Abstract
The unfolded protein response (UPR) controls the levels of molecular chaperones and enzymes involved in protein folding in the endoplasmic reticulum (ER). We recently isolated ATF6 as a candidate for mammalian UPR-specific transcription factor. We report here that ATF6 constitutively expressed as a 90-kDa protein (p90ATF6) is directly converted to a 50-kDa protein (p50ATF6) in ER-stressed cells. Furthermore, we showed that the most important consequence of this conversion was altered subcellular localization; p90ATF6 is embedded in the ER, whereas p50ATF6 is a nuclear protein. p90ATF6 is a type II transmembrane glycoprotein with a hydrophobic stretch in the middle of the molecule. Thus, the N-terminal half containing a basic leucine zipper motif is oriented facing the cytoplasm. Full-length ATF6 as well as its C-terminal deletion mutant carrying the transmembrane domain is localized in the ER when transfected. In contrast, mutant ATF6 representing the cytoplasmic region translocates into the nucleus and activates transcription of the endogenous GRP78/BiP gene. We propose that ER stress-induced proteolysis of membrane-bound p90ATF6 releases soluble p50ATF6, leading to induced transcription in the nucleus. Unlike yeast UPR, mammalian UPR appears to use a system similar to that reported for cholesterol homeostasis.
INTRODUCTION
The endoplasmic reticulum (ER) provides the optimal environment for proper folding and assembly of newly synthesized proteins destined for the secretory pathway (Gething and Sambrook, 1992; Helenius et al., 1992; Huppa and Ploegh, 1998). Eukaryotic cells from yeast to human have developed specific and tight communication systems between the ER and cell nucleus to gear the folding capacity in the ER to the requirements within the ER. Thus, accumulation of unfolded proteins under so-called “ER stress” conditions leads to induction of transcription of genes encoding molecular chaperones and folding enzymes localized in the ER (Lee, 1987; Kozutsumi et al., 1988; Normington et al., 1989; Rose et al., 1989). This transcriptional induction process coupled with intracellular signaling from the ER to the nucleus is called the unfolded protein response (UPR) (McMillan et al., 1994; Shamu et al., 1994).
The most proximal event in the UPR is the sensing step of ER stress, which is considered to be mediated by Ire1p/Ern1p, a transmembrane protein kinase in the ER originally isolated by genetic screening in Saccharomyces cerevisiae (Cox et al., 1993; Mori et al., 1993). Although the detailed mechanism is unknown, Ire1p is activated via oligomerization and autophosphorylation upon accumulation of unfolded proteins in the ER (Shamu and Walter, 1996; Welihinda and Kaufman, 1996). Ire1p is likely to be negatively regulated by Ptc2p, a serine/threonine-specific protein phosphatase (Welihinda et al., 1998). Recent cloning of the mammalian homologue of yeast Ire1p revealed the presence of at least two different molecular species in mammalian cells (Tirasophon et al., 1998; Wang et al., 1998). Overexpression of either type of mammalian Ire1p constitutively activated transcription of ER chaperone genes as in the case of yeast Ire1p, suggesting similarity in the sensing system between yeast and mammals. Interestingly, mammalian ER contains one more transmembrane protein kinase called PEK/PERK, the lumenal domain of which shows significant homology to that of Ire1p (Shi et al., 1998; Harding et al., 1999). PEK/PERK is capable of phosphorylating the α subunit of eukaryotic translation initiation factor 2 and thus appears to be responsible for ER stress-induced translational attenuation (Prostko et al., 1992; reviewed by Kaufman, 1999).
The most distal step in the UPR is the transcription step that depends on the specific interaction of a trans-acting factor with a cis-acting element present in the promoter regions of UPR target genes. In S. cerevisiae, the cis-acting unfolded protein response element (UPRE) was originally identified as a sequence necessary and sufficient for induction of the KAR2 gene encoding the yeast homologue of a major ER chaperone, GRP78/BiP (Mori et al., 1992; Kohno et al., 1993). We recently showed that KAR2 UPRE contains an E box-like partially palindromic sequence separated by a spacer of one nucleotide (CAGCGTG) that is essential for its function (Mori et al., 1996). Subsequent characterization of UPRE sequences responsible for induction of other ER stress-responsive genes led us to propose that this unique feature of UPRE explains why only a specific set of ER proteins are induced in yeast UPR (Mori et al., 1998).
In contrast to yeast UPR, multiple cis-acting elements appeared to be involved in the mammalian UPR (Wooden et al., 1991). Recently, however, we and others independently found that a unique sequence with a consensus of CCAAT-N9-CCACG is commonly present in promoter regions of almost all ER stress-responsive mammalian genes (Yoshida et al., 1998; Roy and Lee, 1999). This novel cis-acting element was designated the ER stress response element (ERSE). By extensive mutational analysis, we demonstrated that ERSE is necessary and sufficient for induction of at least three major ER chaperones (GRP78, GRP94, and calreticulin) (Yoshida et al., 1998). The CCAAT portion of ERSE was confirmed to be a binding site for the general transcription factor NF-Y/CBF as shown previously (Roy and Lee, 1995; Roy et al., 1996). Thus, we proposed that the CCACG portion of ERSE exactly 9 bp downstream of the CCAAT portion provides specificity of the mammalian UPR (Yoshida et al., 1998). The CCACG may correspond to a half site of the palindromic sequence found in the yeast UPRE.
In yeast, Hac1p/Ern4p was identified as the UPR-specific transcription factor (Cox and Walter, 1996; Mori et al., 1996; Nikawa et al., 1996). Hac1p contains a basic leucine zipper (bZIP) motif and specifically binds to the E box-like palindromic sequence in UPRE. Yeast cells lacking Hac1p show the same phenotype as those lacking Ire1p; cells are unable to induce transcription of ER chaperone genes in response to ER stress. Subsequent analysis conducted in Walter’s and our laboratories revealed an unexpected connection between the most proximal event in the ER and the most distal event in the nucleus, namely the unconventional splicing system for HAC1 mRNA. HAC1 mRNA is constitutively expressed, but its translation is tightly blocked because of the presence of an intron of 252 nucleotides within the Hac1p-coding region. The HAC1 intron is specifically removed by a splicing event that is activated by signaling from the ER. Spliced mRNA is translated, and the Hac1p thus synthesized activates transcription of UPR target genes through the UPRE (Cox and Walter, 1996; Chapman and Walter, 1997; Kawahara et al., 1997, 1998). Enzymes responsible for this unconventional mRNA splicing were identified by Walter and colleagues. The splicing reaction is initiated by Ire1p, a sensor molecule of ER stress by itself; both 5′- and 3′ splice sites are cleaved by the action of the C-terminal endonuclease domain of Ire1p, which is postulated to be activated upon ER stress (Sidrauski and Walter, 1997). The splicing reaction is completed by Rlg1p (Sidrauski et al., 1996), a tRNA ligase located in the nucleus (Clark and Abelson, 1987). This unique system allows yeast cells to produce transcription factor Hac1p only when they sense ER stress and require increased amounts of ER chaperones to cope with unfolded proteins accumulated in the ER (reviewed by Sidrauski et al., 1998).
Recently, we screened for putative human ERSE-binding proteins and obtained the bZIP protein ATF6 as a candidate (Yoshida et al., 1998). The amino acid sequence of the ATF6 basic region shows significant similarity with that of yeast Hac1p. In contrast to Hac1p, however, ATF6 is constitutively expressed as a 90-kDa protein, and ATF6 mRNA is not spliced in response to ER stress (Zhu et al., 1997; Yoshida et al., 1998). Here, we analyzed the mechanism of activation of ATF6 by ER stress and revealed an important step that connects the event in the ER with that in the nucleus.
MATERIALS AND METHODS
Plasmid Construction
Recombinant DNA techniques were performed according to standard procedures (Sambrook et al., 1989). The ATF6 expression plasmid pCGN-ATF6 (Zhu et al., 1997) was kindly provided by Dr. R. Prywes (Columbia University, New York, NY). This plasmid contains the full-length ATF6 cDNA at the XbaI site of the mammalian expression vector pCGN and thus expresses ATF6 protein tagged with the influenza virus hemagglutinin (HA) epitope at the N terminus under the control of the cytomegalovirus promoter [referred to here as pCGN-ATF6 (670)]. pCGN-ATF6 (402), pCGN-ATF6 (373), and pCGN-ATF6 (366) encoding truncated forms of ATF6 were constructed by PCR-mediated amplification of the region corresponding to amino acids 1–402, 1–373, and 1–366 of ATF6, respectively, together with a stop codon (TAG) followed by insertion of the resultant fragments into the XbaI site of pCGN after their sequences had been confirmed.
Cell Culture and Transient Transfection
HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin sulfate. Cells were maintained in a 5% CO2 incubator at 37°C. Transfection was carried out by the standard calcium phosphate method (Sambrook et al., 1989) as described previously (Yoshida et al., 1998) with some modifications. Briefly, HeLa cells plated at ∼10% confluency 1 d before transfection were incubated with calcium phosphate–DNA complex for 16 h at 37°C. After washing with PBS, cells were cultured in fresh medium for 24 h before extraction for immunoblotting or fixation for immunofluorescence analysis unless otherwise specified. Transfection efficiency depended on the amount of DNA used: usually ∼5% with 1 μg of DNA and ∼50% with 10 μg of DNA. To induce UPR, cells were treated with 2 μg/ml tunicamycin (Sigma, St. Louis, MO), 300 nM thapsigargin (Sigma), or 1 mM dithiothreitol for various periods as indicated.
Purification of Anti-ATF6 Antibody
Rabbit anti-B03N antiserum raised against the N-terminal region of ATF6 (amino acids 6–307) fused to Escherichia coli maltose-binding protein (Yoshida et al., 1998) was purified by removing materials bound to CH-Sepharose 4B (Amersham Pharmacia Biotech, Buckinghamshire, UK) on which soluble proteins extracted from E. coli cells producing large amounts of maltose-binding protein from the plasmid pMAL-c2 (New England BioLabs, Beverly, MA) had been immobilized. The flow-through fraction highly specific to ATF6 (see Figure 1) was used as anti-ATF6 antibody.
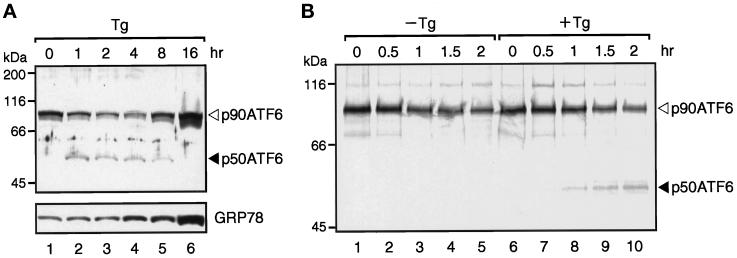
Figure 1.
Direct conversion of p90ATF6 to p50ATF6 in thapsigargin-treated HeLa cells. (A) Immunoblotting analysis of ATF6. HeLa cells cultured in 60-mm dishes until 60% confluency were incubated in the presence of 300 nM thapsigargin (Tg) for the indicated periods. Cells were washed with PBS, scraped with a rubber policeman, and lysed in 100 μl of 1× Laemmli’s SDS sample buffer. After boiling for 5 min, 5-μl aliquots of each sample were subjected to SDS-PAGE (10% gel) and analyzed by immunoblotting with anti-ATF6 antibody or anti-KDEL antibody, which recognizes GRP78. The positions of p90ATF6 and p50ATF6 are indicated by the open and closed arrowheads, respectively. The positions of prestained SDS-PAGE molecular weight standards (Bio-Rad, Hercules, CA) are also shown. (B) Pulse–chase analysis of ATF6. HeLa cells cultured in 60-mm dishes were pulse labeled for 30 min with [35S]methionine and [35S]cysteine and then chased for the indicated periods in the absence (−Tg) or presence (+Tg) of 300 nM thapsigargin as described in MATERIALS AND METHODS. It should be noted that thapsigargin was not added during the pulse labeling of the cells. 35S-Labeled proteins were extracted and immunoprecipitated with anti-ATF6 antibody, and the immunoprecipitates were subjected to SDS-PAGE (7.5% gel). Radioactivities of 35S-labeled p90ATF6 and p50ATF6 were visualized using a BAS-2000 BioImaging Analyzer (Fuji Photo film, Stanford, CT).
Immunoblotting
Immunoblotting analysis was carried out according to the standard procedure (Sambrook et al., 1989) as described previously (Yoshida et al., 1998) using an enhanced chemiluminescence Western blotting detection system kit (Amersham Pharmacia Biotech). Mouse anti-KDEL monoclonal antibody (clone 10C3), mouse anti-HSP70 monoclonal antibody (clone C92F3A-5), and rabbit antisera against the N or C terminus of calnexin were obtained from StressGen Biotechnologies (Victoria, British Columbia, Canada). Rabbit anti-HA epitope polyclonal antibody (Y-11) and goat anti-lamin B polyclonal antibody (M-20) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Pulse–Chase Analysis of ATF6
HeLa cells cultured in 60-mm dishes until ∼80% confluency were incubated for 20 min in methionine- and cysteine-free Dulbecco’s modified Eagle’s medium supplemented with 2 mM glutamine, antibiotics, and 10% dialyzed fetal calf serum. Cells were then pulse labeled for 30 min using 0.5 mCi (18.5 MBq)/plate EXPRE35S35S protein labeling mix (DuPont, Wilmington, DE) dissolved in 1 ml of the above medium. To chase 35S-labeled proteins, cells were washed with fresh complete medium three times and incubated in fresh complete medium for various periods. After removal of medium, cells were lysed by incubation for 10 min on ice in 0.1 ml of lysis buffer (1× Tris-buffered saline containing 1% SDS, 1 mM PMSF, and 5 μg/ml leupeptin). The lysates were boiled for 5 min and, after addition of 0.4 ml of dilution buffer (1× Tris-buffered saline containing 2% Triton X-100), clarified by centrifugation at 15,000 × g for 10 min. The supernatant was incubated with 50 μl of normal rabbit serum (Sigma) for 3 h at 4°C, and the mixture was rotated overnight at 4°C after addition of 50 μl of 50% protein A-Sepharose 4 Fast Flow (Amersham Pharmacia Biotech). The mixture was clarified by brief centrifugation, and 10 μl of anti-ATF6 antibody were added to the resulting supernatant. After standing for 3 h at 4°C, 50 μl of 50% protein A-Sepharose 4 Fast Flow were added, and the mixture was rotated for 2 h at 4°C. The resin was then washed as described by Franzusoff et al. (1991), and the immunoprecipitates were subjected to SDS-PAGE.
Indirect Immunofluorescence
HeLa cells were grown directly on slide glasses in 90-mm dishes. Cells were fixed with PLP solution (2% p-formaldehyde, 10 mM sodium metaperiodate, and 75 mM l-lysine in PBS) for 10 min at room temperature, permeabilized with ice-cold acetone for 10 min, and then reacted with various primary antibodies for 1 h at 37°C. Rabbit anti-ATF6 antibody was diluted by 50-fold with 1% BSA-TBST solution (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% Tween 20, and 1% BSA) before use. Rabbit anti-HA epitope antibody and mouse anti-KDEL antibody were diluted with 1% BSA-TBST solution to concentrations of 4 and 20 μg/ml, respectively. Primary antibodies were visualized by incubation for 1 h at 37°C with either 10 μg/ml FITC-conjugated goat anti-rabbit immunoglobulin G antibody (Vector Laboratories, Burlingame, CA) or 50 μg/ml rhodamine-conjugated goat anti-mouse immunoglobulin G antibody (ICN Pharmaceuticals, Aurora, OH). The cells were counterstained with 0.3 μg/ml DAPI in PBS-glycerol (1:9). Photographs were taken using a 465- to 495-nm excitation filter and a 515- to 555-nm emission filter for fluorescein isothiocyanate and a 525- to 540-nm excitation filter and a 605- to 655-nm emission filter for rhodamine to avoid mutual interference.
RESULTS
ER Stress Induces Processing of ATF6
We and others have shown that ATF6 is constitutively expressed in HeLa cells as a 90-kDa protein (p90ATF6) (Zhu et al., 1997; Yoshida et al., 1998). Interestingly, however, we found that the p90ATF6 level decreased, and instead a new band of 50 kDa (p50ATF6) appeared in ER-stressed cells before the induction of GRP78, a major target protein of the mammalian UPR (Yoshida et al., 1998). As the anti-B03N antiserum used in the previous study reacted with multiple proteins present in HeLa cell extracts, we purified it as described in MATERIALS AND METHODS. The purified antibody recognized p90ATF6 almost exclusively on immunoblotting of extracts of unstressed HeLa cells (Figure 1A, lane 1). We thus used this as anti-ATF6 antibody, which should recognize the N-terminal region of ATF6 (amino acids 6–307, indicated by the thick line in Figure 4B).
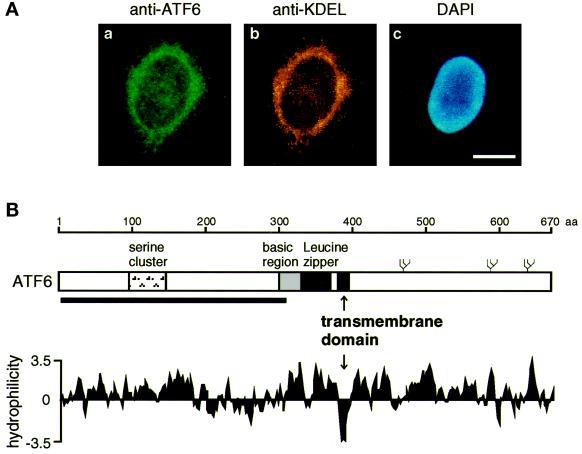
Figure 4.
Identification of an internal hydrophobic stretch that anchors ATF6 in the ER membrane. (A) Indirect immunofluorescence analysis. Unstressed HeLa cells were fixed and stained with anti-ATF6 antibody (a), anti-KDEL antibody (b), or DAPI (c) as described in MATERIALS AND METHODS. Bar, 10 μm. (B) Schematic structure of ATF6 consisting of 670 amino acids. The positions of the serine cluster, basic region, and leucine zipper (Zhu et al., 1997; Yoshida et al., 1998) as well as the transmembrane domain identified in this report are indicated. The thick line below the sequence represents the region (amino acids 6–307) fused with E. coli maltose-binding protein to raise anti-ATF6 antibody (Yoshida et al., 1998). Three potential glycosylation sites are also shown schematically (ψ). The hydropathy index was calculated by the method of Kyte and Doolittle (1982).
We reexamined the time course of the conversion of p90ATF6 to p50ATF6 using the purified antibody in HeLa cells treated with thapsigargin, which causes ER stress by inhibiting the ER Ca2+-ATPase (Li et al., 1993). Consistent with our previous report (Yoshida et al., 1998), p90ATF6 level decreased, and p50ATF6 appeared from 1 h after thapsigargin treatment, whereas an increase in the level of GRP78 was detected from 4 h (Figure 1A). To examine whether p90ATF6 was directly converted to p50ATF6 upon ER stress, we performed pulse–chase experiments (Figure 1B). ATF6 was immunoprecipitated from HeLa cells pulse labeled for 30 min with [35S]methionine and [35S]cysteine and then chased for various periods. In the absence of thapsigargin in the chase medium, p90ATF6 level decreased with a half-life of ∼2 h, and no band corresponding to p50ATF6 was detected (Figure 1B, lanes 1–5). In the presence of thapsigargin, p50ATF6 appeared after the chase (Figure 1B, lanes 6–10), suggesting that ATF6 is synthesized as a precursor protein (p90ATF6) in unstressed cells and processed into a mature form (p50ATF6) in response to ER stress.
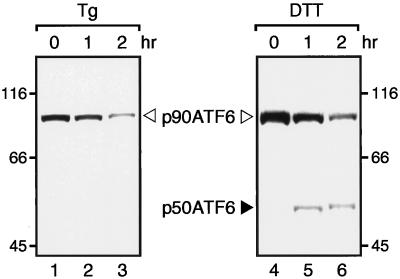
To further confirm whether ER stress did not induce de novo synthesis of p50ATF6, we examined the effects of blocking new protein synthesis on the conversion of p90ATF6 to p50ATF6. It was recently shown that PERK, a transmembrane protein kinase involved in ER stress-induced translational attenuation, was activated by thapsigargin treatment of COS-1 cells even in the absence of protein synthesis (Harding et al., 1999). In marked contrast, p50ATF6 was not produced when HeLa cells (Figure 2, lanes 1–3) or COS-1 cells (our unpublished observation) were pretreated for 30 min with cycloheximide, an inhibitor of protein synthesis, before thapsigargin treatment. The reason for the difference is currently unknown. Importantly, however, we found that the conversion of p90ATF6 to p50ATF6 could occur in the absence of protein synthesis under a certain condition such as dithiothreitol treatment, which disrupts disulfide bonding and thus malfolds proteins in the ER directly (Figure 2, lanes 4–6). All of the above results strongly indicate that the appearance of p50ATF6 in ER-stressed cells is due to proteolytic cleavage of preexisting p90ATF6.
Figure 2.
Conversion of p90ATF6 to p50ATF6 in the absence of new protein synthesis in dithiothreitol-treated HeLa cells. HeLa cells cultured in 60-mm dishes until 80% confluency were pretreated for 30 min with 10 μg/ml cycloheximide and then treated for the indicated periods with 300 nM thapsigargin (Tg) or 1 mM dithiothreitol (DTT) in the presence of 10 μg/ml cycloheximide. Immunoblotting analysis of ATF6 was carried out essentially as described in the legend to Figure 1A.
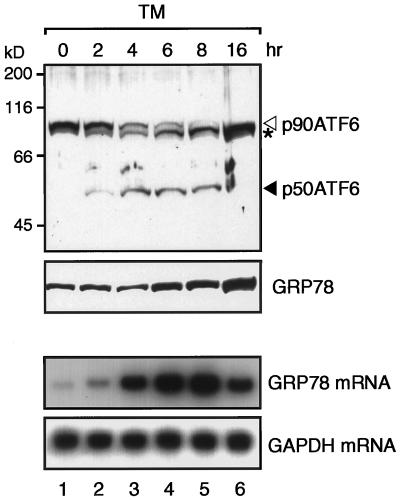
We also reexamined the time course of the processing of ATF6 in HeLa cells treated with tunicamycin, which causes ER stress by inhibiting N-glycosylation of newly synthesized proteins (Lee, 1987; Kozutsumi et al., 1988). Consistent with our previous report (Yoshida et al., 1998), p90ATF6 level decreased, and p50ATF6 appeared from 2 h after tunicamycin treatment, whereas an increase in GRP78 level was detected from 6 h (Figure 3). Although the processing of ATF6 was significantly slower in tunicamycin-treated cells than that in thapsigargin-treated cells, appearance of p50ATF6 accompanied the increase in level of GRP78 mRNA (Figure 3), suggesting that the proteolytic conversion of p90ATF6 to p50ATF6 is a key regulatory step in mammalian UPR.
Figure 3.
Processing of ATF6 in tunicamycin-treated HeLa cells. HeLa cells cultured in 100-mm dishes until 60% confluency were incubated in the presence of 2 μg/ml tunicamycin (TM) for the indicated periods. Proteins were extracted and analyzed by immunoblotting as described in the legend to Figure 1A. The asterisk denotes an unglycosylated form of p90ATF6. Total RNA was also extracted and analyzed by Northern blot hybridization using radiolabeled probes specific to GRP78 or GAPDH as described previously (Yoshida et al., 1998).
ATF6 Contains a Hydrophobic Stretch That Anchors in the ER Membrane
The experiments shown in Figures 1–3 revealed the production of a novel band migrating slightly faster than p90ATF6 in addition to p50ATF6 in tunicamycin-treated cells but not in thapsigargin- or dithiothreitol-treated cells, suggesting that p90ATF6 might be glycosylated and thus associated with the ER. In fact, when unstressed HeLa cells were examined by indirect immunofluorescence analysis (Figure 4A), fine reticular structures surrounding the nucleus were observed using purified anti-ATF6 antibody, and the staining pattern was very similar to that obtained with anti-KDEL antibody, which recognizes two major ER chaperones (GRP78 and GRP94) in HeLa cells. Furthermore, hydropathy analysis identified a hydrophobic stretch of 21 amino acids (amino acids 378–398) near the center of the ATF6 molecule, which is long enough to span the membrane once (Figure 4B). These results suggested that p90ATF6 is a transmembrane protein in the ER.
p90ATF6 Is a Type II Transmembrane Glycoprotein in the ER
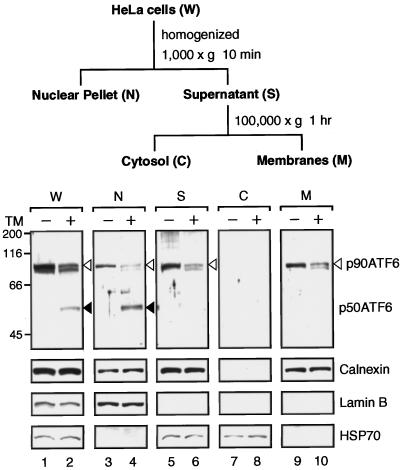
We conducted fractionation experiments using untreated HeLa cells and those treated with tunicamycin for 4 h as starting materials to monitor recovery of p90ATF6 and p50ATF6, respectively (Figure 5). After the first low-speed centrifugation of the homogenate, p50ATF6 was recovered exclusively in the nuclear pellet similarly to the nuclear protein lamin B (Figure 5, lane 4), whereas the majority of p90ATF6 remained in the supernatant (Figure 5, lane 5). After the second high-speed centrifugation, all p90ATF6 was recovered in the membrane fraction (Figure 5, lane 9). The distribution pattern of p90ATF6 was almost identical to that of calnexin, a transmembrane-type chaperone localized in the ER (Wada et al., 1991), but completely different from those of lamin B (Moir et al., 1995) and the cytosolic protein HSP70 (Welch and Suhan, 1986), revealing that p90ATF6 is indeed associated with membranes. It should be noted that p90ATF6 remaining in tunicamycin-treated cells as well as a protein band migrating slightly faster than p90ATF6 (lane 2) were distributed in each fraction similarly to p90ATF6 in untreated cells.
Figure 5.
Distribution of p90ATF6 and p50ATF6 in fractions of HeLa cells. HeLa cells cultured in 175-cm2 flasks until 80% confluency were incubated in the absence (−) or presence (+) of 2 μg/ml tunicamycin (TM) for 4 h. Cells were harvested, disrupted using a Dounce-type homogenizer, and then centrifuged at 1000 × g for 10 min to obtain the nuclear pellet (N) essentially as described by Dignam et al. (1983). The resulting supernatant (S) was further centrifuged at 100,000 × g for 1 h to separate the soluble cytosolic fraction (C) from insoluble membrane fraction (M). Aliquots of the indicated fraction as well as unfractionated HeLa cells (whole; W) corresponding to 0.5 × 105 cells were subjected to SDS-PAGE (10% gel) and analyzed by immunoblotting with anti-ATF6 antibody or various other antibodies as indicated. The positions of p90ATF6 and p50ATF6 are marked as in Figure 1.
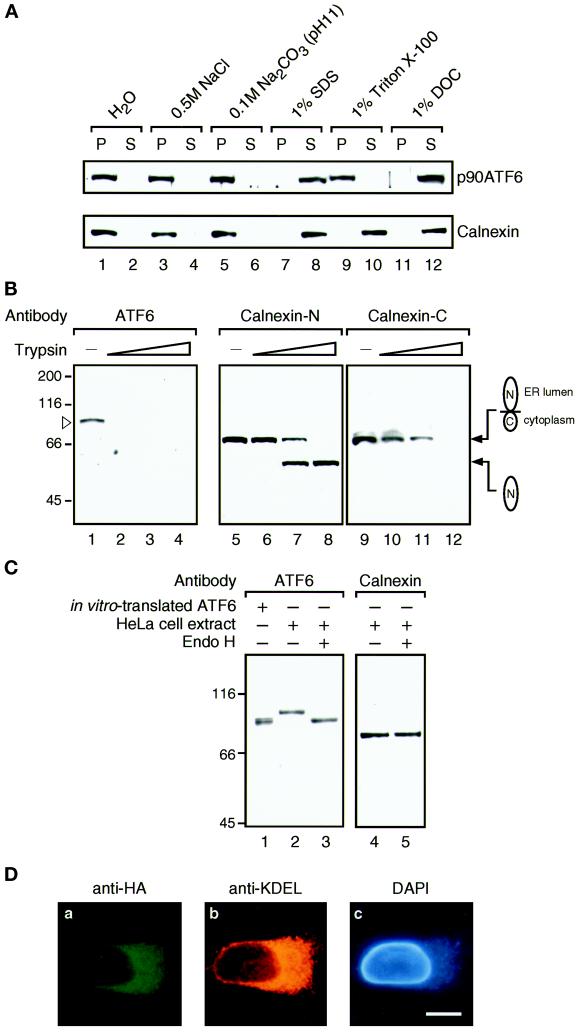
To determine whether p90ATF6 is a peripheral or integral membrane protein, differential solubilization experiments were carried out (Figure 6A). The supernatant fraction obtained after low-speed centrifugation of untreated HeLa cell homogenate was treated with various reagents and then centrifuged at high speed. Neither p90ATF6 nor calnexin was released from membranes by 0.5 M NaCl or 0.1 M Na2CO3, pH 11, treatment, which should extract peripheral membrane proteins. In contrast, both proteins were released into the supernatant by detergents such as 1% SDS and 1% sodium deoxycholate, whereas only calnexin was extracted by 1% Triton X-100; Triton X-100 may cause aggregation of p90ATF6 as in the case of yeast Ire1p (Mori et al., 1993). Thus, p90ATF6 is an integral membrane protein.
Figure 6.
p90ATF6 is an integral membrane glycoprotein with its N terminus located in the cytoplasm. (A) Differential solubilization of p90ATF6. The 1000 × g supernatant fraction prepared from unstressed HeLa cells as described in the legend to Figure 5 was mixed with 0.1 vol of one of the following solutions: H2O, 5 M NaCl, 1 M Na2CO3, pH 11, 10% SDS, 10% Triton X-100, or 10% sodium deoxycholate (DOC). After incubation for 15 min at room temperature, mixtures were centrifuged at 100,000 × g for 1 h to separate supernatant (S) from pellet (P), followed by SDS-PAGE (10% gel) and immunoblotting analysis using anti-ATF6 antibody or anti-N terminus of calnexin antibody. (B) Topology of p90ATF6. The 1000 × g supernatant fraction prepared from unstressed HeLa cells (50 μg of proteins) was incubated with increasing amounts of trypsin (0 μg for lanes 1, 5, and 9; 0.1 μg for lanes 2, 6, and 10; 0.3 μg for lanes 3, 7, and 11; and 1.0 μg for lanes 4, 8, and 12) for 15 min at room temperature. Digestion was terminated by addition of an equal volume of 2× Laemmli’s SDS sample buffer followed by boiling for 5 min. Samples were subjected to SDS-PAGE (10% gel) and analyzed by immunoblotting with anti-ATF6 antibody (lanes 1–4), anti-N terminus of calnexin antibody (Calnexin-N; lanes 5–8), or anti-C terminus of calnexin antibody (Calnexin-C; lanes 9–12). The position of p90ATF6 is marked by the open arrowhead. The positions of full-length calnexin and its truncated form lacking the cytoplasmic domain are shown schematically. (C) Glycosylation of p90ATF6. The 1000 × g supernatant fraction prepared from unstressed HeLa cells (2.5 μg of proteins) was boiled in the presence of 1% SDS and 1% 2-mercaptoethanol for 5 min. After addition of 2 volumes of 150 mM sodium citrate buffer, pH 5.5, the samples were incubated for 20 h at 37°C in the absence (lanes 2 and 4) or presence (lanes 3 and 5) of 0.25 mU endoglycosidase H (Endo H) obtained from ICN (Costa Mesa, CA). Samples as well as in vitro–translated ATF6 (lane 1) prepared as described by Yoshida et al. (1998) were subjected to SDS-PAGE (7.5% gel) and analyzed by immunoblotting with anti-ATF6 antibody (lanes 1–3) or anti-N terminus of calnexin antibody (lanes 4 and 5). (D) Indirect immunofluorescence analysis of transfected cells. HeLa cells on slide glasses transiently transfected with pCGN-ATF6 (670) (see Figure 8A for its schematic structure) were fixed and stained with anti-HA epitope antibody (a), anti-KDEL antibody (b), or DAPI (c). Bar, 10 μm.
Next, the orientation of p90ATF6 in membranes was examined by trypsin treatment (Figure 6B). Calnexin, a type I transmembrane protein, was used as a control and digestion of calnexin was monitored using two different antibodies specific to either the N-terminal region present in the ER lumen (Calnexin-N) or the C-terminal region located in the cytoplasm (Calnexin-C). At the concentration of trypsin that decreased the amount of full-length calnexin, anti-Calnexin-N antibody (Figure 6B, lanes 7 and 8) but not anti-Calnexin-C antibody (Figure 6B, lanes 11 and 12) detected a fragment the size of which matched that of the N-terminal region of calnexin, indicating that proteins or segments in the lumen were protected from digestion by trypsin as expected. Under these conditions, p90ATF6 disappeared at the lowest concentration of trypsin used, and no fragments of ∼50 kDa were detected by anti-ATF6 antibody raised against the N-terminal region of ATF6 (Figure 6B, lane 2). These results strongly indicated that the N-terminal domain of ATF6 containing the bZIP region is oriented facing the cytoplasm.
Examination of the amino acid sequence of the lumenal domain of ATF6 indicated the presence of three potential glycosylation sites as schematically presented in Figure 4B. To determine whether p90ATF6 is actually glycosylated, unstressed HeLa cell extracts were denatured and treated with endoglycosidase H followed by immunoblotting analysis (Figure 6C). p90ATF6 treated with endoglycosidase H (Figure 6C, lane 3) migrated faster than untreated p90ATF6 (Figure 6C, lane 2) but migrated at the same position as in vitro–translated full-length ATF6 (Figure 6C, lane 1), whereas the mobility of calnexin was unaffected by endoglycosidase H treatment (Figure 6C, lanes 4 and 5); calnexin contains no potential glycosylation sites. The sensitivity of the carbohydrate moiety to digestion with endoglycosidase H implied that p90ATF6 stays in the ER and is not subjected to retrograde transport from the Golgi apparatus.
We further determined the localization of p90ATF6 after transfection. pCGN-ATF6 (670) expressed full-length ATF6 tagged with the HA epitope at the N terminus (see MATERIALS AND METHODS). Immunofluorescence analysis of transfected HeLa cells using anti-HA epitope antibody revealed that expression of ATF6 (670) was restricted to perinuclear structures, which were also stained with anti-KDEL antibody (Figure 6D, compare a with b and c), demonstrating that p90ATF6 was localized in the ER even when overexpressed. Taken together, we concluded that p90ATF6 is a type II transmembrane glycoprotein localized in the ER.
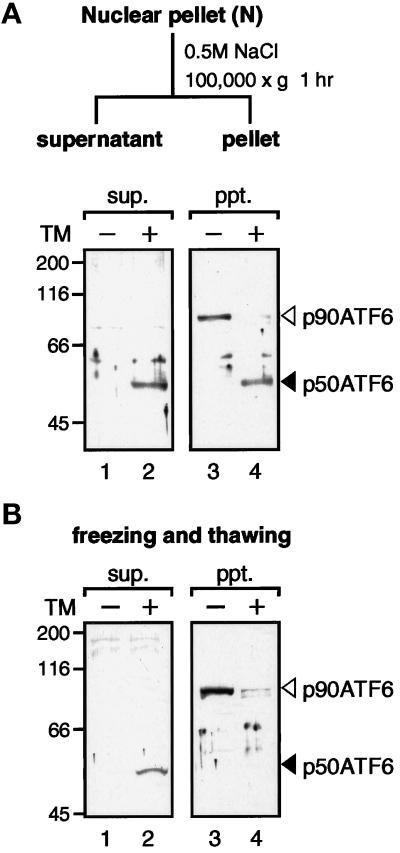
p50ATF6 Is a Soluble Nuclear Protein
In contrast to the tight association of p90ATF6 with ER membranes, p50ATF6 detected in ER-stressed cells was recovered exclusively in the nuclear pellet fraction (Figure 5, lane 4). As a small portion of p90ATF6 was also recovered in the same fraction (Figure 5, lane 3), we further extracted the nuclear pellet with high salt buffer (Figure 7A). As a result, more than half of p50ATF6 was released into the supernatant (Figure 7A, compare lane 2 with lane 4), whereas all p90ATF6 remained in the pellet (Figure 7A, compare lane 1 with lane 3). Furthermore, when HeLa cells were extracted by repeated freezing and thawing (Figure 7B), all p50ATF6 was recovered in the supernatant (Figure 7B, compare lane 2 with lane 4), whereas all p90ATF6 was recovered in the pellet (Figure 7B, compare lane 1 with lane 3). From these results, we concluded that p50ATF6 is a soluble nuclear protein in marked contrast to p90ATF6.
Figure 7.
Solubility of p90ATF6 and p50ATF6. (A) Fractionation of nuclear pellet. The nuclear pellet fraction prepared as described in the legend to Figure 5 was washed with PBS three times and resuspended in nuclear extraction buffer (20 mM HEPES-KOH, pH 7.6, 25% glycerol, 0.5 M NaCl, 1.5 mM MgCl2, 1 mM EDTA, 5 μg/ml pepstatin A, 5 μg/ml leupeptin, and 2 μg/ml aprotinin). After rotating for 1 h at 4°C, the samples were centrifuged at 100,000 × g for 1 h to separate the supernatant (sup.) from the pellet (ppt.). Aliquots of the indicated fractions were subjected to SDS-PAGE (10% gel) and analyzed by immunoblotting with anti-ATF6 antibody. The positions of p90ATF6 and p50ATF6 are marked as in Figure 1. (B) Effect of freezing and thawing. HeLa cells cultured in 60-mm dishes until 80% confluency were incubated in the absence (−) or presence (+) of 2 μg/ml tunicamycin (TM) for 4 h. Cells were washed with PBS, scraped with a rubber policeman, and centrifuged at 1000 × g for 5 min. After three cycles of freezing and thawing of cell pellets suspended in 50 μl of PBS, samples were centrifuged at 15,000 × g for 10 min to separate the supernatant (sup.) from the pellet (ppt.), which was then resuspended in 50 μl of PBS. Aliquots of each supernatant and pellet corresponding to 1 × 105 cells were subjected to SDS-PAGE (10% gel) and analyzed by immunoblotting with anti-ATF6 antibody.
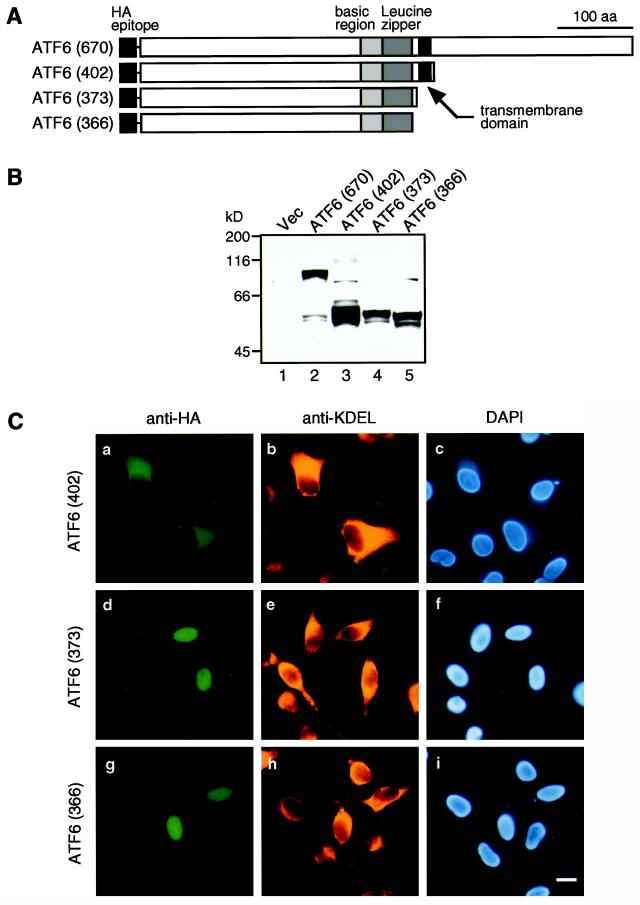
Nonetheless, we could not determine the localization of endogenous p50ATF6 by immunofluorescence analysis. We reproducibly observed decreased staining of the ER with anti-ATF6 antibody 4 h after tunicamycin treatment but hardly detected increased staining of the nucleus, probably because the small amounts of p50ATF6 produced diffused out in the nucleus, the volume of which was much larger than that of the ER (see Figure 4A). Because of this technical problem, it was also not possible to visualize the change in localization of ATF6 from the ER to the nucleus after tunicamycin or thapsigargin treatment of HeLa cells transfected with pCGN-ATF6 (670) and thus overproducing full-length ATF6. It should be noted that in thapsigargin-treated cells the amounts of p90ATF6 were much greater than those of p50ATF6 at all time points (Figure 1A). Similarly, the total amounts of p90ATF6 plus unglycosylated p90ATF6 were much greater than those of p50ATF6 in tunicamycin-treated cells at all time points (Figure 3). To circumvent this difficulty, we overexpressed various ATF6 mutants and examined their localization in transiently transfected HeLa cells (Figure 8).
Figure 8.
Expression and localization of various mutant forms of ATF6 in transfected HeLa cells. (A) Schematic structures of ATF6 derivatives analyzed. Full-length ATF6 cDNA, ATF6 (670), and three C-terminal deletion mutants were inserted into the mammalian expression vector pCGN. The positions of the HA epitope, basic region, leucine zipper, and transmembrane domain are indicated. (B) Immunoblotting analysis of transfected cells. HeLa cells in 60-mm dishes were transiently transfected with 1 μg of pCGN vector alone (Vec) or each of the ATF6 expression plasmids as indicated. Total proteins were extracted from transfected cells directly with 1× Laemmli’s SDS sample buffer followed by boiling for 5 min. Samples were subjected to SDS-PAGE (10% gel) and analyzed by immunoblotting with anti-ATF6 antibody. (C) Indirect immunofluorescence analysis of transfected cells. HeLa cells on slide glasses transiently transfected with pCGN-ATF6 (402) (a–c), pCGN-ATF6 (373) (d–f), and pCGN-ATF6 (366) (g–i) were fixed and stained with anti-HA epitope antibody (a–g), anti-KDEL antibody (b–h), or DAPI (c–i). Bar, 10 μm.
The N-terminal Fragment of ATF6 Representing p50ATF6 Translocates into the Nucleus
The above results indicated that ER stress triggers processing of ATF6, which not only decreases its molecular mass from 90 to 50 kDa but also alters its subcellular localization. We hypothesized that such dual outcomes could result from proteolysis of ATF6 in the C-terminal region; p50ATF6 reacts with anti-ATF6 antibody raised against the N-terminal region of ATF6. We therefore constructed various C-terminal deletion mutants and examined the localization of expressed proteins (see schematic structures depicted in Figure 8A). The boundaries of three functional domains should be noted; the basic region (R308-Y330), the leucine zipper (L334-X6-A-X6-L-X6-L-X6-V-X6-L369), and the transmembrane domain (V378-L398). Thus, ATF6 (402) lacked the majority of the C-terminal lumenal domain but retained the transmembrane domain, whereas two mutants, ATF6 (373) and ATF6 (366), lacked both lumenal and transmembrane domains. ATF6 (373) contained the entire bZIP region. ATF6 (366) contained the entire basic region and majority of the leucine zipper region. All mutant proteins were tagged with the HA epitope at the N terminus (Figure 8A).
Before localization analysis, we examined by immunoblotting whether these mutant forms of ATF6 were actually expressed in transfected HeLa cells (Figure 8B). Simultaneously, this analysis provided useful information on the cleavage site in p90ATF6. The doublet protein bands detected at ∼50 kDa in cells transfected with pCGN-ATF6 (670) (Figure 8B, lane 2) served as a molecular size marker for p50ATF6; these proteins were considered to represent p50ATF6, which was produced by constitutively activated proteolysis of the HA-tagged ATF6 (670) overexpressed in transfected cells (see DISCUSSION for explanation). The faster-migrating band might have lost the HA tag. Comparison of the mobilities of various C-terminal deletion mutants on SDS-PAGE revealed that the size of p50ATF6 was close to those of ATF6 (373) and ATF6 (366) (Figure 8B, lanes 4 and 5, respectively), suggesting that p90ATF6 may be cleaved between the bZIP and transmembrane domains to produce p50ATF6 when the cellular UPR was activated. We thus used ATF6 (373) and ATF6 (366) as representatives of p50ATF6 in the following experiments.
Immunofluorescence analysis showed that ATF6 (402) was localized in the ER (Figure 8C, compare a with b and c), indicating that the C-terminal lumenal domain is dispensable for the association with ER membranes. In marked contrast, ATF6 mutants lacking both lumenal and transmembrane domains entered the nucleus; the nucleus was clearly stained with anti-HA epitope antibody in cells overexpressing ATF6 (373) and ATF6 (366) (Figure 8C, compare d and g with e, f, h, and i). These results demonstrated that the localization of ATF6 is determined by the presence or absence of the transmembrane domain in the molecule and strongly suggested that the ATF6 molecule released from ER membranes (p50ATF6) can translocate into the nucleus.
ATF6 Mutants Representing p50ATF6 Enhance the Levels of Endogenous GRP78 by Activating Transcription
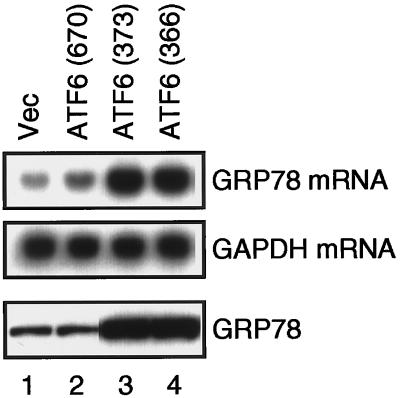
We finally examined the effects of overexpression of ATF6 mutants on the levels of endogenous UPR targets at both mRNA and protein levels (Figure 9). The level of GRP78 mRNA was slightly enhanced by overexpression of full-length ATF6, ATF6 (670), compared with the vector control (Figure 9, compare lane 2 with lane 1), although ATF6 (670) was primarily localized in the ER (Figure 6D; see DISCUSSION for explanation). Most importantly, the levels of both GRP78 mRNA and GRP78 were markedly enhanced by overexpression of ATF6 (373) and ATF6 (366) (Figure 9, lanes 3 and 4), both of which were localized in the nucleus (Figure 8C). We concluded that these two ATF6 mutants representing p50ATF6 directly activated transcription of the GRP78 genes in the nucleus, leading to enhanced levels of GRP78 in the ER.
Figure 9.
Effects of overexpression of full-length ATF6 and two nuclear localization mutants on the levels of endogenous GRP78. HeLa cells in 60-mm dishes were transiently transfected with 10 μg of pCGN vector alone (Vec) or each of the ATF6-expression plasmids as indicated, the structures of which are shown schematically in Figure 8A. Total RNA was prepared 48 h after transfection and analyzed by Northern blot hybridization using radiolabeled probes specific to GRP78 or GAPDH as described previously (Yoshida et al., 1998). Proteins were extracted 48 h after transfection by repeated freezing and thawing followed by sonication. The amounts of proteins recovered in the supernatant after centrifugation at 15,000 × g for 10 min were determined using a Bio-Rad protein assay kit. Aliquots of 30 μg were subjected to SDS-PAGE (10% gel) and analyzed by immunoblotting with anti-KDEL antibody.
DISCUSSION
Eukaryotic cells possess multiple intracellular signaling pathways from the ER to the nucleus, each modulating gene expression in response to changes in or surrounding the ER (reviewed by Pahl and Baeuerle, 1997). One of these, the UPR, deals with homeostasis of the folding capacity in the ER. Without this system, both yeast cells (Cox et al., 1993; Mori et al., 1993, 1996; Nikawa et al., 1996) and mammalian cells (Li and Lee, 1991; Little and Lee, 1995; Morris et al., 1997; Liu et al., 1998) are unable to survive under ER stress conditions that cause continuous accumulation of unfolded proteins in the ER.
In this study, we analyzed the mechanism of activation of ATF6 by ER stress, which we recently isolated as a candidate for a mammalian UPR-specific transcription factor. We showed that constitutively expressed 90-kDa protein (p90ATF6) is directly converted to a 50-kDa protein (p50ATF6) specifically in ER-stressed cells before the induction of GRP78, a major target protein of the mammalian UPR (Figures 1–3). The results presented here revealed the most important consequence of this conversion, that is, alteration of subcellular localization (see our current model depicted in Figure 10). Fractionation (Figure 5), differential solubilization (Figure 6A), topological studies (Figure 6B), glycosylation analysis (Figure 6C), and immunoflurescence analysis (Figures 4A and 6D) clearly demonstrated that p90ATF6 is a type II transmembrane glycoprotein embedded in the ER membrane. In contrast, p50ATF6 is a soluble nuclear protein (Figures 5 and 7). When overexpressed, ATF6 mutants representing p50ATF6 were accumulated in the nucleus (Figure 8) and constitutively activated transcription of the GRP78 gene, leading to enhanced levels of GRP78 in the ER (Figure 9). We thus propose that p50ATF6 represents an active and mature form of the mammalian transcription factor ATF6, and its production is mediated by ER stress-induced proteolysis of p90ATF6, which is synthesized as a precursor protein embedded in the ER membrane. Upon ER stress, p50ATF6 is released from the ER membrane, allowing it to enter the nucleus. In the nucleus, p50ATF6 containing a bZIP domain activates transcription of ER chaperone genes such as GRP78 through ERSE in collaboration with the general transcription factor NF-Y; we recently found that ATF6 recognizes ERSE and directly interacts with NF-Y (our unpublished observation).
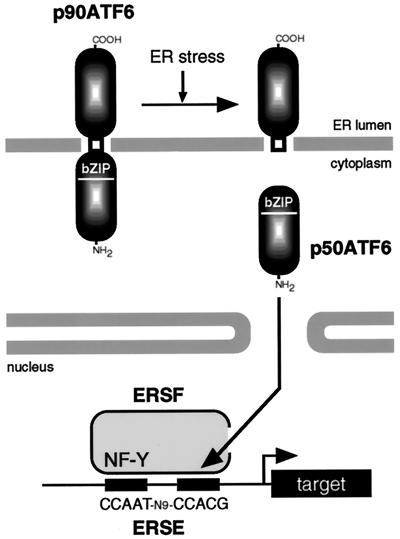
Figure 10.
Model for ER stress-induced processing of ATF6. ATF6 is constitutively synthesized as a precursor protein (p90ATF6) that anchors in the ER membrane through the single transmembrane domain near the center of the molecule. ER stress-induced proteolysis of p90ATF6 releases the N-terminal fragment (p50ATF6) containing bZIP, although the precise cleavage site is unknown. p50ATF6 translocates into the nucleus and interacts with the general transcription factor NF-Y to form a complex designated here as ER stress response factor (ERSF). ERSF activates transcription through ERSE (CCAAT-N9-CCACG) present in the promoter regions of mammalian UPR target genes.
Interestingly, p90ATF6 appears to be turned over fairly quickly; its half-life within the cell is ∼2 h (Figure 1). This rapid turnover rate allowed the cells to restore p90ATF6 at 16 h after thapsigargin treatment (Figure 1A). Similarly, p90ATF6 was restored at 16 h after tunicamycin treatment, although it was unglycosylated (Figure 3). Therefore, p90ATF6 itself might serve as a sensor molecule of ER stress; under the conditions that cause accumulation of unfolded proteins in the ER, p90ATF6 would not be able to fold properly, and this may somehow activate proteolytic processing of p90ATF6, resulting in production of p50ATF6. In this connection, it is noteworthy that overexpression of full-length ATF6, ATF6 (670), constitutively activated transcription of the GRP78 gene, albeit only slightly (Figure 9). We reasoned that overproduction of ATF6 (670), a transmembrane protein in the ER, is sensed as ER stress by the cell probably because normal levels of ER chaperones are insufficient for proper folding of exogenous proteins expressed at high levels. As a result, portions of endogenous p90ATF6 and exogenous ATF6 (670) are subjected to proteolytic processing constitutively, resulting in enhanced transcription of ER chaperone genes by constitutively produced p50ATF6 through ERSE in the nucleus. Indeed, p50ATF6-like doublet protein bands were detected in extracts of cells transfected with pCGN-ATF6 (670) (Figure 8B, lane 2). This also explains why various promoters of ER chaperone genes were constitutively activated when full-length ATF6 was overexpressed in HeLa cells (Yoshida et al., 1998).
We showed here that the mammalian UPR uses a system very similar to that previously identified for cholesterol homeostasis, another well-investigated intracellular signaling pathway from the ER to the nucleus (Yokoyama et al., 1993; Wang et al., 1994; Sakai et al., 1996, reviewed by Brown and Goldstein, 1997). Cholesterol metabolism is primarily regulated at the level of transcription, and sterol regulatory element binding proteins (SREBPs) mediate transcriptional activation of genes involved in cholesterol biosynthesis as well as receptor-mediated endocytosis of cholesterol-containing lipoproteins from plasma. Under normal conditions, SREBPs are bound to membranes of the ER and nuclear envelope through two hydrophobic stretches separated by a spacer of 31 amino acids and located around the center of the molecule. Depletion of sterols from culture medium activates proteolytic cleavage of SREBPs at the two sites in a sequential manner, allowing entrance of the N-terminal fragment into the nucleus. This released N-terminal fragment contains all of the functional domains necessary for active transcription factors such as DNA-binding (basic helix–loop–helix), dimerization (leucine zipper), and transactivation (acid blob) domains and thus enhances transcription of target genes in the nucleus.
In contrast to SREBPs, the precise cleavage site of ATF6 is as yet unknown. The region from amino acid 378 to 398 functions as a transmembrane domain because it is the only hydrophobic segment found in ATF6 (Figure 4B). The calculated molecular weight of the N-terminal region (1–377) is 41,161 and is thus significantly smaller than the size of p50ATF6 estimated from the mobility on SDS-PAGE. However, yeast Hac1p behaves as a 41-kDa protein on SDS-PAGE despite its calculated molecular weight of 26,903, presumably because of high contents of basic amino acids (Kawahara et al., 1997). Similarly, full-length ATF6 behaves as a 90-kDa protein on SDS-PAGE despite its calculated molecular weight of 74,597 (Zhu et al., 1997; Yoshida et al., 1998). Comparison of the mobilities of various C-terminal deletion mutants with that of p50ATF6 on SDS-PAGE (Figure 8B) suggested that ATF6 is cleaved between the bZIP and transmembrane domains, although we cannot exclude the possibility that ATF6 is cleaved within the transmembrane domain, similarly to cleavage at site 2 in SREBP2 (Sakai et al., 1996), or cleaved around the boundary between the cytoplasmic and transmembrane domains, similarly to Notch-1 (Chan and Jan, 1998; Schroeter et al., 1998). We are currently carrying out mutational analysis to determine the precise cleavage site. Such information will be useful for identification of the protease(s) responsible for proteolysis of ATF6.
We have identified a key regulatory step that connects events in the ER with those in the nucleus in mammalian UPR, and our observations have raised an important question: i.e., whether ER stress-induced proteolysis of ATF6 is regulated by mammalian Ire1p, a putative sensor molecule of ER stress identified recently (Tirasophon et al., 1998; Wang et al., 1998). The detection of endonuclease activity in the C-terminal tail region of human Ire1p suggests the presence of a mammalian mRNA splicing system similar to that for yeast HAC1 mRNA (Tirasophon et al., 1998), but ATF6 mRNA is not spliced (Yoshida et al., 1998). This raises the question of whether there are any substrates of such a mammalian splicing system, and if so, what is the consequence of such mRNA splicing. In yeast, transcriptional induction of ER chaperones is coupled to phospholipid biosynthesis; yeast cells lacking either Ire1p or Hac1p require exogenous inositol for growth (Nikawa and Yamashita, 1992; Cox et al., 1993; Mori et al., 1993, 1996; Nikawa et al., 1996; Sidrauski et al., 1996). It has been proposed that yeast UPR coordinates the synthesis of ER chaperones and ER membranes via the Ire1p–Hac1p pathway (Cox et al., 1997). This provides a basis for one intriguing speculation that in mammalian cells ER membrane biosynthesis may be controlled differently from transcriptional regulation of ER chaperone genes. Thus, the mammalian mRNA splicing system may be specialized to adjust the production of phospholipids according to the requirements within the ER. On the other hand, mammalian Ire1p may regulate the synthesis of ER chaperones by phosphorylating a putative protease(s), which activates ATF6. Answers to these important questions will further extend our understanding of the molecular mechanism of the mammalian UPR.
ACKNOWLEDGMENTS
We thank Masako Nakayama, Mayumi Ueda, Hideaki Kanazawa, Rika Takahashi, Seiji Takahara, and Tomoko Yoshifusa for technical assistance.
Abbreviations used:
- bZIP
basic leucine zipper
- ER
endoplasmic reticulum
- ERSE
ER stress response element
- HA
influenza virus hemagglutinin
- SREBP
sterol regulatory element binding protein
- UPR
unfolded protein response
- UPRE
unfolded protein response element
REFERENCES
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Chan Y-M, Jan YN. Roles for proteolysis and trafficking in Notch maturation and signal transduction. Cell. 1998;94:423–426. doi: 10.1016/s0092-8674(00)81583-4. [DOI] [PubMed] [Google Scholar]
- Chapman RE, Walter P. Translational attenuation mediated by an mRNA intron. Curr Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- Clark MW, Abelson J. The subnuclear localization of tRNA ligase in yeast. J Cell Biol. 1987;105:1515–1526. doi: 10.1083/jcb.105.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Franzusoff A, Rothblatt J, Schekman R. Analysis of polypeptide transit through yeast secretory pathway. Methods Enzymol. 1991;194:662–674. doi: 10.1016/0076-6879(91)94048-h. [DOI] [PubMed] [Google Scholar]
- Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Helenius A, Marquardt T, Braakman I. The endoplasmic reticulum as a protein folding compartment. Trends Cell Biol. 1992;2:227–231. doi: 10.1016/0962-8924(92)90309-b. [DOI] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. The eS-Sence of -SH in the ER. Cell. 1998;92:145–148. doi: 10.1016/s0092-8674(00)80907-1. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes & Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Yanagi H, Yura T, Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T, Yanagi H, Yura T, Mori K. Unconventional splicing of HAC1/ERN4 mRNA required for the unfolded protein response: sequence-specific and nonsequential cleavage of the splice sites. J Biol Chem. 1998;273:1802–1807. doi: 10.1074/jbc.273.3.1802. [DOI] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee AS. Coordinated regulation of a set of genes by glucose and calcium ionophores in mammalian cells. Trends Biochem Sci. 1987;12:20–23. [Google Scholar]
- Li WW, Alexandre S, Cao X, Lee AS. Transactivation of the grp78 promoter by Ca2+ depletion. A comparative analysis with A23187 and the endoplasmic reticulum Ca2+-ATPase inhibitor thapsigargin. J Biol Chem. 1993;268:12003–12009. [PubMed] [Google Scholar]
- Li X, Lee AS. Competitive inhibition of a set of endoplasmic reticulum protein genes (GRP78, GRP94, and ERp72) retards cell growth and lowers viability after ionophore treatment. Mol Cell Biol. 1991;11:3446–3453. doi: 10.1128/mcb.11.7.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little E, Lee AS. Generation of a mammalian cell line deficient in glucose-regulated protein stress induction through targeted ribozyme driven by a stress-inducible promoter. J Biol Chem. 1995;270:9526–9534. [PubMed] [Google Scholar]
- Liu H, Miller E, van de Water B, Stevens JL. Endoplasmic reticulum stress proteins block oxidant-induced Ca2+ increases and cell death. J Biol Chem. 1998;273:12858–12862. doi: 10.1074/jbc.273.21.12858. [DOI] [PubMed] [Google Scholar]
- McMillan DR, Gething MJ, Sambrook J. The cellular response to unfolded proteins: intercompartmental signaling. Curr Opin Biotechnol. 1994;5:540–545. doi: 10.1016/0958-1669(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Goldman RD. The dynamic properties and possible functions of nuclear lamins. Int Rev Cytol. 1995;162B:141–182. doi: 10.1016/s0074-7696(08)62616-9. [DOI] [PubMed] [Google Scholar]
- Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signaling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein-response element in Saccharomyces cerevisiae. J Biol Chem. 1998;273:9912–9920. doi: 10.1074/jbc.273.16.9912. [DOI] [PubMed] [Google Scholar]
- Mori K, Sant A, Kohno K, Normington K, Gething MJ, Sambrook JF. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JA, Dorner AJ, Edwards CA, Hendershot LM, Kaufman RJ. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J Biol Chem. 1997;272:4327–4334. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Akiyoshi M, Hirata S, Fukuda T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 1996;24:4222–4226. doi: 10.1093/nar/24.21.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- Pahl HL, Baeuerle PA. Endoplasmic reticulum-induced signal transduction and gene expression. Trends Cell Biol. 1997;7:50–55. doi: 10.1016/S0962-8924(96)10050-7. [DOI] [PubMed] [Google Scholar]
- Prostko CR, Brostrom MA, Malara EM, Brostrom CO. Phosphorylation of eukaryotic initiation factor (eIF) 2 alpha and inhibition of eIF-2B in GH3 pituitary cells by perturbants of early protein processing that induce GRP78. J Biol Chem. 1992;267:16751–16754. [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Roy B, Lee AS. Transduction of calcium stress through interaction of the human transcription factor CBF with the proximal CCAAT regulatory element of the grp78/BiP promoter. Mol Cell Biol. 1995;15:2263–2274. doi: 10.1128/mcb.15.4.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Lee AS. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 1999;27:1437–1443. doi: 10.1093/nar/27.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Li WW, Lee AS. Calcium-sensitive transcriptional activation of the proximal CCAAT regulatory element of the grp78/BiP promoter by the human nuclear factor CBF/NF-Y. J Biol Chem. 1996;271:28995–29002. doi: 10.1074/jbc.271.46.28995. [DOI] [PubMed] [Google Scholar]
- Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signaling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Cox JS, Walter P. The unfolded-protein-response pathway in yeast. Trends Cell Biol. 1994;4:56–60. doi: 10.1016/0962-8924(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Chapman R, Walter P. The unfolded protein response: an intracellular signaling pathway with many surprising features. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes & Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada I, Rindress D, Cameron PH, Ou WJ, Doherty JJd, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJ. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Wang X-Z, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch WJ, Suhan JP. Cellular and biochemical events in mammalian cells during and after recovery from physiological stress. J Cell Biol. 1986;103:2035–2052. doi: 10.1083/jcb.103.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Kaufman RJ. The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p(Ern1p) are required for kinase activation. J Biol Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Green SR, Kaufman RJ. Protein serine/threonine phosphatase Ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol Cell Biol. 1998;18:1967–1977. doi: 10.1128/mcb.18.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooden SK, Li LJ, Navarro D, Qadri I, Pereira L, Lee AS. Transactivation of the grp78 promoter by malfolded proteins, glycosylation block, and calcium ionophore is mediated through a proximal region containing a CCAAT motif which interacts with CTF/NF-I. Mol Cell Biol. 1991;11:5612–5623. doi: 10.1128/mcb.11.11.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins; involvement of basic-leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Zhu C, Johansen FE, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]