Abstract
Clinically ineffective transplatin [trans-diamminedichloridoplatinum(II)] is used in the studies of the structure-pharmacological activity relationship of platinum compounds. In addition, a number of transplatin analogs exhibit promising toxic effects in several tumor cell lines including those resistant to conventional antitumor cisplatin. Moreover, transplatin-modified oligonucleotides have been shown to be effective modulators of gene expression. Owing to these facts and because DNA is also considered the major pharmacological target of platinum complexes, interactions between transplatin and DNA are of great interest. We examined, using biophysical and biochemical methods, the stability of 1,3-GNG intrastrand cross-links (CLs) formed by transplatin in short synthetic oligodeoxyribonucleotide duplexes and natural double-helical DNA. We have found that transplatin forms in double-helical DNA 1,3-GNG intrastrand CLs, but their stability depends on the sequence context. In some sequences the 1,3-GNG intrastrand CLs formed by transplatin in double-helical DNA readily rearrange into interstrand CLs. On the other hand, in a number of other sequences these intrastrand CLs are relatively stable. We show that the stability of 1,3-GNG intrastrand CLs of transplatin correlates with the extent of conformational distortion and thermodynamic destabilization induced in double-helical DNA by this adduct.
INTRODUCTION
Almost 40 years have elapsed since the accidental discovery of the anticancer activity of cisplatin (cis-diamminedichloridoplatinum(II)) (Fig. 1 A) (1). Despite its success, cisplatin has several disadvantages. The drawbacks coupled with cisplatin toxicity have been the impetus for the development of improved platinum drugs. The design of new platinum drugs with improved antitumor effects depends on understanding existing platinum agents with a view toward developing new modes of attack (2,3). Therefore, it is of great interest to understand details of the biophysical and biochemical mechanisms underlying the biological efficacy of the platinum compounds and to define their structure-pharmacological relationships. Although a lot of effort has been devoted to understanding the mechanism of antitumor effects of cisplatin, not all aspects of the mechanism are entirely clear.
FIGURE 1.
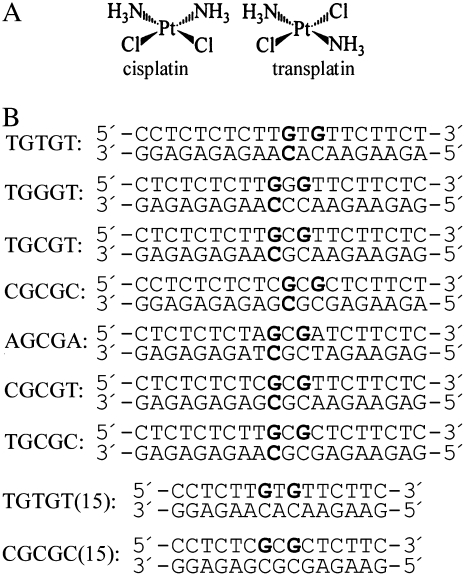
(A) Structures of the platinum complexes. (B) Sequences of the synthetic oligodeoxyribonucleotides used in this study with their abbreviations. The top and bottom strands of each pair are designated top and bottom, respectively, in the text. The bold letters in the top strands of the duplexes indicate the location of the 1,3-intrastrand cross-link after modification of the oligodeoxyribonucleotides by transplatin in the way described in the experimental section; the bold letters in the bottom strands of these duplexes indicate the location of the sites that were involved in the interstrand cross-links after the linkage isomerization reaction was completed (see the text).
Structure-activity studies often use inactive compounds such as trans-diamminedichloridoplatinum(II) (transplatin, the geometric isomer of cisplatin) (Fig. 1 A) (4). This compound has been used widely to investigate the mechanism of action of cisplatin. In this approach, one searches for differences between active and inactive compounds that may be responsible for the pharmacological effect. Hence, the clinical inactivity of the trans isomer of cisplatin was considered for a relatively long time a paradigm for the classical structure–activity relationships of platinum drugs (5). However, a series of bifunctional trans-platinum(II) complexes have been synthesized that show anticancer activity distinct from cisplatin (sometimes even more efficient than cisplatin itself and its analogs) and these complexes bind to DNA in a manner distinctly different from that of cisplatin (6–10).
It is generally accepted that DNA is the primary pharmacological target of platinum antitumor drugs, although other biological targets may also be important in the cisplatin mechanism (11). Platinum antitumor drugs bind efficiently to DNA forming a variety of adducts that block replication and transcription and induce cell death (8,12,13). The mechanistic explanation of transplatin inactivity has been based on the type of DNA adducts formed by this isomer, as well as on its chemical reactivity that can render the complex susceptible to deactivation before its delivery to the tumor site. Notably, a distinct difference between cisplatin and its trans isomer is that transplatin is chemically more reactive than cisplatin and more susceptible to deactivation. Whereas the major adducts of cisplatin are intrastrand cross-links (CLs) between neighboring purine residues, transplatin-DNA adducts are interstrand CLs (∼12% in linear DNA) (14) and a relatively large portion of adducts (50%) remains monofunctional(15).
Transplatin is sterically restricted in the type of intrastrand CLs it can feasibly produce; that is, it is unable to cross-link neighboring bases in a DNA strand like cisplatin. It can crosslink two bases on the same strand only if they are separated by at least one intervening base, forming mostly 1,3-GNG CLs (where G = guanine and N = adenine, cytosine, or thymine) (14–16). In contradiction to this finding, it has been suggested (17,18) that these intrastrand adducts of transplatin are only stable in single-stranded DNA, whereas they rearrange in double-helical DNA into interstrand CLs (17,19). It has also been suggested (18) that in cells transplatin forms only a small amount of interstrand adducts and no intrastrand CLs.
The conclusion that intrastrand CLs are not formed in the reaction between transplatin and double-helical DNA (17,19) was drawn on the basis of the findings that 1,3-GNG intrastrand CLs readily formed by transplatin in single-stranded DNA rearrange into interstrand CLs after hybridization of these platinated single-strands with their complementary counterparts. However, these studies were only carried out with a relatively small number of sequence contexts. In addition, the phenomenon that the linkage isomerization reaction (rearrangement of intrastrand CLs of transplatin into interstrand adducts) is exclusively triggered by the formation of a double helix has been proposed as a basis for concept of the use of transplatin-modified oligonucleotides to block the cellular machinery specifically and irreversibly, i.e., as modulators of gene expression (19,20). Also interestingly, the replacement of an ammine ligand(s) in transplatin by a heterocyclic ligand(s) results in the distinctively enhanced stability of the intrastrand CLs in several sequence contexts and concomitantly in cytotoxicity in tumor cell lines including those resistant to cisplatin (21–25). Thus, there is an interest in understanding more deeply the properties of intrastrand CLs of transplatin and to clarify some discrepancies that appear in the literature concerning the stability of these adducts and the frequency of their occurrence in double-helical DNA.
Therefore, we reexamined in this study, using various biophysical and biochemical methods, the stability of 1,3-GNG intrastrand CLs in double-helical DNA formed by transplatin in a number of other sequence contexts than were those used in previous studies. We have found that the stability of these intrastrand adducts of transplatin depends on the sequence context and correlates with the extent of conformational distortion and thermodynamic destabilization induced in double-helical DNA by this adduct.
MATERIALS AND METHODS
Starting materials
Transplatin (Fig. 1 A) was obtained from Sigma (Prague, Czech Republic). The stock solution of transplatin was prepared at a concentration of 5 × 10−4 M in 10 mM NaClO4 in the dark at 25°C. Plasmid pUC19 (2686 bp) was isolated according to standard procedures. The synthetic oligodeoxyribonucleotides (Fig. 1 B) were purchased from VBC-genomics (Vienna, Austria) or DNA Technology (Aarhus, Denmark). The purity of the oligonucleotides was verified by high-pressure liquid chromatography (HPLC) or gel electrophoresis. Restriction endonucleases and the Klenow fragment from DNA polymerase I (exonuclease minus, mutated to remove the 3′→5′ proofreading domain) and T4 polynucleotide kinase were purchased from New England BioLabs (Beverly, MA). Sequenase 2 was from USB Corporation (Cleveland, OH). Acrylamide, bis(acrylamide), agarose, o-phenatroline (OP), and NaCN were from Merck KgaA (Darmstadt, Germany). Dimethyl sulfate (DMS), KMnO4, diethyl pyrocarbonate (DEPC), KBr, KHSO5, and thiourea were from Sigma. [γ-32P]-deoxyriboadenosine triphosphate ([γ-32P]dATP) was from MP Biomedicals, LLC (Irvine, CA). Dimethyl sulfate (DMS) was from Sigma.
Platination reactions
The single-stranded oligonucleotides (the top strands of the duplexes shown in Fig. 1 B) were reacted in stoichiometric amounts with transplatin. The platinated oligonucleotides were repurified by ion-exchange fast protein liquid chromatography (FPLC). It was verified by platinum FAAS and by the measurements of the optical density that the modified oligonucleotides contained one platinum atom. It was also verified using DMS footprinting of platinum on DNA (14,26) that one platinum molecule was coordinated to two guanines at their N7 position in the top strands of the duplexes shown in Fig. 1 B. The platinated top strands were allowed to anneal with unplatinated complementary strands (the bottom strands shown in Fig. 1 B) in 0.2 M NaClO4. Other details are in the text (vide infra) or have been described previously (14,27,28).
Rearrangement of intrastrand cross-links of transplatin
The platinated single-stranded oligodeoxyribonucleotides (the top strands of the 20-bp duplexes shown in Fig. 1 B) (20 μM) were allowed to anneal with the unplatinated complementary (bottom) strands in 0.2 M NaClO4, 5 mM Tris-HCl buffer (pH 7.5) and 0.1 mM EDTA at 20°C for 30 min, and for 2 h at 4°C. Samples of these duplexes (2 μM) were further incubated at 37°C in the same medium; at various time intervals, aliquots were withdrawn and analyzed by electrophoresis in 12% PAA/8 M urea gel. The bases involved in the interstrand CLs were determined by Maxam-Gilbert footprinting (14,22).
Digestion by o-phenanthroline-copper complex and chemical modifications
o-Phenanthroline-copper complex (OP-Cu) digestions were carried out according to the procedure described previously (29) with minor modifications (30). Platinated and unplatinated oligonucleotides were 5′-end labeled with T4 polynucleotide kinase with the appropriate [γ-32P]dNTP. After digestion with OP-Cu, transplatin was removed after reaction of the DNA with the probe by incubation with 0.2 M NaCN (pH 11) at 45°C for 10 h in the dark.
Modifications by KMnO4, DEPC, and KBr/KHSO5 were carried out as described previously (27,31–33) at 4°C. The duplex strands were 5′-end labeled with [γ−32P]ATP. In the case of the platinated oligonucleotides, transplatin was removed after reaction of the DNA with the probe by incubation with 0.2 M NaCN (pH 11) at 45°C for 10 h in the dark.
Isothermal titration calorimetry
The standard isothermal titration calorimetry (ITC) buffer for these studies contained 50 mM NaCl with 10 mM phosphate buffer (Na2HPO4/NaH2PO4 [pH 7.0]). Sufficient quantities of ITC solutions were prepared to carry out a set of titrations of the 50 μM solution of the bottom strands of the duplexes TGTGT(15) or CGCGC(15) (for their sequences, see Fig. 1 B) into the 5 μM solution of the top strand of nonmodified TGTGT(15) or CGCGC(15) or the same duplexes containing a single, site-specific 1,3-GNG intrastrand CL of transplatin at 4°C. Molar extinction coefficients for the single-stranded unmodified or platinated oligonucleotides (related to the strands that were 15 nucleotides long) used in the ITC experiments were determined as described in our previous articles (34–38), i.e., by phosphate analysis (39). Stock solutions of the strands for ITC studies were prepared in the ITC buffer and were exhaustively dialyzed against this buffer. It was verified that the enthalpies of ITC injections of each individual oligomer into buffer, of buffer into buffer, and of excess oligomer into a solution of the duplex were all the same as water into water injections, within error. From this data, it was concluded that the effects of any solvent mismatching are negligible. Titrations were carried out on a VP-ITC instrument (MicroCal LLC, Northampton, MA). For each titration, the top strand of the nonmodified TGTGT(15) or CGCGC(15) duplex or the same duplex containing a single, site-specific 1,3-GNG intrastrand CL of transplatin was loaded into a 1.4-ml sample cell, and the complementary oligomer (bottom strand of the duplex TGTGT(15) or CGCGC(15)) was loaded into the 300 μL injection syringe. The stirring rate of the injection syringe was 400 rpm, and the samples were equilibrated thermally before each titration until the baseline had leveled off and the rms noise was <0.015 μcal × s−1. A typical titration consisted of 50 injections of 5 μL each, with 3 min between injections. The data from individual titrations were analyzed by using the Origin 5.0 software package (Origin, Northampton, MA) to extract the relevant thermodynamic parameters [the enthalpy change (ΔH), the entropy change (ΔS), the stoichiometry (n), and the equilibrium constant (K) for strand association].
Replication mapping of the 1,3-GNG intrastrand cross-links
Replication with Sequenase 2 and electrophoretic analysis of the products of the DNA polymerization reaction were carried out according to the protocols recommended by Amersham, Pharmacia and in Burstyn et al. (40) and Intini et al. (41). The (AflIII/EcoRI) restriction fragment (410 bp) from plasmid pUC19, which had been used as a template for Sequenase 2, was modified with transplatin in 10 mM NaClO4 at 37°C for 24 h in the dark. The level of platination of DNA corresponded to rb = 0.001 (rb is defined as the number of platinum atoms coordinated per nucleotide residue in DNA). Other details are in the text.
Other physical methods
Absorption spectra were measured with a Varian Cary 4000 UV-VIS spectrophotometer equipped with a thermoelectrically controlled cell holder and quartz cells with the pathlength of 1 cm. Purification of oligonucleotides with the aid of HPLC was carried out on a Waters HPLC system consisting of Waters 262 Pump, Waters 2487 UV detector, and Waters 600S Controller with MonoQ HR 5/5 column. The FAAS measurements were carried out on a Varian AA240Z Zeeman atomic absorption spectrometer equipped with a GTA 120 graphite tube atomizer. For FAAS analyses, DNA was dissolved in 0.1 M HCl. The gels were visualized by using a BAS 2500 FUJIFILM bioimaging analyzer, and the radioactivity associated with each band was quantitated with the AIDA image analyzer software (Raytest, Germany).
RESULTS
Stability of the 1,3-GNG intrastrand cross-links of transplatin
Previous studies (17,42) have shown that the 1,3-GNG intrastrand CL of transplatin is stable within single-stranded DNA under physiological conditions. It has also been shown (17,42) that within double-helical DNA, the stability of the CL formed by transplatin in some selected sequence contexts is reduced markedly and is even notably smaller than that of the interstrand CLs (preferentially formed by this platinum compound between guanine and complementary cytosine residues (14)). Consequently, the pairing of single-stranded DNA containing a 1,3-GNG intrastrand CL of transplatin in these selected sequence contexts with their complementary DNA strands results in a rearrangement of these intrastrand adducts into interstrand CLs (17,42).
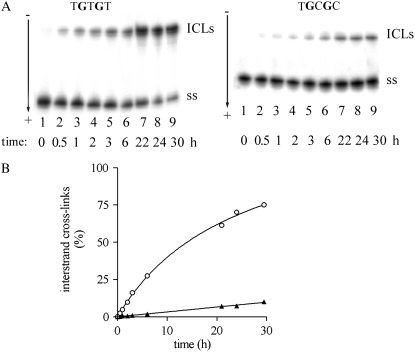
The stability of 1,3-GNG intrastrand CLs of transplatin was investigated using 20-mer oligodeoxyribonucleotides (the top strands of the duplexes shown in Fig. 1 B) that were radioactively labeled at their 5′-ends and platinated so that they contained single and central, site-specific 1,3-GTG, GGG, or GCG intrastrand CL. The sequences of these duplexes (Fig. 2) were chosen to substantially complement the spectrum of sequence contexts in which these CLs were formed and used in the previous studies (17,42) of the stability of 1,3-intrastrand CLs of transplatin. The single-stranded oligonucleotides containing this CL or the corresponding duplexes (Fig. 2) were incubated in 0.2 M NaClO4 at 37°C. At various time intervals, aliquots were withdrawn and analyzed by gel electrophoresis under denaturing conditions (shown for the duplexes TGTGT and TGCGC in Fig. 3, A and B, respectively). The 1,3-GNG intrastrand adducts of transplatin in all single-stranded oligonucleotides (the top strands of the duplexes shown in Fig. 1 B) were inert over a long period of time (>5 days) (not shown). It was verified by DMS footprinting that no rearrangement of the 1,3-intrastrand CL occurred within this period. In contrast, the adduct formed by transplatin after pairing the platinated single-stranded oligonucleotides with their complementary strands was labile, consistent with the previous results (17,42). As a function of time, the radioactivity associated with the band corresponding to the 1,3-intrastrand CL decreased with a concomitant increase of the radioactivity associated with the new, more slowly migrating species (shown for the duplexes TGTGT and TGCGC containing the 1,3-GTG intrastrand CLs of transplatin in Fig. 2 A). This new band migrated at approximately the same rate as the 20-bp duplex TGTGT containing a single interstrand CL (this cross-link resulted from the reaction between the oligonucleotide and transplatin carried out as described (14)). No other bands were detected. Similar results were obtained at higher concentrations of the platinated duplex, which excluded an interduplex reaction. This result was interpreted to mean that the 1,3-intrastrand CL was transformed into an interstrand CL (17,42). After 24 h of incubation of the duplexes TGTGT and TGGGT containing a 1,3-intrastrand CL of transplatin, 70% and 54% of the 1,3-intrastrand CLs were transformed into the interstrand CLs (Table 1 and shown for the CL in the TGTGT duplex in Fig. 2 B). On the other hand, the yield of this rearrangement reaction involving the formation of a 1,3-intrastrand CL of transplatin in the sequences TGCGT, CGCGC, AGCGA, CGCGT, TGCGT was radically lower in the range of only 7%–26% (Table 1 and shown for the duplex TGCGC in Fig. 2 B).
FIGURE 2.
Rearrangement of the 1,3-intrastrand cross-links formed by transplatin in the duplexes TGTGT and TGCGC. The top strands of these duplexes were modified so that they contained a single, site-specific 1,3-GNG intrastrand cross-link. These platinated single-stranded oligodeoxyribonucleotides (20 μM) were allowed to anneal with the unplatinated complementary (bottom) strands in 0.2 M NaClO4, 5 mM Tris-HCl buffer (pH 7.5), and 0.1 mM EDTA at 20°C for 30 min, and for 2 h at 4°C. Samples of these duplexes (2 μM) were further incubated at 37°C in the same medium; at various time intervals, the aliquots were withdrawn and analyzed by electrophoresis in 12% PAA/8 M urea gel. (A) Autoradiograms of the gels of the duplex TGTGT (left) and TGCGC (right), both modified by transplatin radioactively labeled at the 5′-end of its top strand. Incubation times in minutes are indicated under each lane. Lane 1 refers to the 5′-end labeled single-stranded top (platinated) strand. (B) Plot of the percentages of 1,3-intrastrand CL of transplatin in TGTGT (○) or TGCGC (▴) versus time. These percentages were calculated from the ratio of the radioactivity in each lane in panel A associated with the band corresponding to the lower bands in panel A to the sum of the radioactivities associated with both bands (multiplied by 100). For other details, see the text.
FIGURE 3.
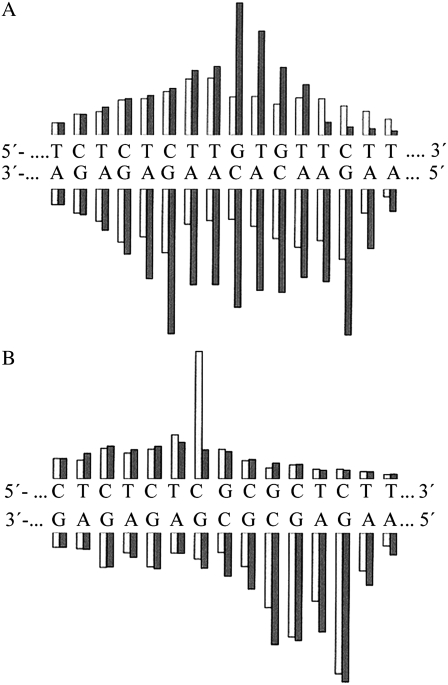
Quantitation of Op-Cu digestion of TGTGT (A), CGCGC (B) duplexes, and their analogs containing a single, site-specific 1,3-GNG intrastrand cross-link. The autoradiograms (not shown) were quantitated by microdensitometry. The black bars represent digestion of the unplatinated duplexes, and the light bars represent digestions of the platinated duplexes. The heights of the bars are proportional to the band intensities on the autoradiograms.
TABLE 1.
Rearrangement of single, site-specific 1,3-GNG intrastrand cross-links of transplatin in 20-bp duplexes into interstrand cross-links
| Intrastrand cross-linked sequence* | Interstrand CLs after 24 h at 37°C (%) | t50% (h)† | Reference |
|---|---|---|---|
| TGAGT | 91 | 6 | (42) |
| TGTGT | 70 | 12 | (42) and this work |
| 5′-CGTGT | >50 | 6 | (42) |
| 5′-TGTGC | >50 | 18 | (42) |
| 5′-GGTGT | >50 | 4 | (17) |
| AGAGA | >50 | 1.6 | (17) |
| TGAGT | >50 | 4.8‡ | (17) |
| TGGGT | 43 | >24 | this work |
| TGCGT | 29 | >24 | this work |
| CGCGC | 26 | >24 | this work |
| AGCGA | 18 | >24 | this work |
| 5′-CGCGT | 14 | >24 | this work |
| TGCGC | 7 | >24 | this work |
The platinated duplexes (2 μM) were incubated in a medium of 0.2 M NaClO4, 5 mM Tris-HCl, pH 7.5 at 37°C.
The central nucleotide sequence of the top strand of the 20-bp duplex at which the 1,3-GNG intrastrand cross-link of transplatin was formed. Gs in bold represent the platinated sites in the intrastrand cross-link.
The time at which the rearrangement reached 50%.
This value was obtained for the rearrangement reaction at 40°C.
The bases in the interstrand CLs resulting from the rearrangement of the 1,3-intrastrand CLs were identified from Maxam-Gilbert footprinting in the same way as described in previous articles (17,42) (not shown). The results indicated that the interstrand CLs of transplatin in all duplexes tested in this work were always formed between the platinated 5′ G of the 1,3-GNG intrastrand CL and the complementary C.
Chemical probing of conformational distortions
We tested an eventuality that an important factor contributing to differences in the rate of rearrangement of intrastrand CLs formed by transplatin in various sequence contexts into interstrand adducts is the character and extent of conformational distortion and resulting local destabilization of double-stranded DNA. We characterized conformational alterations induced in DNA by intrastrand CLs of transplatin in the 20-bp duplexes TGTGT or CGCGC containing a central, site-specific CL treating these duplexes with several chemical agents that are used as tools for monitoring the existence of conformations other than canonical B-DNA. The duplexes TGTGT and CGCGC were chosen to represent groups of duplexes in which the yield of the linkage isomerization reaction is high and very low, respectively (Table 1). In addition, to minimize the rearrangement of intrastrand CLs of transplatin to interstrand CLs during reactions of intrastrand crosslinked duplexes with chemical probes, which could obscure analysis focused on intrastrand CLs, the intrastrand crosslinked duplexes were kept at 4°C for 12 h; at this temperature rearrangement of the 1,3-GNG intrastrand CLs is reduced markedly. It was verified by gel electrophoresis (vide supra) that the 1,3-GTG intrastrand CL formed in the duplexes TGTGT and CGCGC was stable at 4°C so that only 1.3% and 0.7% of intrastrand crosslinked duplexes TGTGT and CGCGC, respectively, were transformed into interstrand crosslinked within 12 hr (not shown). Reactions with chemical nuclease or chemical probes were carried out at 4°C, but the times of these reactions were only 5–30 min (see Materials and Methods). Therefore, it was also verified by gel electrophoresis that only a negligible fraction (<1%) of duplexes containing a 1,3-GTG intrastrand CL formed in the duplexes TGTGT or CGCGC rearranged to interstrand crosslinked duplexes within 30 min (not shown).
OP-Cu is a chemical nuclease that nicks nucleic acids by oxidation attack on the sugar moiety (43,44). The cleavage efficiency of OP-Cu is dependent on the secondary structure of DNA. A comparison of the cleavage patterns of unplatinated TGTGT duplex and those modified with a 1,3-intrastrand CL of transplatin shows a markedly lower reactivity of the central nucleotide sequence containing the CL (Fig. 3 A). In contrast, the cleavage pattern of unplatinated CGCGC duplex and of those modified with a 1,3-intrastrand CL of transplatin also shows an altered reactivity of the central nucleotide sequence containing the CL (Fig. 3 B), but different from that seen for the central nucleotide sequence containing the CL in the duplex TGTGT. We interpret the results obtained with OP-Cu nuclease to mean that the geometry of the double helix is altered over several base pairs around the 1,3-intrastrand CL in both sequence contexts, but differently.
The other agents used as chemical probes of DNA conformation also include KMnO4, bromine, or DEPC as probes for thymine, cytosine, and adenine/guanine residues, respectively (27,31–33,45). These chemical probes react, under the conditions used, with base residues in single-stranded DNA and distorted double-stranded DNA, but not with the base residues in intact, double-stranded DNA (27,31–33,45). For this analysis, we used exactly the same methodology as in our recent studies dealing with DNA adducts of various antitumor platinum drugs. Thus, the details of this experiment can be found in those articles (27,46), and representative gels showing piperidine-induced specific strand cleavage at KMnO4-modified, KBr/KHSO5-modified, and DEPC-modified bases in the TGTGT and CGCGC duplexes unplatinated or containing a single 1,3-GNG intrastrand CL of transplatin are shown in the Supplementary Material, Fig. S1.
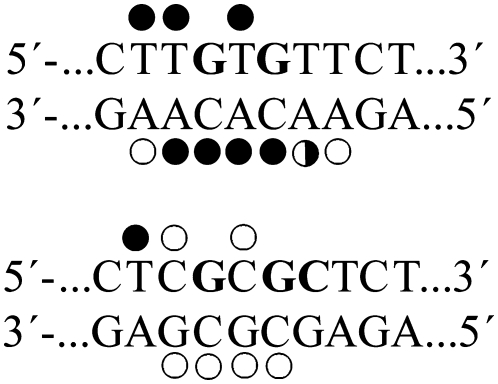
The results are schematically summarized in Fig. 4. The pattern and degree of reactivity toward the chemical probes were different for the 1,3-GNG intrastrand CLs formed by transplatin in the sequences TGTGT and CGCGC. The extent of the distortion was markedly larger in the TGTGT sequence than in the CGCGC sequence (Fig. 4), indicating the sequence-dependent character of the conformational distortion induced by the 1,3-GNG intrastrand CL of transplatin.
FIGURE 4.
Summary of the reactivity of chemical probes with the duplexes TGTGT and CGCGC containing a 1,3-GNG intrastrand cross-link of transplatin. Solid, half-solid, and open circles designate strong, medium, or weak reactivity, respectively.
Isothermal titration calorimetry
Calorimetric technique was used to characterize the influence of the 1,3-GNG intrastrand CLs of transplatin on the thermal stability and energetics of the site-specifically platinated 15-bp DNA duplexes containing the central sequences TGTGT and CGCGC (duplexes TGTGT(15) and CGCGC(15), for their sequence, see Fig. 1 B). Such thermodynamic data can show how the platinum CL influences duplex stability, a property that has been shown to play a significant role in cellular processes, such as recognition of DNA damage by DNA-binding proteins and repair of this damage, i.e., the processes that may modulate potency of antitumor drugs including metal-based cytostatics. Recently, differential scanning calorimetry (DSC) was used to characterize the influence of different CLs of platinum antitumor drugs on the thermal stability and energetics of 15–20-bp DNA duplexes site-specifically modified by these drugs (34–36,47,48). We decided to expand these studies to oligodeoxyribonucleotide duplexes containing unique 1,3-GNG intrastrand CLs formed by transplatin in the TGTGT and CGCGC sequences. DSC makes it possible to measure excess heat capacity versus temperature profiles for the thermally induced transitions of nonmodified DNA duplexes and the duplexes containing a unique adduct of the metal-based drug. Such thermograms are usually recorded with a heating rate of 60°C/h and after reaching the maximum temperature of 95°C the samples are cooled at the same rate to the starting temperature of 25°C (34,47–49). This implies that the duplexes containing the unique adduct are exposed to higher temperatures for a relatively long period. Hence, DSC does not seem to be suitable for analysis of the duplexes containing 1,3-GNG intrastrand CLs of transplatin since higher temperatures may facilitate rearrangement of this CL to interstrand CL; a more prominent fraction of interstrand crosslinked duplexes in a sample of intrastrand crosslinked duplexes would obscure analysis of the intrastrand adducts. A suitable alternative is ITC, which makes it possible to study the thermodynamic parameters of the duplex formation from its two complementary single strands over a range of temperatures including low temperatures, such as 4°C, at which the stability of the 1,3-GNG intrastrand CLs is expected to be markedly enhanced. It was verified by gel electrophoresis (vide supra) that the 1,3-GTG intrastrand CL formed in the duplex TGTGT(15) was stable at 4°C, so that only <1% of intrastrand crosslinked duplexes were transformed into interstrand crosslinked (not shown).
Fig. 5 shows the ITC profiles of duplex formation from the nonmodified bottom strand of the duplexes TGTGT(15) and CGCGC(15) titrated into complementary nonmodified top strands, or into the same strands containing a single 1,3-GNG intrastrand CL of transplatin at 4°C. It was verified that the melting temperature of the duplexes was significantly higher than this temperature and that the 1,3-intrastrand CLs of transplatin formed in both 15-bp duplexes or in their single-stranded top strands were stable for >12 h. The ITC profiles were analyzed as described in Materials and Methods to obtain the results listed in Table 2. All thermodynamic parameters discussed in this work refer to the duplex formation process. Differences in the dissociation thermodynamics due to the presence of the adduct are presented as “ΔΔ” parameters. These parameters are computed by subtracting the appropriate value measured for the control, unmodified duplex from the value measured for the duplex containing a single, site-specific intrastrand platinum CL.
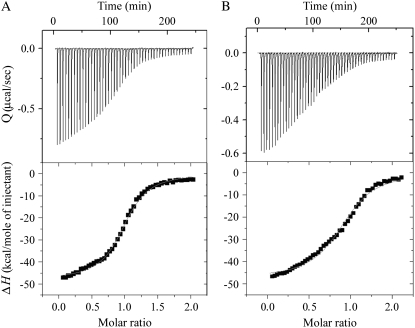
FIGURE 5.
Isothermal titration calorimetry. ITC binding isotherms for the association of the top strand of the 15 bp TGTGT(15) (A) and CGCGC(15) (B) duplexes containing a single, site-specific 1,3-GNG intrastrand cross-link of transplatin with the complementary (nonmodified) strand (bottom strands of the duplexes TGTGT(15) and CGCGC(15), respectively) at 4°C in 10 mM phosphate buffer (pH 7.0) containing 50 mM NaCl. The upper panels show total heat released on injecting 5 μL aliquots of the 50 μM bottom strand into a 1.4 ml reaction cell containing the 5 μM top strand. The lower panels show the resultant binding isotherm (solid squares) obtained by integrating the peak areas of each injection. The continuous line represents the nonlinear least squares fit of the affinity (K), enthalpy (ΔH), and stoichiometry (n) to a single-site binding model. For other details, see the text.
TABLE 2.
Calorimetrically-derived thermodynamic parameters for the formation of the 15-bp duplexes nonmodified or containing a single, site-specific 1,3-GNG intrastrand cross-link of transplatin at 4°C
| ΔH†
|
ΔS†
|
ΔG4°C†
|
K‡
|
||
|---|---|---|---|---|---|
| Duplex* | kcal/mol duplex | cal/K·mol duplex | kcal/mol duplex | M−1 | n‡ |
| TGTGT(15) | −64.9 | −193 | −11.5 | 8.57 × 108 | 1.07 |
| TGTGT(15) · transPt | −47.9 (17) | −144 (49) | −8.0 (3.5) | 3 × 106 | 1.01 |
| CGCGC(15) | −55.4 | −165 | −9.7 | 5.7 × 107 | 1.09 |
| CGCGC(15) · transPt | −48.0 (7.4) | −145 (20) | −7.8 (1.9) | 1.5 × 107 | 1.01 |
The ΔH and ΔS values are averages derived from three independent experiments. The experimental uncertainties of the parameters are as follows: ΔH (2%), ΔS (3%), ΔG4 (3%). “ΔΔ” parameters are in parentheses (these parameters are computed by subtracting the appropriate value measured for the control, unmodified duplex from the value measured for the duplex containing a single, site-specific transplatin adduct).
G's in bold represent platinated guanine residues.
ΔH, ΔS, and ΔG4 denote, respectively, the enthalpy, entropy, and free energy (at 4°C) of duplex formation.
K and n denote, respectively, the association constant and binding stoichiometry for strand association.
Inspection of these thermodynamic parameters shows that the exothermic formation of a single 1,3-intrastrand adduct in the duplexes TGTGT(15) or CGCGC(15) by transplatin resulted in a large decrease in the change in enthalpy of duplex formation by 17.0 and 7.4 kcal/mol, respectively. In other words, the 1,3-GNG intrastrand CL of transplatin enthalpically destabilizes the duplex relative to its nonmodified counterpart. In addition, the formation of a 1,3-GNG intrastrand CL by transplatin in both sequence contexts resulted in a substantial increase in the entropy of the duplexes TGTGT(15) or CGCGC(15) of 49 or 20 cal/K·mol (TΔΔS = 13.6 and 5.5 kcal/mol at 4°C), respectively. In other words, the 1,3-GNG intrastrand CL of transplatin increases the entropy of the platinated duplexes and in this way entropically stabilizes the duplexes. Thus, the 17.0 or 7.4 kcal/mol enthalpic destabilization of the TGTGT(15) or CGCGC(15) duplexes due to the 1,3-GNG intrastrand CL of transplatin is partially, but not completely, compensated by the entropic stabilization of the duplexes induced by this adduct of 13.8 or 5.5 kcal/mol at 4°C, respectively. The net result of these enthalpic and entropic effects is that the formation of a 1,3-GNG intrastrand CL with the duplexes TGTGT(15) or CGCGC(15) at 4°C induces a decrease in duplex thermodynamic stability (ΔΔG4°C) of −3.2 or −1.9 kcal/mol, respectively, with this destabilization being enthalpic in origin. In this respect, the 1,3-GNG intrastrand CL formed by transplatin in the TGTGT sequence thermodynamically destabilized DNA considerably more than that formed in the CGCGC sequence.
Replication mapping of the 1,3-GNG intrastrand cross-links in the 410-bp DNA fragment
Further investigations were aimed at finding the sites in natural DNA in which intrastrand CLs were formed during the reaction with transplatin. Previous work has shown that the in vitro DNA synthesis by DNA polymerases on a DNA template modified by various platinum complexes is terminated at the level of the adducts (50–52). A similar approach was used in this work.
The primer extension footprinting assay was used to determine the sites of intrastrand CLs of transplatin, their sequence selectivity and stability. The AflIII/EcoRI fragment of pUC19 plasmid (410-bp with a random natural nucleotide sequence) was incubated with transplatin for 24 h at 37°C; the level of the modification was relatively low and corresponded to rb = 0.001. To determine the location of the intrastrand CLs, mapping analyses were carried out using two types of the 410-bp fragments modified by transplatin. One type of fragment was globally modified by transplatin and was used without any further treatment, i.e., contained all types of DNA adducts formed by transplatin. The other type contained exclusively intrastrand CLs [the interstrand crosslinked fragments were removed from the sample globally modified by transplatin by separation on a denaturing agarose gel; monofunctional adducts were removed from fragments containing no interstrand CL by reaction with thiourea (14–16)].
After alkaline denaturation, the DNA samples were primed with 20-mer primer (Fig. 1 B) and new DNA was synthesized by Sequenase 2 in the presence of [α-32P]dATP and unlabeled dNTPs. The products of DNA polymerization were separated by electrophoresis on a 6% polyacrylamide/8 M urea denaturing gel in parallel to a sequence ladder obtained using an untreated control DNA.
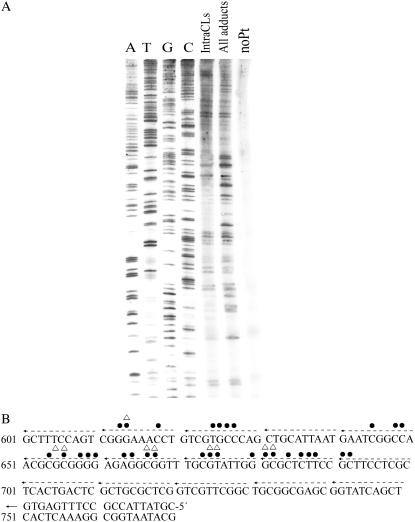
The strong and medium intensity bands observed specifically for the template containing exclusively intrastrand CLs (Fig. 6 A, lane Intra CLs) were taken to indicate the sites of preferential formation of the intrastrand CLs of transplatin. Importantly, they all occurred at or in immediate proximity to the sequences, such as GGG or GCG (positions 612–614, 653–657, 665–667, 672–674, 681–683 (Fig. 6 B)), i.e., at the sequences in which the yield of the rearrangement of intrastrand CLs of transplatin into interstrand CLs was low (Table 1). Importantly, no bands were observed at the sequences, such as GTG or GAG (positions 623–625, 660–664), in which the yield of the rearrangement of intrastrand CLs of transplatin into interstrand CLs was relatively high (Table 1). On the other hand, many additional strong or medium intensity bands were observed for the template containing all types of transplatin adducts (i.e., besides intrastrand also interstrand CLs and monofunctional adducts) indicating small sequence selectivity of DNA binding of transplatin (G and C stop sites) (Fig. 6 A, lane All adducts, and B) in accord with the results published previously (14).
FIGURE 6.
(A) Autoradiogram of a 6% polyacrylamide/8 M urea sequencing gel showing inhibition of DNA synthesis by Sequenase 2 on transplatin-modified (AflIII/EcoRI) fragments 410 bp long. After reaction with transplatin at 37°C in 10 mM NaClO4 for 24 h, rb = 0.001, one half of the sample was left so that it contained all types of DNA adducts of transplatin. The fragments contained in the other half were further separated by gel electrophoresis, so that all interstrand cross-linked fragments were removed and the fraction of fragments containing no interstrand cross-links was still treated with thiourea to remove monofunctional adducts. Thus, the other half of the fragments treated in this way contained exclusively intrastrand cross-links (for other details, see the text). Lanes: noPt, relative to the unplatinated template; All adducts, relative to the DNA globally modified by transplatin containing all types of transplatin adducts; IntraCLs, relative to the DNA containing exclusively intrastrand cross-links; A, T, G, and C are relative to chain terminated marker DNAs. (B) Schematic diagram showing a portion of the sequence used to monitor inhibition of DNA synthesis by transplatin. The arrow indicates the start site of Sequenase 2. • and Δ, stop signals from panel A. •, all types of adducts; Δ, intrastrand cross-links. Numbers correspond to nucleotide numbering on the pUC19 nucleotide sequence map.
DISCUSSION
Clinically ineffective transplatin is often used in studies investigating the structure-pharmacological activity relationship of platinum compounds (4). In addition, a number of transplatin analogs exhibit antitumor effects comparable to cisplatin or even promising toxic effects in several cisplatin resistant tumor cell lines (6,53). Also importantly, transplatin-modified oligonucleotides have been shown to be effective modulators of gene expression (19,20,54). These facts along with the generally accepted view that DNA is the major pharmacological target of platinum antitumor complexes are impetus for the studies of interactions between transplatin and DNA.
Bifunctional platinum compounds, such as transplatin, cisplatin and their analogs, form on DNA various types of CLs. These CLs are formed in a two-step process (55). In the first step, the monofunctional adducts are formed preferentially at guanine residues that subsequently close to the CLs. Several studies (17,19,56) have concluded that the major bifunctional adducts formed by transplatin in double-helical DNA are interstrand CLs and that intrastrand CLs are not formed. It has been proposed (17,56) that stable intrastrand CLs (1,3-GNG intrastrand CLs) are only formed in the reaction between single-stranded DNA and transplatin and that these intrastrand CLs rearrange into interstrand CLs as soon as the platinated single-stranded DNA is paired with its complementary strand. The conclusion that transplatin forms in double-helical DNA no intrastrand CLs was drawn on the basis of the results showing instability of the 1,3-intrastrand CLs formed by transplatin only in a relatively small number of selected sequence contexts (17,56,57). On the other hand, earlier work (14–16) has indicated that transplatin forms directly in double-helical DNA beside monofunctional adducts and a relatively small amount of interstrand CLs (∼12%) also a considerable amount of 1,3-GNG intrastrand CLs.
We show in this study that transplatin forms in double-helical DNA 1,3-GNG intrastrand CLs, but their stability depends on the sequence context (Table 1). We confirm that in the nucleotide sequences tested in the previous work (17,56,57) the 1,3-GNG intrastrand CLs of transplatin readily rearrange in double-helical DNA into interstrand CLs. On the other hand, we also show that in a number of other sequences these CLs are relatively stable. This conclusion is drawn on the basis of the results obtained not only with short synthetic oligodeoxyribonucleotide duplexes (Fig. 2, Table 1), but also with natural DNA with a random sequence (Fig. 6).
We also tested the hypothesis that the sequence-dependent stability of 1,3-GNG intrastrand CLs of transplatin in double-helical DNA and their propensity to rearrange into interstrand CLs is dependent on the extent of local conformational distortion and consequent destabilization of double-helical DNA by this intrastrand adduct. In other words, we examined whether a more extensive local destabilization of double-helical DNA due to the 1,3-GNG intrastrand CL may facilitate its rearrangement into interstrand CLs. An analysis of conformational distortions induced in DNA by 1,3-GNG intrastrand CLs of transplatin carried out in this study showed substantial differences in the character of these distortions if the CL was formed in the TGTGT or CGCGC sequence (Figs. 3 and 4), i.e., in the sequences in which stability of the intrastrand CLs in double-helical DNA is low or high, respectively (Table 1). The analysis by chemical probes of DNA conformation showed (Fig. 4) that the distortion induced in the sequence TGTGT extended over at least 7 bp, whereas the distortion induced in the sequence CGCGC was less extensive and markedly less pronounced.
Consistent with this observation were the results of the ITC analysis (Fig. 5 and Table 2). The association constants, K, for the formation of duplexes TGTGT(15) and CGCGC(15) containing 1,3-GNG intrastrand CLs of transplatin were 286 or 4 times lower, respectively, than the K value for the formation of the control (nonmodified) duplexes (Table 2). Hence, the 1,3-intrastrand CLs formed in both TGTGT or CGCGC sequences thermodynamically destabilized DNA, with this destabilization being enthalpic in origin. Importantly, the formation of the interstrand CL of transplatin induces no marked changes not only in enthalpy, but also in transition entropy (38). Thus, in contrast to intrastrand CLs, interstrand CLs of transplatin structurally perturb DNA in a way that does not lead to the pronounced thermodynamic destabilization consistent with conformational analysis of the interstrand CL of transplatin carried out by the methods of molecular biophysics (27). Therefore, it seems reasonable to suggest that a driving force behind the process involving rearrangement of the 1,3-GNG intrastrand CLs formed by transplatin in double-helical DNA to interstrand CLs is thermodynamic destabilization imposed on DNA by the 1,3-GNG intrastrand CL formed by transplatin in a respective nucleotide sequence context.
In aggregate, the 1,3-GTG intrastrand CL formed in the TGTGT sequence destabilized DNA considerably more than the CL formed in the CGCGC sequence, in which this CL exhibits a markedly higher stability (Table 1). Hence, the results of the ITC analysis are consistent with the view and support the hypothesis that the reduced propensity of the 1,3-GNG intrastrand CL of transplatin in the CGCGC sequence to rearrange into an interstrand CL is associated with its markedly lowered efficiency to destabilize thermodynamically DNA in contrast to the CL formed in the TGTGT sequence.
The data in this study provide new information on DNA interactions of clinically ineffective transplatin, which is used frequently in studies of structure-pharmacological relationship of platinum complexes as a counterpart of antitumor cisplatin. In addition, this study has provided valuable information for the design of new antitumor analogs of transplatin. Notably, a number of new analogs of transplatin, which we believe shows promising toxicity in various tumor cell lines, form in double-helical DNA a large number of stable intrastrand CLs. For instance, trans-[PtCl2(NH3)(piperidine)], trans-[PtCl2(NH3)(piperazine)], trans-[PtCl2(NH3)(4-hydroxymethylpyridine)], trans-[PtCl2(4-picoline)(piperidine)], and trans-[PtCl2(4-picoline)(piperazine)] · HCl form in DNA after 24 h at 37°C 60%–90% stable intrastrand CLs (23–25). Thus, it cannot be excluded that the capability of these transplatinum complexes to form in double-helical DNA a relatively high number of stable intrastrand CLs might be one of the factors significantly contributing to the activation of trans geometry in platinum coordination complexes. This work suggests that an enhanced frequency of intrastrand CLs yielded by these new transplatinum complexes might be a consequence of the fact that intrastrand CLs of these new analogs of transplatin thermodynamically destabilize DNA in far less sequence contexts than inefficient parent transplatin. Another eventuality might be that interstrand CLs of these new transplatinum complexes distort and consequently thermodynamically destabilize DNA markedly more than the same adducts of parent transplatin, which could also make the rearrangement of intrastrand CLs into interstrand CLs difficult. The work aimed at testing these intriguing hypotheses, positive testing of which would rationalize the design of new antitumor drugs based on transplatin analogs, is in progress in our laboratory and the results will be reported in further communication. Our findings may also be useful for the design of transplatin-modified oligonucleotides as modulators of gene expression emphasizing a need to carefully design nucleotide sequence contexts in these oligonucleotides for the platination.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
J.K. is the international research scholar of the Howard Hughes Medical Institute. The authors acknowledge that their participation in the EU COST Action D39 enabled them to exchange regularly the most recent ideas in the field of anticancer metallodrugs with several European colleagues.
This research was supported by the Ministry of Education of the CR (MSMT LC06030, 6198959216, ME08017, OC08003), the Academy of Sciences of the Czech Republic (1QS500040581, KAN200200651, AV0Z50040507 and AV0Z50040702), the Grant Agency of the Academy or Sciences of the CR (IAA400040803) and the Grant Agency of the Ministry of Health of the Czech Republic (NR8562-4/2005).
Editor: Jonathan B. Chaires.
References
- 1.Rosenberg, B., L. Van Camp, J. E. Trosko, and V. H. Mansour. 1969. Platinum compounds: a new class of potent antitumor agents. Nature. 222:385–386. [DOI] [PubMed] [Google Scholar]
- 2.Vrana, O., V. Brabec, and V. Kleinwächter. 1986. Polarographic studies on the conformation of some platinum complexes: relations to anti-tumour activity. Anticancer Drug Des. 1:95–109. [PubMed] [Google Scholar]
- 3.Farrell, N., L. R. Kelland, J. D. Roberts, and M. Van Beusichem. 1992. Activation of the trans geometry in platinum antitumor complexes: a survey of the cytotoxicity of trans complexes containing planar ligands in murine-L1210 and human tumor panels and studies on their mechanism of action. Cancer Res. 52:5065–5072. [PubMed] [Google Scholar]
- 4.Brabec, V., V. Kleinwächter, J. L. Butour, and N. P. Johnson. 1990. Biophysical studies of the modification of DNA by antitumor platinum coordination complexes. Biophys. Chem. 35:129–141. [DOI] [PubMed] [Google Scholar]
- 5.Reedijk, J. 1996. Improved understanding in platinum antitumor chemistry. Chem. Commun. 801–806.
- 6.Farrell, N. 1996. Current status of structure-activity relationships of platinum anticancer drugs: activation of the trans geometry. In Metal Ions in Biological Systems. A. Sigel and H. Sigel, editors. Marcel Dekker, New York, Basel, Hong Kong. 603–639. [PubMed]
- 7.Kelland, L. R., S. Y. Sharp, C. F. O. Neill, F. I. Raynaud, P. J. Beale, and I. R. Judson. 1999. Mini-review: discovery and development of platinum complexes designed to circumvent cisplatin resistance. J. Inorg. Biochem. 77:111–115. [PubMed] [Google Scholar]
- 8.Brabec, V. 2002. DNA modifications by antitumor platinum and ruthenium compounds: their recognition and repair. Prog. Nucleic Acid Res. Mol. Biol. 71:1–68. [DOI] [PubMed] [Google Scholar]
- 9.Natile, G., and M. Coluccia. 2004. Antitumor active trans-platinum compounds. In Metal Ions in Biological Systems. A. Sigel and H. Sigel, editors. Marcel Dekker, New York, Basel. 209–250. [PubMed]
- 10.Brabec, V., and J. Kasparkova. 2005. Modifications of DNA by platinum complexes: relation to resistance of tumors to platinum antitumor drugs. Drug Resist. Updat. 8:131–146. [DOI] [PubMed] [Google Scholar]
- 11.Jamieson, E. R., and S. J. Lippard. 1999. Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 99:2467–2498. [DOI] [PubMed] [Google Scholar]
- 12.Siddik, Z. H. 2003. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 22:7265–7279. [DOI] [PubMed] [Google Scholar]
- 13.Wang, D., and S. J. Lippard. 2005. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 4:307–320. [DOI] [PubMed] [Google Scholar]
- 14.Brabec, V., and M. Leng. 1993. DNA interstrand cross-links of trans-diamminedichloroplatinum(II) are preferentially formed between guanine and complementary cytosine residues. Proc. Natl. Acad. Sci. USA. 90:5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eastman, A., M. M. Jennerwein, and D. L. Nagel. 1988. Characterization of bifunctional adducts produced in DNA by trans-diamminedichloroplatinum(II). Chem. Biol. Interact. 67:71–80. [DOI] [PubMed] [Google Scholar]
- 16.Eastman, A., and M. A. Barry. 1987. Interaction of trans-diamminedichloroplatinum(II) with DNA: formation of monofunctional adducts and their reaction with glutathione. Biochemistry. 26:3303–3307. [DOI] [PubMed] [Google Scholar]
- 17.Boudvillain, M., R. Dalbies, C. Aussourd, and M. Leng. 1995. Intrastrand cross-links are not formed in the reaction between transplatin and native DNA: relation with the clinical inefficiency of transplatin. Nucleic Acids Res. 23:2381–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leng, M., A. Schwartz, and M. J. Giraud-Panis. 2000. Transplatin-modified oligonucleotides as potential antitumor drugs. In Platinum-Based Drugs in Cancer Therapy. L. R. Kelland and N. P. Farrell, editors. Humana Press, Totowa, New Jersey. 63–85.
- 19.Giraud-Panis, M.-J., and M. Leng. 2000. Transplatin-modified oligonucleotides as modulators of gene expression. Pharmacol. Ther. 85:175–181. [DOI] [PubMed] [Google Scholar]
- 20.Aupeix-Scheidler, K., S. Chabas, L. Bidou, J.-P. Rousset, M. Leng, and J.-J. Toulme. 2000. Inhibition of in vitro and ex vivo translation by a transplatin-modified oligo(2′-O-methylribonucleotide) directed against the HIV-1 gag-pol frameshift signal. Nucleic Acids Res. 28:438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasparkova, J., O. Novakova, N. Farrell, and V. Brabec. 2003. DNA binding by antitumor trans-[PtCl2(NH3)(thiazole)]. Protein recognition and nucleotide excision repair of monofunctional adducts. Biochemistry. 42:792–800. [DOI] [PubMed] [Google Scholar]
- 22.Brabec, V., K. Neplechova, J. Kasparkova, and N. Farrell. 2000. Steric control of DNA interstrand cross-link sites of trans platinum complexes: specificity can be dictated by planar nonleaving groups. J. Biol. Inorg. Chem. 5:364–368. [DOI] [PubMed] [Google Scholar]
- 23.Kasparkova, J., V. Marini, Y. Najajreh, D. Gibson, and V. Brabec. 2003. DNA binding mode of the cis and trans geometries of new antitumor nonclassical platinum complexes containing piperidine, piperazine or 4-picoline ligand in cell-free media. Relations to their activity in cancer cell lines. Biochemistry. 42:6321–6332. [DOI] [PubMed] [Google Scholar]
- 24.Stehlikova, K., J. Kasparkova, O. Novakova, A. Martinez, V. Moreno, and V. Brabec. 2006. Recognition of DNA modified by trans-[PtCl2NH3(4-hydroxymethylpyridine)] by tumor suppressor protein p53 and character of DNA adducts of this cytotoxic complex. FEBS J. 273:301–314. [DOI] [PubMed] [Google Scholar]
- 25.Najajreh, Y., J. Kasparkova, V. Marini, D. Gibson, and V. Brabec. 2005. Structural characterization and DNA interactions of new cytotoxic transplatin analogues containing one planar and one nonplanar heterocyclic amine ligand. J. Biol. Inorg. Chem. 10:722–731. [DOI] [PubMed] [Google Scholar]
- 26.Comess, K. M., C. E. Costello, and S. J. Lippard. 1990. Identification and characterization of a novel linkage isomerization in the reaction of trans-diamminedichloroplatinum(II) with 5′-d(TCTACGCGTTCT). Biochemistry. 29:2102–2110. [DOI] [PubMed] [Google Scholar]
- 27.Brabec, V., M. Sip, and M. Leng. 1993. DNA conformational distortion produced by site-specific interstrand cross-link of trans-diamminedichloroplatinum(II). Biochemistry. 32:11676–11681. [DOI] [PubMed] [Google Scholar]
- 28.Brabec, V., J. Reedijk, and M. Leng. 1992. Sequence-dependent distortions induced in DNA by monofunctional platinum(II) binding. Biochemistry. 31:12397–12402. [DOI] [PubMed] [Google Scholar]
- 29.Yoon, C., M. D. Kuwabara, R. Law, R. Wall, and D. S. Sigman. 1988. Sequence-dependent variability of DNA structure. J. Biol. Chem. 263:8458–8463. [PubMed] [Google Scholar]
- 30.Schwartz, A., L. Marrot, and M. Leng. 1989. Conformation of DNA modified at a d(GG) or a d(AG) site by the antitumor drug cis-diamminedichloroplatinum(II). Biochemistry. 28:7975–7978. [DOI] [PubMed] [Google Scholar]
- 31.Bailly, C., D. Gentle, F. Hamy, M. Purcell, and M. J. Waring. 1994. Localized chemical reactivity in DNA associated with the sequence-specific bis intercalation of echinomycin. Biochem. J. 300:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, S. A., and C. J. Burrows. 1996. Cytosine-specific chemical probing of DNA using bromide and monoperoxysulfate. Nucleic Acids Res. 24:5062–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailly, C., and M. J. Waring. 1997. Diethylpyrocarbonate and osmium tetroxide as probes for drug-induced changes in DNA conformation in vitro. In Drug-DNA Interaction Protocols. K. R. Fox, editor. Humana Press, Totowa, New Jersey. 51–79. [DOI] [PubMed]
- 34.Hofr, C., N. Farrell, and V. Brabec. 2001. Thermodynamic properties of duplex DNA containing a site-specific d(GpG) intrastrand crosslink formed by an antitumor dinuclear platinum complex. Nucleic Acids Res. 29:2034–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofr, C., and V. Brabec. 2001. Thermal and thermodynamic properties of duplex DNA containing site-specific interstrand cross-link of antitumor cisplatin or its clinically ineffective trans isomer. J. Biol. Chem. 276:9655–9661. [DOI] [PubMed] [Google Scholar]
- 36.Bursova, V., J. Kasparkova, C. Hofr, and V. Brabec. 2005. Effects of monofunctional adducts of Platinum(II) complexes on thermodynamic stability and energetics of DNA duplexes. Biophys. J. 88:1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novakova, O., H. Chen, O. Vrana, A. Rodger, P. J. Sadler, and V. Brabec. 2003. DNA interactions of monofunctional organometallic ruthenium(II) antitumor complexes in cell-free media. Biochemistry. 42:11544–11554. [DOI] [PubMed] [Google Scholar]
- 38.Hofr, C., and V. Brabec. 2005. Thermal stability and energetics of 15-mer DNA duplex interstrand cross-linked by trans-diamminedichloroplatinum(II). Biopolymers. 77:222–229. [DOI] [PubMed] [Google Scholar]
- 39.Murphy, J. H., and T. L. Trapane. 1996. Concentration and extinction coefficient determination for oligonucleotides and analogs using a general phosphate analysis. Anal. Biochem. 240:273–282. [DOI] [PubMed] [Google Scholar]
- 40.Burstyn, J. N., W. J. HeigerBernays, S. M. Cohen, and S. J. Lippard. 2000. Formation of cis-diamminedichloroplatinum(II) 1,2-intrastrand cross-links on DNA is flanking-sequence independent. Nucleic Acids Res. 28:4237–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Intini, F. P., A. Boccarelli, V. C. Francia, C. Pacifico, M. F. Sivo, G. Natile, D. Giordano, P. De Rinaldis, and M. Coluccia. 2004. Platinum complexes with imino ethers or cyclic ligands mimicking imino ethers: synthesis, in vitro antitumor activity, and DNA interaction properties. J. Biol. Inorg. Chem. 9:768–780. [DOI] [PubMed] [Google Scholar]
- 42.Dalbies, R., D. Payet, and M. Leng. 1994. DNA double helix promotes a linkage isomerization reaction in trans-diamminedichloroplatinum(II)-modified DNA. Proc. Natl. Acad. Sci. USA. 91:8147–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigman, D. S., and C. H. B. Chen. 1990. Chemical nucleases—new reagents in molecular biology. Annu. Rev. Biochem. 59:207–236. [DOI] [PubMed] [Google Scholar]
- 44.Sigman, D. S. 1990. Chemical nucleases. Biochemistry. 29:9097–9105. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen, P. E. 1990. Chemical and photochemical probing of DNA complexes. J. Mol. Recognit. 3:1–24. [DOI] [PubMed] [Google Scholar]
- 46.Kasparkova, J., N. Farrell, and V. Brabec. 2000. Sequence specificity, conformation, and recognition by HMG1 protein of major DNA interstrand cross-links of antitumor dinuclear platinum complexes. J. Biol. Chem. 275:15789–15798. [DOI] [PubMed] [Google Scholar]
- 47.Pilch, D. S., S. U. Dunham, E. R. Jamieson, S. J. Lippard, and K. J. Breslauer. 2000. DNA sequence context modulates the impact of a cisplatin 1,2-d(GpG) intrastrand cross-link an the conformational and thermodynamic properties of duplex DNA. J. Mol. Biol. 296:803–812. [DOI] [PubMed] [Google Scholar]
- 48.Malina, J., C. Hofr, L. Maresca, G. Natile, and V. Brabec. 2000. DNA interactions of antitumor cisplatin analogs containing enantiomeric amine ligands. Biophys. J. 78:2008–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novakova, O., J. Kasparkova, V. Bursova, C. Hofr, M. Vojtiskova, H. Chen, P. J. Sadler, and V. Brabec. 2005. Conformation of DNA modified by monofunctional Ru(II) arene complexes: recognition by DNA-binding proteins and repair. Relationship to cytotoxicity. Chem. Biol. 12:121–129. [DOI] [PubMed] [Google Scholar]
- 50.Comess, K. M., J. N. Burstyn, J. M. Essigmann, and S. J. Lippard. 1992. Replication inhibition and translesion synthesis on templates containing site-specifically placed cis-diamminedichloroplatinum(II) DNA adducts. Biochemistry. 31:3975–3990. [DOI] [PubMed] [Google Scholar]
- 51.Novakova, O., J. Kasparkova, J. Malina, G. Natile, and V. Brabec. 2003. DNA-protein cross-linking by trans-[PtCl2(E-iminoether)2]. A concept for activation of the trans geometry in platinum antitumor complexes. Nucleic Acids Res. 31:6450–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriarity, B., O. Novakova, N. Farrell, V. Brabec, and J. Kasparkova. 2007. 1,2-GG intrastrand cross-link of antitumor dinuclear bifunctional platinum compound with spermidine linker inhibits DNA polymerization more effectively than the cross-link of conventional cisplatin. Arch. Biochem. Biophys. 459:264–272. [DOI] [PubMed] [Google Scholar]
- 53.Coluccia, M., and G. Natile. 2007. Trans-platinum complexes in cancer therapy. Anti-Cancer Agents Med. Chem. 7:111–123. [DOI] [PubMed] [Google Scholar]
- 54.Gee, J. E., I. Robbins, A. C. VanderLaan, J. H. VanBoom, C. Colombier, M. Leng, A. M. Raible, J. S. Nelson, and B. Lebleu. 1998. Assessment of high-affinity hybridization, RNase H cleavage, and covalent linkage in translation arrest by antisense oligonucleotides. Antisense Nucleic Acid Drug Dev. 8:103–111. [DOI] [PubMed] [Google Scholar]
- 55.Bancroft, D. P., C. A. Lepre, and S. J. Lippard. 1990. Pt-195 NMR kinetic and mechanistic studies of cis-diamminedichloroplatinum and trans-diamminedichloroplatinum(II) binding to DNA. J. Am. Chem. Soc. 112:6860–6871. [Google Scholar]
- 56.Dalbies, R., M. Boudvillain, and M. Leng. 1995. Linkage isomerization reaction of intrastrand cross-links in trans-diamminedichloroplatinum(II)-modified single-stranded oligonucleotides. Nucleic Acids Res. 23:949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leng, M., D. Locker, M. J. Giraud-Panis, A. Schwartz, F. P. Intini, G. Natile, C. Pisano, A. Boccarelli, D. Giordano, and M. Coluccia. 2000. Replacement of an NH3 by an iminoether in transplatin makes an antitumor drug from an inactive compound. Mol. Pharmacol. 58:1525–1535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.