find regulators of dFoxO activity in Drosophila S2 cells. By using a combination of transcriptional activity and localization assays we identified several enzymes that modulate dFoxO transcriptional activity, intracellular localization and/or protein stability. Importantly, several currently known dFoxO regulators were found in the screening, confirming the validity of our approach. In addition, several interesting new regulators were identified, including protein kinase C and glycogen synthase kinase 3β, two proteins with important roles in insulin signaling. Furthermore, several mammalian orthologs of the proteins identified in Drosophila also regulate FOXO activity in mammalian cells. Our results contribute to a comprehensive understanding of FoxO regulatory processes.
Keywords: dFoxO, insulin signaling, PKC
Forkhead box class O (FoxO) transcription factors are members of the forkhead box transcription factor superfamily, with orthologs in various species such as mammals (1, 2), C.elegans (3), Zebrafish (4) and Drosophila (5, 6). FoxO proteins possess a wide range of cellular functions ranging from the induction of apoptosis and cell cycle control, to the oxidative stress response and lifespan determination. In addition, FoxOs are well defined targets downstream of the conserved insulin/TOR and JNK signaling networks having an important role in the regulation of processes as diverse as cellular growth, stress resistance, and energy homeostasis (7, 8). Consequently, FoxOs have several characterized bona fide target genes involved in metabolism and growth, such as g6pase (9), pepck (10), 4e-bp (5, 6), insulin receptor (6, 11), and myc (12).
To date, several proteins are known to interact with FoxO transcription factors, regulating their intracellular localization and/or activity, and the number of newly identified regulators is rapidly increasing (13). One of the best documented is the AKT/PKB -kinase, which phosphorylates FoxO in three conserved Ser/Thr residues, leading to FoxO cytoplasmic retention and transcriptional inactivation (14–17). In the cytoplasm, FoxO is ubiquitinylated and targeted for degradation (18, 19). Upon growth factor depletion, FoxO is predominantly nuclear. Interestingly, FoxO transcriptional activity can also be modulated in the nucleus (20, 21). Furthermore, some subpopulations of growing cells possess nuclear inactive FoxO, implying that additional layers of control exist in the nucleus (22). Given the variety of cellular functions where FoxOs are implicated, and the observation that FoxOs act as a converging point for many different signaling pathways guiding the cellular response to varying nutritional conditions and stress factors, it is likely that FoxO transcription factors are regulated through many different mechanisms.
To identify proteins modulating FoxO activity, we turned to the powerful system of Drosophila S2 cells. Here, we describe the first genome wide screen of Drosophila kinases and phosphatases aiming to identify novel regulators of dFoxO transcriptional activity. By using a combination of transcriptional reporter assays, Western blot analysis and high-throughput microscopy, we were able to identify several enzymes that regulate dFoxO intracellular localization, protein stability, and/or transactivation. Furthermore, some of the identified modifiers were found to act similarly in mammalian cells stressing out the conserved nature of these interactions. Our results add new insights to the complexity of the regulatory network around FoxO.
Results
Primary Screening.
We used a kinase and phosphatase RNAi library to search for modulators of dFoxO transcriptional activity in Drosophila S2 cell culture (23). The RNAi library comprised 251 and 86 known and predicted kinases and phosphatases, respectively. When applicable, the multiple transcripts of a given gene were individually targeted. The strategy we used in the primary screen was to use a reporter construct having a synthetic promoter consisting of four repeats of a dFoxO recognition element (4xFRE) (6). This promoter construct was cloned upstream of an ORF for EGFP (24). Upon dsRNA treatment, followed by induction of dFoxO expression, cells were scored for their EGFP intensity, which represents activity from the dFoxO responsive promoter. A control vector expressing Red Fluorescent Protein (RFP) was used to normalize the transcriptional and translational efficiency. For each dsRNA treatment, 6,000 cells were quantified for their EGFP and RFP intensities by high-throughput microscopy. The normalized EGFP intensities from each treatment were averaged and converted into Z scores. Genes having an average Z score of 1.3 or higher, corresponding to P ≤ 0.1 confidence level, were selected for the secondary screen. This low stringency was possible due to the relatively low number of genes screened. A scheme describing this strategy can be seen in supporting information (SI) Fig. S1.
Thirty-eight positive hits were identified in the primary screen (Table S1). Importantly, several known dFoxO regulators were detected, confirming the validity of our approach. dFoxO regulators, such as the mst-like kinase Hippo (25), dJNK (26), MNB/DYRK1 (27), and dTOR (20) changed significantly EGFP/RFP ratio (Table S1). In addition, dsRNAs against PDK1, AKT, and PTEN, all known insulin signaling components, also affected EGFP/RFP ratio. Interestingly, we found 26 kinases and five phosphatases previously unknown to interact with FoxO that regulated dFoxO activity (as measured by the EGFP reporter). These were selected for further characterization and are hereafter referred as primary hits.
Secondary Screening.
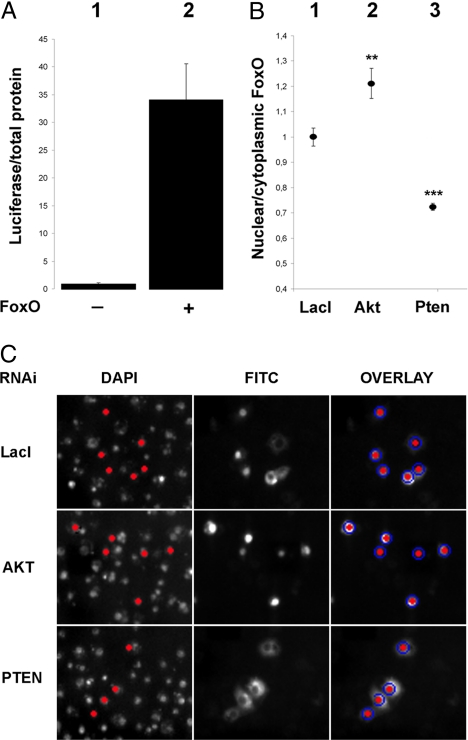
To confirm that the 31 new proteins found in the first screen are indeed dFoxO regulators, a secondary screen was developed where the readout of transcriptional activity was accompanied by simultaneous detection of dFoxO intracellular localization and measurement of dFoxO protein abundance. New dsRNA molecules were designed for each target with the purpose of avoiding possible off-target effects. To measure dFoxO transcriptional activity, we used a luciferase reporter under the control of the Drosophila Insulin Receptor (dInR) promoter (6). The reporter construct was chosen based on its very high signal to noise ratio, which enables to easily score transcriptional response derived exclusively from the overexpressed dFoxO protein. Fig. 1A, bar 1, shows that in the absence of overexpressed dFoxO expression, background levels of the dInR reporter are negligible. In contrast, after CuSO4 addition, which activates dFoxO expression from the metallothionein promoter, luciferase activity increases 30-fold (Fig. 1A, bar 2).
Fig. 1.
Secondary screen controls. (A) Overexpression of the WT dFoxO in S2 cells induces the expression of a luciferase reporter under the dInR promoter. (B) Conditions for the dFoxO subcellular localization detection were tested with known insulin signaling components AKT and PTEN. (C) Representative high-throughput microscope images are shown. The nuclei were identified by DAPI staining and selected for analysis based on the nuclear FITC intensity (dFoxO) exceeding the background level (red nuclei). The nuclear FITC intensity was measured from the area of DAPI staining (red nuclei) and from the surrounding cytoplasm (blue circles). **, P < 0.01; ***, P < 0.001.
To detect and quantify dFoxO intracellular localization, we scored nucleus vs. cytoplasmic dFoxO localization by using high-throughput microscopy (Fig. 1 B and C). As controls, we used dsRNAs against two well known insulin signaling pathway components PTEN (which drives dFoxO localization in the cytoplasm) and AKT (which causes dFoxO accumulation in the nucleus) (Fig. 1 B and C). Finally, to determine whether dsRNA treatments affect dFoxO stability, dFoxO protein levels were measured by western. In all experiments, the treatments were compared with the negative control Escherichia coli lacI gene dsRNA.
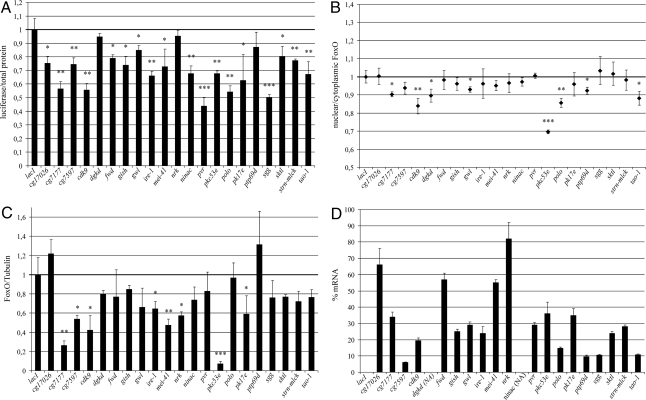
Transcriptional activity, subcellular localization, and protein abundance were scored for all 31 suspected dFoxO regulators, and the results are shown in Fig. 2. The transcriptional assay confirmed that knocking down 18 out of the original 31 primary hits reduced dFoxO transcriptional activity, consistent with a role as true dFoxO activators. The list includes well known kinases like PKC and POLO (Fig. 2A). Knocking down eight primary hits also affected dFoxO localization (Fig. 2B) by reducing the amount of dFoxO in the nucleus (Fig. 2B), indicating that these proteins regulate dFoxO activity primarily by modulating its nuclear/cytoplasmic localization, in a similar way to the AKT/PTEN system. Examples of this kind of regulators are CDK9 and TAO1 kinases (Fig. 2B). In addition to nuclear/cytoplasmic localization, FoxO activity is also regulated by proteosomal degradation (18, 19). Therefore, we investigated whether levels of dFoxO protein were affected by dsRNA treatments against the 31 primary hits. Fig. 2C shows that dFoxO protein levels were significantly reduced by dsRNA against eight targets, including PKC53E. Finally, we used qPCR to make sure the mRNA for each target was expressed in S2 cells and to measure the efficiency of the dsRNA treatments (Fig. 2D). In summary, we identified 21 kinases/phosphatases that modulated the dFoxO transcriptional activity, protein abundance and/or subcellular localization. These proteins are grouped based on their regulatory function on dFoxO in Fig. S2.
Fig. 2.
Characterized hits from the secondary screen. (A) dFoxO activity. (B) dFoxO intracellular localization. (C) dFoxO abundance. (D) Knock down efficiency. LacI was used as a control. Knockdown efficiency could not be estimated for ninac and dgkd due to their low level of expression in S2 cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine whether changes in dFoxO activity/localization were induced through the insulin signaling pathway, we determined phosphorylation of AKT Ser-505 for all hits in Fig. 2. No change was observed in AKT Ser-505 phosphorylation (Fig. 3H and data not shown), indicating that the observed effects in dFoxO localization and/or activity are related to dFoxO regulation through signaling pathways different from the canonical insulin/AKT signaling pathway. An additional experiment was performed to confirm this interpretation. dFoxO phosphorylation by AKT is readily detectable by a change of mobility in SDS/PAGE (6). Therefore, we performed Western blot analysis against dFoxO in conditions where we would detect a shift of mobility produced by AKT. However, knocking down any of the 21 hits did not alter dFoxO eletrophoretic mobility, whereas dsRNA treatments for PTEN and AKT affected dFoxO mobility as expected (Fig. S3). Therefore, we rule out the possibility that the effects seen with positive hits in the secondary screen are due to activation/repression of the insulin signaling pathway through AKT.
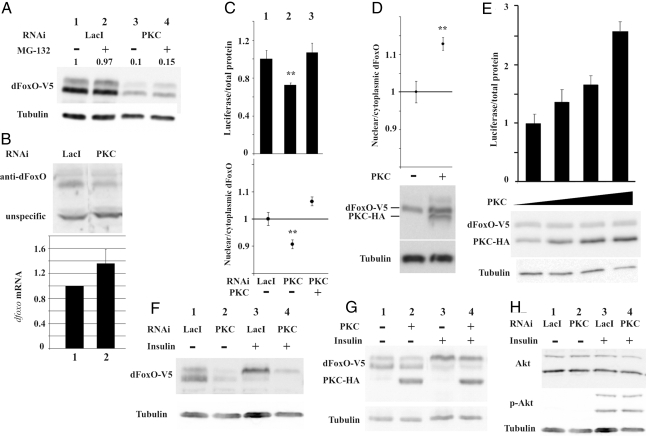
Fig. 3.
PKC53E is a modulator of dFoxO activity. (A) Western blot showing overexpressed dFOXO-V5 in S2 cells treated with RNAi against LacI (lanes 1 and 2) or PKC53E (lanes 3 and 4), in the presence of MG132 (lanes 2 and 4). (B) (Upper) Endogenous dFoxO protein is reduced after PKC53E knockdown (lane 2). (Lower) dFoxO mRNA levels. (C) A dsRNA targeted to the PKC53E 3′ UTR decreases the dFoxO driven luciferase reporter activity and the amount of nuclear dFoxO (lane 2). Overexpression of the PKC53E is able to rescue this phenotype (lane 3). (D and E). Overexpression of the PKC53E in S2 cells increases the proportion of nuclear dFoxO and the luciferase activity. (F and G). Insulin induces a band shift of dFoxO (lanes 1 and 3). This band shift was not affected upon PKC53E RNAi (lanes 2 and 4 in F) or overexpression (lanes 2 and 4 in G). (H) The level of AKT Ser-505 phosphorylation induced by insulin was not affected upon PKC53E RNAi (lanes 2 and 4). **, P < 0.01.
We identified two genes, diacylglycerol kinase d (dgkd) and protein tyrosine phosphatase 69D (ptp69d), which, interestingly, affected dFoxO localization but did not affect transcriptional activity as measured with the dInR promoter reporter (Fig. 2). It is possible that both proteins regulate dFoxO transcription on specific promoters in conjunction with other activators, and such factors are missing in Drosophila S2 cells, thus explaining the negative result obtained with our luciferase reporter assay. Alternatively, these proteins could affect dFoxO stability without altering its transcriptional activity, having a net effect on dFoxO accumulation in the nucleus.
dPKC Is a Regulator of dFoxO.
To further verify our screening results, we chose protein kinase C 53E (PKC53E) for further characterization. PKC53E is a member of the well characterized AGC protein kinase family and is implicated in wide range of cellular functions (28). PKCs have important roles in insulin signaling, and, in our screening, PKC53E dsRNA treatment results in a shift in dFoxO subcellular localization from the nucleus to the cytoplasm, which is accompanied by a decrease in luciferase reporter activity and a reduction in the levels of dFoxO protein (Fig. 2). The notable reduction in dFoxO protein level was found to be only partially dependent on the proteosomal degradation machinery as the proteosomal inhibitor MG-132 modestly augmented the dFoxO abundance upon PKC knockdown (Fig. 3A, lanes 3 and 4). No effect was seen with ammonium chloride, suggesting that lysosomal degradation is not involved (data not shown). Importantly, the reduction of dFoxO protein levels upon PKC knockdown was also observed with endogenous dFoxO (Fig. 3B, lane 2), whereas dfoxo mRNA levels increase, suggesting a compensatory mechanism. To confirm that the observed phenotype is indeed the result of PKC53E ablation, we performed a rescue experiment. A dsRNA molecule was designed to target the PKC53E 3′ UTR. As expected, this dsRNA produced a significant reduction in luciferase activity (Fig. 3C, bar 2). Interestingly, this phenotype was fully rescued by overexpression of wild-type PKC53E (Fig. 3C, bar 3). Furthermore, dFoxO localization shift toward the cytoplasm was fully rescued by overexpressing PKC53E (Fig. 3C, lane 3). These results demonstrate that the reduction of dFoxO activity observed with dsRNA against PKC53E is a consequence of PKC53E loss of function. We also independently overexpressed wild-type PKC53E in S2 cells and found that dFoxO accumulated into the nucleus resulting in an increase of transcription as measured by the luciferase reporter (Fig. 3 D and E). Importantly, we could not detect any difference after PKC53E knockdown or overexpression in the AKT-dependent dFoxO mobility shift (Fig. 3 F and G), or differences in AKT Ser-505 phosphorylation (Fig. 3H), demonstrating that the regulation of dFoxO by PKC53E is independent of AKT.
The FoxO Regulatory Network is Conserved in Mammals.
The regulation of insulin signaling by FOXO transcription factors is well conserved between Drosophila and mammals (7). Interestingly, most of the hits identified in this screen have a characterized mammalian ortholog (see Table S2). Therefore, we determined whether mammalian FOXOs are regulated by this group of effectors too.
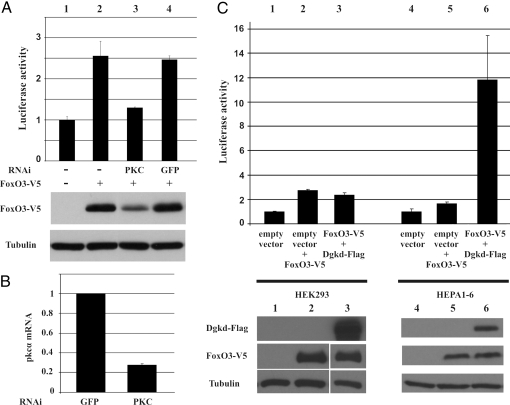
First, we used RNAi to knockdown the closest human ortholog of Drosophila PKC53E, PKCα, in human embryonic kidney cells (HEK293) and simultaneously measured the activity of FOXO3a by using a luciferase reporter under the control of human insulin receptor promoter (11). Similar to what was observed in Drosophila S2 cells, the knockdown of PKCα resulted in reduced luciferase activity accompanied with decreased FOXO3a protein levels (Fig. 4A, lane 3). Control experiments showed efficient knockdown of PKCα mRNA (Fig. 4B). Next, based on the availability of characterized expression plasmids, we chose eight orthologs for further analysis. We expressed the cDNAs in mammalian cells together with human FOXO3a and recorded the transcriptional activity of the human InR-luciferase reporter. Cotransfection of this set of kinases increased FOXO3a activity, with variable effect depending on the specific kinase. Interestingly, FOXO3a activity was substantially increased in mouse hepatoma cells (HEPA1–6), but only slightly, and not for all kinases, in HEK293 cells, suggesting tissue specific regulation of FOXO3a by this group of kinases. An example is illustrated in Fig. 4C. Whereas coexpression of FOXO3a and the human Diacylglycerol kinase δ2 (DGKδ2) in HEK293 cells did not affect the luciferase reporter expression (compare lanes 2 and 3 in Fig. 4C), coexpression in HEPA1–6 cells resulted in ≈6-fold increase in reporter activity compared with the empty vector control (compare lanes 5 and 6 in Fig. 4C). Similar results were obtained with the other overexpressed kinases (Fig. S4). These results demonstrate that several of the regulators identified from this screening also control FOXO activity in mammals.
Fig. 4.
PKCα and DGKδ2 regulate FOXO3a activity in mammalian cells. (A) FOXO3a activity and protein levels are reduced in PKCα knockout (lane 3). Lane 4, GFP control. (B) PKCα knockdown efficiency. (C) Coexpression of DGKδ2 with FOXO3a in HEK293 cells (lanes 1–3) and HEPA1–6 cells (lanes 4–6). Luciferase activity is measured with a human InR luciferase reporter.
Discussion
By using a combination of transcriptional reporter and localization assays, we have discovered 21 dFoxO regulators. Some positive hits from our screen had an effect in dFoxO activity, localization, and protein stability, whereas other hits affected only transcriptional activity, suggesting that more mechanisms beyond subcellular localization and degradation are used by cells to regulate dFoxO activity. In addition to the 18 proteins that affected dFoxO transcriptional activity, our screening produced three more hits. Two of them seem to affect only dFoxO localization (dgkd and ptp69d), and one, neurospecific receptor kinase (nrk), affected exclusively dFoxO protein stability. It is possible that these proteins regulate dFoxO transcription on specific promoters in conjunction with other activators and that such factors are missing in Drosophila S2 cells. This would explain their lack of effect on the dInR promoter. Alternatively, they could affect dFoxO stability resulting in a net effect of dFoxO protein accumulation in the nucleus.
Initially, our screening strategy was designed to identify both positive and negative regulators of dFoxO activity; however, no dFoxO repressors were found. Putative dFoxO repressors were present in our primary hit list of 31 targets, but those were later excluded in the secondary screen. This surprising observation suggests that our screen may be biased against dFoxO repressors. dFoxO is a well known inhibitor of protein biosynthesis in vivo (29), so under conditions of increased dFoxO activity, we expect a reduction of general translation that could affect GFP and luciferase translation too. Therefore, we hypothesize that in the case of enhanced dFoxO activity it is possible that the concomitant inhibition of protein biosynthesis overruled a slight increase in reporter accumulation. This would explain the lack of dFoxO repressors among the targets of our screen. Moreover, the design of our screening based on S2 cells excludes the identification of regulatory mechanisms specific for other cell types, and instances where dFoxO is acting as a cofactor thereby regulating transcription indirectly.
Our results demonstrate that Drosophila PKC53E isoform is a dFoxO activator. Similar results were obtained in mammalian cells pointing out that the interaction is conserved. PKC isoforms have very important roles in insulin signaling, and each of the isoforms has been shown to be activated by insulin stimulation or conditions important for effective insulin stimulation (30). Importantly, PKC isoforms can both activate or inhibit insulin signaling: Atypical PKC isoforms are required for insulin-stimulated glucose transport in muscle and adipocytes (31). In contrast, certain conventional and novel PKC isoforms are known to antagonize insulin signaling in vertebrates (32, 33). This interaction is implicated in the pathogenesis of free fatty acid mediated insulin resistance (reviewed in ref. 34). Drosophila possesses six PKC isoforms whose role in this context has not yet been addressed. PKC53E homolog is closest to human conventional PKCα (35). Interestingly, it has been shown that PKCα inhibits insulin signaling through binding and phosphorylation of IRS1 (33). Thus, PKCα would serve as a constitutively active inhibitory regulator of the insulin cascade through its association with IRS1. On stimulation with insulin, PKCα would dissociate from IRS1, thus releasing this protein from its down-regulated state. This would open the “gate” for transmission of the insulin signal. We previously found that dFoxO/FOXO1 increases insulin sensitivity by up-regulating insulin receptor transcription (11). The observation that Drosophila PKCα activates dFoxO adds an additional twist in the complex regulatory network that dFoxO has on insulin signaling. Interestingly, in our experimental system AKT dependent dFoxO bandshift and AKT Ser-505 phosphorylation was not affected by PKC53E, indicating that PKC53E regulation of dFoxO is independent of AKT signaling.
Another well known enzyme implicated in the control of metabolism identified as a regulator of dFoxO transcriptional activity is the Drosophila ortholog of Glycogen synthase kinase 3β (GSK-3β, Shaggy). GSK-3β is a regulator of glucose metabolism through the phosphorylation and inhibition of glycogen synthase, the rate limiting enzyme of glycogen deposition. GSK-3β is inhibited by AKT (36), so it was not surprising to see that GSK-3β activates dFoxO. GSK-3β protein level and activity is elevated in type II diabetic skeletal muscle cells reflecting the impairment of whole body glucose uptake characteristic to this disease (37). In addition, selective inhibition of GSK-3β by lithium chloride represses the expression of g6pase and pepck in rat hepatoma cells (38), both known targets of FoxO (9, 10). Taken together, these observations suggest that some of the metabolic effects of GSK-3β are achieved by directly modulating dFoxO activity.
An interesting dFoxO regulator is Polo-like kinase. Polo-like kinases (Plks) are known regulators of cell cycle progression (39). In addition, Plks have a role in the protection against cellular stress through the transcription factor HSF1 (40). Recently it was proposed that an intricate tradeoff between lifespan and cancer results from opposing effects of enzymes regulating FoxO and p53 activity (41). Plks are known to inhibit p53 transcriptional activity (42), so our results raise the possibility that Plks mediate the common but opposing regulators of p53 and FoxO. Interestingly, FoxOs are necessary in the completion of the cell cycle, which is partly mediated by cell cycle dependent activation of Plk expression by FOXO3a (22). Our results show that Drosophila dFoxO is regulated by Polo, suggesting the existence of a positive feedback mechanism that has a role in achieving periodic M-phase gene expression and proper cell cycle exit.
dFoxO localization was affected by eight modulators; however, band shifts demonstrated that none of these proteins phosphorylated dFoxO in the three conserved Ser/Thr amino acids known to regulate nuclear/cytoplasmic status through AKT (6). This observation raises the possibility that some of the newly identified dFoxO regulators could affect dFoxO nuclear/cytoplasmic localization by phosphorylating dFoxO in additional residues that do not alter its electrophoretic mobility, or that dFoxO regulation by these proteins is indirect. Further studies will be needed to clarify this point.
In summary, we have identified 21 dFoxO modulators. Our results underscore the complexity underlying dFoxO regulation and establish dFoxO as a transcription factor controlled exquisitely by an intricate network of kinases and phosphatases achieving a perfect balance of activity. This balance ensures the correct execution of key cellular processes in metabolism, response to stress, and life span.
Materials and Methods
Plasmid Constructs.
We used the following plasmid constructs: pMt-dFoxO WT-V5 (6), pGL3-dInR (6), and pGL-InRprom (11). Clone GH03188 containing full-length PKC53E was cloned into pMtV5-HisA (Invitrogen). The V5 tag was replaced to HA by PCR. pGL3–4xFRE-EGFP plasmid was produced by subcloning the EGFP ORF into the pGL3–4xFRE vector (6). The RFP ORF was cloned into the pMtV5-HisA giving the pMt-RFP-V5 vector. To construct the pcDNA-FOXO3a-V5 plasmid human FOXO3a was amplified by PCR and ligated to pcDNA-V5-His (Invitrogen). All constructs were confirmed by sequencing. The plasmids expressing the mammalian kinases are identified in Table S2.
Cell Culture, RNAi, and Transfection.
Drosophila S2 cells were maintained, treated with dsRNA, and transfected in M3 medium (Sigma) supplemented with insect medium supplement (Sigma), 2% FBS, penicillin, and streptomycin. We used 5 μg/ml human insulin, 20 μM MG-132, and 20 mM NH4Cl. The dsRNA library of kinases and phosphatases was constructed according to the list of genes provided in ref. 23. Primer sequences used to produce the dsRNA molecules are available upon request. A T7 promoter sequence was added and in vitro transcription was performed with T7 Megascript (Ambion). In the secondary screen, before the dsRNA treatment, cells were diluted to 1 × 107/ml. One hundred microliters of the suspension was mixed with 10 μg of the dsRNA and incubated for 30 min. Subsequently, 900 μl of fresh medium was added and the cells were then grown for 1 extra day before transfection. S2 cell transfections were performed with Effectene transfection reagent (Qiagen). After 5 h, 600 μM CuSO4 was added. The cells were grown further for 48 h, and luciferase activity was measured (Promega). Luciferase values were normalized to the total protein content of the lysates measured by Bradford reagent (Bio-Rad). Human embryonic kidney cells (HEK293) or mouse hepatoma cells (HEPA1–6) were transfected with Fugene HD reagent (Roche) and 100 ng of each expression plasmid and 50 ng of reporter plasmid per well. Three days after transfection, luciferase activity was measured. For RNAi experiments, plasmids expressing 29-mer hairpin RNAs (HuSH; Origene) were transfected as described above. The antibodies used in this study were anti-Akt, anti-Aktser505 (Cell Signaling Technology), anti-α-tubulin (Sigma), anti-V5 (Invitrogen), anti-HA (Covance Research Products), anti-FLAG (Sigma), or anti-dFoxO (6). The secondary antibodies were anti-mouse-HRP, anti-rabbit-HRP (Upstate Biotechnology), or anti-mouse Alexa Fluor 488 (Invitrogen).
High-Throughput Microscopy.
Cells were attached to 0.5 mg/ml Concavalin A (in dH2O)-treated optical Packard 96-well view plates, fixed with 3.7% formaldehyde and scanned for their EGFP and RFP intensity, using a Cellomics Arrayscan 4.5 system. In localization studies, the cells were stained with anti-V5 (Invitrogen) and secondary anti-mouse Alexa Fluor 488 (Invitrogen) and stained by DAPI. The cells were identified by DAPI and the intensity of nuclear and cytoplasmic Alexa Fluor 488 was measured from 5,000–10,000 cells, depending on the cell density and transfection efficiency of the given treatment. The exact programs used by the Cellomics Arrayscan microscope are available upon request.
Western Blot, Determination of dFoxO Protein Levels, and Quantitative RT-PCR.
The cell lysates used for the luciferase measurements were run on 8% SDS/PAGE and blotted against anti-V5 (Invitrogen) for transfected dFoxO and anti-α-tubulin (Sigma) followed by secondary anti-mouse-HRP (Upstate). The band intensities were quantified by the Las-3000 CCD camera (Fujifilm). The relative abundance of the transfected dFoxO was obtained by dividing the dFoxO intensity by the Tubulin intensity. Measurements were done in triplicate. Quantitative RT-PCR was done with total RNA isolated from cells by RNeasy RNA extraction kit (Qiagen), treated with DNaseI (Promega) and converted to cDNA by M-MuLV reverse transcriptase (Fermentas). Quantification was performed using the SYBR green methodology in the ABI Prism 7000 sequence detection platform (Applied Biosystems). The results were analyzed by the comparative CT method and normalized to Drosophila actin or human β-actin genes. All of the qPCR primers are available on request. Statistical significance for all studies was calculated by Student's t test. All of the experiments were done in triplicate, and error bars represent SD.
Supplementary Material
Acknowledgments.
We thank Minna Taipale for providing the RNAi library and the Molecular Imaging Unit staff in the University of Helsinki and Taneli Häme for their assistance; Mikko Frilander, Osamu Shimmi, Linda Partridge, Henri Jasper, and the reviewers for their critical suggestions; and F. Sakane, M-A. de Matteis, K. Kohno, T. Pawson, S. Dias, G.G. Gundersen, Y. Kanaho and M. Cobb for sending constructs. This work was supported by the Helsinki Graduate School in Biotechnology and Molecular Biology (J.M.) and grants from the Finnish Diabetes Association, Novo Nordisk, and the Juselius Foundation (to O.P.) and Academy of Finland (to J.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803022105/DCSupplemental.
References
- 1.Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- 2.Borkhardt A, et al. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23) Oncogene. 1997;14:195–202. doi: 10.1038/sj.onc.1200814. [DOI] [PubMed] [Google Scholar]
- 3.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 4.Biggs WH, III, Cavenee WK, Arden KC. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm Genome. 2001;12:416–425. doi: 10.1007/s003350020002. [DOI] [PubMed] [Google Scholar]
- 5.Junger MA, et al. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab. 2005;16:183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Puig O, Tjian R. Nutrient availability and growth: Regulation of insulin signaling by dFOXO/FOXO1. Cell Cycle. 2006;5:503–505. doi: 10.4161/cc.5.5.2501. [DOI] [PubMed] [Google Scholar]
- 9.Schmoll D, et al. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 10.Nakae J, Kitamura T, Silver DL, Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest. 2001;108:1359–1367. doi: 10.1172/JCI12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puig O, Tjian R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005;19:2435–2446. doi: 10.1101/gad.1340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7:21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 14.Biggs WH, III, Meisenhelder J, Hunter T, Cavenee WK. Arden KC Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 16.Kops GJ, et al. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 17.Tang ED, Nunez G, Barr FG, Guan KL. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- 18.Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci USA. 2003;100:11285–11290. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plas DR, Thompson CB. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem. 2003;278:12361–12366. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- 20.Luong N, et al. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Tsai WC, Bhattacharyya N, Han LY, Hanover JA, Rechler MM. Insulin inhibition of transcription stimulated by the forkhead protein Foxo1 is not solely due to nuclear exclusion. Endocrinology. 2003;144:5615–5622. doi: 10.1210/en.2003-0481. [DOI] [PubMed] [Google Scholar]
- 22.Alvarez B, Martinez A, Burgering BM, Carrera AC. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature. 2001;413:744–747. doi: 10.1038/35099574. [DOI] [PubMed] [Google Scholar]
- 23.Morrison DK, Murakami MS, Cleghon V. Protein kinases and phosphatases in the Drosophila genome. J Cell Biol. 2000;150:F57–F62. doi: 10.1083/jcb.150.2.f57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 25.Lehtinen MK, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 26.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 27.Woods YL, et al. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem J. 2001;355:597–607. doi: 10.1042/bj3550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Idris I, Gray S, Donnelly R. Protein kinase C activation: Isozyme-specific effects on metabolism and cardiovascular complications in diabetes. Diabetologia. 2001;44:659–673. doi: 10.1007/s001250051675. [DOI] [PubMed] [Google Scholar]
- 29.Marr MT, D'Alessio JA, Puig O, Tjian R. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 2007;21:175–183. doi: 10.1101/gad.1506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampson SR, Cooper DR. Specific protein kinase C isoforms as transducers and modulators of insulin signaling. Mol Genet Metab. 2006;89:32–47. doi: 10.1016/j.ymgme.2006.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farese RV, Sajan MP, Standaert ML. Atypical protein kinase C in insulin action and insulin resistance. Biochem Soc Trans. 2005;33:350–353. doi: 10.1042/BST0330350. [DOI] [PubMed] [Google Scholar]
- 32.Leitges M, et al. Knockout of PKC alpha enhances insulin signaling through PI3K. Mol Endocrinol. 2002;16:847–858. doi: 10.1210/mend.16.4.0809. [DOI] [PubMed] [Google Scholar]
- 33.Yu C, et al. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J Biol Chem. 2002;277:50230–50236. doi: 10.1074/jbc.M200958200. [DOI] [PubMed] [Google Scholar]
- 34.Dey D, Basu D, Roy SS, Bandyopadhyay A, Bhattacharya S. Involvement of novel PKC isoforms in FFA induced defects in insulin signaling. Mol Cell Endocrinol. 2006;246:60–64. doi: 10.1016/j.mce.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 35.Shieh BH, Parker L, Popescu D. Protein kinase C (PKC) isoforms in Drosophila. J Biochem. 2002;132:523–527. doi: 10.1093/oxfordjournals.jbchem.a003252. [DOI] [PubMed] [Google Scholar]
- 36.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 37.Nikoulina SE, et al. Potential role of glycogen synthase kinase-3 in skeletal muscle insulin resistance of type 2 diabetes. Diabetes. 2000;49:263–271. doi: 10.2337/diabetes.49.2.263. [DOI] [PubMed] [Google Scholar]
- 38.Lochhead PA, Coghlan M, Rice SQ, Sutherland C. Inhibition of GSK-3 selectively reduces glucose-6-phosphatase and phosphatase and phosphoenolypyruvate carboxykinase gene expression. Diabetes. 2001;50:937–946. doi: 10.2337/diabetes.50.5.937. [DOI] [PubMed] [Google Scholar]
- 39.Martin BT, Strebhardt K. Polo-like kinase 1: Target and regulator of transcriptional control. Cell Cycle. 2006;5:2881–2885. doi: 10.4161/cc.5.24.3538. [DOI] [PubMed] [Google Scholar]
- 40.Kim SA, Yoon JH, Lee SH, Ahn SG. Polo-like kinase 1 phosphorylates heat shock transcription factor 1 and mediates its nuclear translocation during heat stress. J Biol Chem. 2005;280:12653–12657. doi: 10.1074/jbc.M411908200. [DOI] [PubMed] [Google Scholar]
- 41.van der HA, Burgering BM. Stressing the role of FoxO proteins in lifespan and disease. Nat Rev Mol Cell Biol. 2007;8:440–450. doi: 10.1038/nrm2190. [DOI] [PubMed] [Google Scholar]
- 42.Ando K, et al. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem. 2004;279:25549–25561. doi: 10.1074/jbc.M314182200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.