Abstract
The spindle assembly checkpoint (SAC) ensures faithful chromosome segregation by delaying anaphase onset until all sister kinetochores are attached to bipolar spindles. An RNA interference screen for synthetic genetic interactors with a conserved SAC gene, san-1/MAD3, identified spdl-1, a Caenorhabditis elegans homologue of Spindly. SPDL-1 protein localizes to the kinetochore from prometaphase to metaphase, and this depends on KNL-1, a highly conserved kinetochore protein, and CZW-1/ZW10, a component of the ROD–ZW10–ZWILCH complex. In two-cell–stage embryos harboring abnormal monopolar spindles, SPDL-1 is required to induce the SAC-dependent mitotic delay and localizes the SAC protein MDF-1/MAD1 to the kinetochore facing away from the spindle pole. In addition, SPDL-1 coimmunoprecipitates with MDF-1/MAD1 in vivo. These results suggest that SPDL-1 functions in a kinetochore receptor of MDF-1/MAD1 to induce SAC function.
Introduction
To ensure faithful chromosome segregation, the spindle assembly checkpoint (SAC) monitors the status of kinetochore–microtubule binding and inhibits activity of the anaphase-promoting complex/cyclosome (APC/CCDC20), thereby delaying anaphase onset until all sister kinetochores have properly attached to bipolar mitotic spindles (for review see May and Hardwick, 2006; Musacchio and Salmon, 2007).
The SAC signaling pathway is mediated by highly conserved proteins such as MAD1-3, BUB1, and BUB3, first identified by two independent genetic screens in budding yeast (Hoyt et al., 1991; Li and Murray, 1991). These proteins temporally associate with kinetochores that have not achieved bipolar attachment. MAD1 specifically localizes and recruits MAD2 to microtubule-free kinetochores and facilitates binding of MAD2 and CDC20, an APC/C activator, thereby inhibiting APC/C (Sironi et al., 2001; De Antoni et al., 2005). However, how MAD1 is specifically targeted to unattached kinetochores is yet unanswered. In metazoan cells, the ROD–ZW10–ZWILCH (RZZ) complex is crucial in the SAC pathway (for review see Karess, 2005). The RZZ complex recruits dynein/dynactin to kinetochores (Starr et al., 1998). Although the RZZ complex is required to regulate levels of MAD1 and MAD2 at unattached kinetochores (Kops et al., 2005), it localizes to not only microtubule-free but also tension-free kinetochores (Famulski and Chan, 2007).
In Caenorhabditis elegans, MDF-1/MAD1 is required for SAC-dependent delay of mitotic progression induced by chemical or mutational disruption of microtubules in proliferating germ cells (Kitagawa and Rose, 1999) and early-stage embryogenesis (Nystul et al., 2003; Encalada et al., 2005). However, MDF-1 loss hardly affects cell cycle progression during early embryogenesis.
SAN-1/MAD3 is also essential for microtubule defect– or anoxia-induced mitotic delay in early-stage embryos (Nystul et al., 2003) but not for animal viability in C. elegans; deletion of san-1 causes no severe developmental defects (Stein et al., 2007). In contrast, despite being dispensable during early embryogenesis, C. elegans strains carrying deletion mutations in mdf-1 or mdf-2/MAD2 exhibit severe defects in larval and germ cell development (Kitagawa and Rose, 1999). Lethality of the mdf-1 deletion strain is suppressed by reduction of APC/C activity (Furuta et al., 2000; Kitagawa et al., 2002; Tarailo et al., 2007a), suggesting that MDF-1 regulates APC/CCDC20 activity during development. The defect in metaphase-to-anaphase transition in meiosis I caused by APC/C mutants can be suppressed by hypomorphic mutations in mdf-1, mdf-2, or san-1, suggesting that these SAC components are required to regulate APC/C activity during meiosis (Stein et al., 2007).
An RNAi-based genomewide screen for synthetic genetic interactors with known SAC components identified spdl-1, which is required to activate the SAC pathway, particularly MDF-1–mediated signaling pathways. Our findings suggest that SPDL-1 functions in the SAC pathway as a constituent of the kinetochore receptor for MDF-1 in C. elegans.
Results and discussion
SPDL-1, identified as a san-1/MAD3 synthetic genetic interactor, is required for SAC activation
The RNAi screen for genes whose depletion caused phenotypes that were embryonic lethal or had reduced number of progeny in combination with san-1 deletion (san-1Δ) by using Ahringer's RNAi feeding library (Kamath et al., 2003) identified known components of spindles, kinetochore, and cohesion as well as other SAC components (unpublished data). Thus, san-1 synthetic lethal genes include genes whose depletion activates the SAC and those required for SAC activation. (To avoid confusion caused by the inconsistency of nomenclature among organisms, names for C. elegans genes used in this study are listed in Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200805185/DC1.)
Uncharacterized san-1 synthetic lethal genes were further screened for genes whose depletion by RNAi bypasses the mitotic delay induced by ZYG-1 deficiency in two-cell–stage embryos (see Materials and methods). The mitotic delay in ZYG-1–depleted embryos was bypassed by codepletion of MDF-1 (Fig. 1 A), indicating the delay was SAC dependent. This secondary screen identified C06A8.5 (Fig. 1 A), which encodes a protein-sharing sequence similarity with Spindly family proteins (Fig. S1 B; Cheeseman and Desai, 2008). Although our finding that SAC activation needs C06A8.5 does not support that it behaves as an orthologue of Spindly, which silences the SAC rather than activating it (Griffis et al., 2007), we designated this gene spdl-1 as assigned by the Caenorhabditis Genetic Center to be consistent in gene naming.
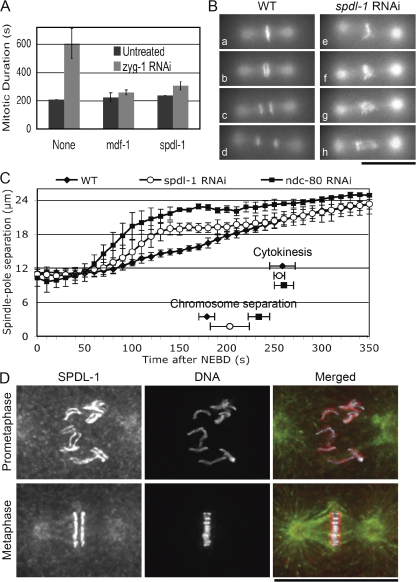
Figure 1.
SPDL-1 is required for proper chromosome segregation and SAC activation. (A) Mitotic duration from NEBD to chromosome decondensation was measured in AB cells of wild-type embryos dissected from adult hermaphrodites soaked with dsRNA of indicated genes alone (Untreated) or in combination with zyg-1 dsRNA (zyg-1 RNAi). Depletion of MDF-1 or SPDL-1 bypassed the ZYG-1 depletion–induced mitotic delay. (B) One-cell–stage embryos expressing GFP–histone H2B and GFP–tubulin were dissected before undergoing first mitosis from adult hermaphrodites injected with buffer (WT) or with spdl-1 dsRNA (spdl-1 RNAi) and studied by time-lapse fluorescence microscopy. Still images of embryos at 10 s before (a and e) and 0 (b and f), 20 (c and g), and 70 s (d and h) after the onset of anaphase (WT) or of anaphase-like separation of chromosome masses (spdl-1 RNAi) are shown. Bar, 20 μm. (C) Kinetics of centrosome separation in embryos dissected from wild-type adult hermaphrodites untreated (WT) or injected with dsRNA of indicated genes. Time at NEBD was set as 0 s. SPDL-1 depletion caused moderate acceleration of centrosome separation. Timing of chromosome separation and onset of cytokinesis are shown. Mean number of data obtained from at least three individual embryos are plotted and standard deviations are shown as error bars. (D) Immunofluorescence images of fixed wild-type embryos at indicated stages of the first mitosis. DNA (white), tubulin (green), and SPDL-1 (red) were stained with DAPI, anti-tubulin, and anti–SPDL-1 antibodies, respectively. SPDL-1 colocalizes to microtubules through mitosis and also temporarily localizes to kinetochores from prometaphase until anaphase. Bar, 20 μm.
RNAi of spdl-1 caused synthetic lethality with san-1Δ but did not affect viability of the mdf-1 mutant allele or the mdf-2Δ allele (Fig. S1 C). This result was in contrast to that obtained for other candidates of the screen, such as HCP-1, BUB-3, or GOA-1, whose depletion was synthetically lethal with not only san-1Δ but also the mutant allele of mdf-1 or mdf-2Δ (Fig. S1 C). MDF-1 depletion was also lethal with san-1Δ but not with the mdf-1 mutant or the mdf-2Δ allele (Fig. S1 C), suggesting that SPDL-1 functions in an MDF-1–mediated pathway.
RNAi by feeding the bacterial clone expressing spdl-1 double-stranded RNA (dsRNA) only partially depleted SPDL-1, with little effect on brood size or number of progeny of wild-type worms, but considerably reduced viability of progeny in the san-1Δ strain (Fig. S1 C). However, microinjection with spdl-1 dsRNA into gonad arms of wild-type adult hermaphrodites caused substantial reduction of SPDL-1 (Fig. S1 D) and the embryonic lethal phenotype at high penetrance (not depicted). Thus, SPDL-1 is indispensable for embryogenesis.
During chromosome segregation in embryos expressing GFP–histone H2B and GFP–β-tubulin, depletion of SPDL-1 caused severe chromosome missegregation at the first mitosis (Fig. 1 B). In SPDL-1–depleted embryos, chromosome congression was incomplete and sister chromatids being separated were often connected by lagging chromosomes. Regardless of the presence of lagging chromosomes, cytokinesis occurred at the same time as in untreated embryos (Fig. 1 C), leading to karyomere formation at a frequency similar to that reported previously (Sonnichsen et al., 2005). Time course analysis revealed that depletion of SPDL-1 induced rapid and premature centrosome separation, which reflects a defect in kinetochore–microtubule attachment (Fig. 1 C), suggesting that SPDL-1 plays a role in proper kinetochore–microtubule attachment in addition to the SAC function. However, the defect was less severe than that caused by depletion of NDC-80, a constituent of the core microtubule-binding site (Fig. 1 C; Desai et al., 2003).
We next tested the genetic interaction of SPDL-1 and the APC/C, the downstream target of the SAC. At 24°C, mat-3/APC8 temperature-sensitive allele, mat-3(or180) (referred to as mat-3ts), causes embryonic arrest at metaphase of meiosis I at high penetrance (Table I; Golden et al., 2000). When mat-3ts embryos were depleted of SPDL-1, they exited meiosis and entered the mitotic cycle with abnormal chromosome segregation, resulting in arrested embryos with multiple cells, which resembles the previously described suppression effect of the mdf-1Δ allele on the mat-3ts phenotype (Tables I and II; Stein et al., 2007). Thus, SPDL-1 depletion bypassed the meiosis I metaphase arrest of mat-3ts, possibly by decreasing the SAC activity to inhibit the APC/C.
Table I. Suppression of the embryonic lethality of mat-3(or180) by RNAi-mediated depletion of SAC genes.
| RNAi | Total | Hatched | Ratio of hatching |
|---|---|---|---|
| % | |||
| pUC19 | 342 | 10 | 2.9 |
| mdf-1 | 263 | 9 | 3.4 |
| san-1 | 330 | 115 | 34.8 |
| bub-3 | 367 | 139 | 37.9 |
| spdl-1 | 331 | 0 | 0 |
Three L4 mat-3(or180) hermaphrodites fed with HT115 bacteria expressing dsRNA of indicated genes were shifted to 24°C for 36 h, and then the total number of eggs laid (Total) and hatching (Hatched) were counted. Ratio of hatched to total embryos (Ratio of hatching) are shown.
Table II. Suppression of the meiotic arrest of mat-3(or180) by RNAi-mediated depletion of SAC genes.
| RNAi | One cell | More than one cell |
Ratio of worms laying embryos with more than one cell |
|---|---|---|---|
| % | |||
| pUC19 | 30 | 0 | 0 |
| mdf-1 | 12 | 18 | 60 |
| spdl-1 | 22 | 8 | 26.7 |
30 L4 mat-3(or180) hermaphrodites fed with HT115 bacteria expressing dsRNA of indicated genes were shifted to 24°C for 24 h, fixed with methanol and acetone, and then stained with 100 ng/ml DAPI. The numbers of worms that lay only embryos in meiotic division (one cell) and those that lay embryos that had entered mitosis (more than one cell) are shown.
The spdl-1 deletion allele spdl-1(ok1515) (referred to as spdl-1Δ) is a recessive lethal allele. The spdl-1Δ homozygotes that were segregated from spdl-1Δ hemizygotes survived through embryogenesis may be due to maternal contribution but showed larval lethal or sterile phenotype (unpublished data). Sterile spdl-1Δ homozygotes had oocytes that prematurely exited meiotic prophase, resulting in endomitosis (Emo phenotype; Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200805185/DC1; Iwasaki et al., 1996). Oocytes having extra chromosomes were also identified (Fig. S2, B and C). Sterility was suppressed by ectopic expression of GFP–SPDL-1 (unpublished data), confirming that the germline defect was caused by loss of SPDL-1 function. mdf-1Δ homozygotes also showed an abnormal number of bivalents in oocytes and the Emo phenotype (Kitagawa and Rose, 1999). As the sterile phenotype of mdf-1Δ homozygotes is suppressed by the temperature-sensitive allele of emb-30/APC4, emb-30(tn377) (referred to as emb-30ts; Furuta et al., 2000; Kitagawa et al., 2002; Tarailo et al., 2007a), we tested whether an emb-30ts mutation suppresses the sterile phenotype of spdl-1Δ homozygotes. In contrast to spdl-1Δ homozygotes that showed sterility at high penetrance, 90% of emb-30ts; spdl-1Δ homozygotes laid eggs (Fig. S2 D). However, these embryos were arrested, suggesting that emb-30ts mutation suppressed the oocyte maturation defect but not the embryonic lethality of spdl-1Δ homozygotes. These results strongly suggest that SPDL-1 plays a role in regulating APC/C activity during oogenesis presumably via its SAC function.
Kinetochore localization of SPDL-1 depends on KNL-1 and CZW-1 but not on NDC-80 or BUB-1
Immunofluorescence microscopy using a SPDL-1–specific antibody (Fig. S2 E) revealed that SPDL-1 localized to microtubules through mitosis and to kinetochores upon nuclear envelope breakdown (NEBD) until anaphase (Fig. 1 D and Fig. S2 F). We confirmed this localization pattern in living embryos expressing GFP–SPDL-1 in the spdl-1Δ background (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200805185/DC1).
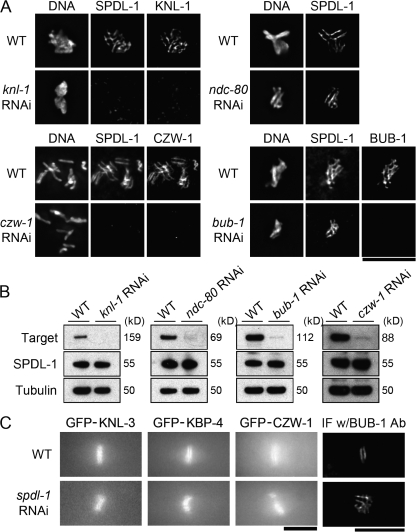
We analyzed the molecular dependency of kinetochore localization of SPDL-1 on other kinetochore proteins. RNAi-mediated depletion of KNL-1 (Desai et al., 2003), a core outer kinetochore component, or CZW-1/ZW10 (Starr et al., 1997), a component of the RZZ complex, significantly reduced the targeting of SPDL-1 on chromosomes (Fig. 2 A) without affecting localization of SPDL-1 on microtubules (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200805185/DC1) and total amount of cellular SPDL-1 in whole-worm lysate (Fig. 2 B), suggesting that both are specifically required for SPDL-1 targeting to kinetochores. However, in embryos depleted substantially of endogenous BUB-1 (Oegema et al., 2001) or NDC-80 (Fig. 2 B and Fig. S3 B), SPDL-1 remained associated with chromosomes (Fig. 2 A and Fig. S3 A).
Figure 2.
Molecular dependency of kinetochore localization of SPDL-1 and other kinetochore proteins. (A) Embryos dissected from adult hermaphrodites untreated (WT) or injected with dsRNA of indicated genes were fixed and analyzed by immunofluorescence microscopy. Images of embryos at metaphase of the first mitosis are shown. KNL-1 and CZW-1 are required but NDC-80 or BUB-1 are not for SPDL-1 targeting to kinetochores. Bar, 20 μm. (B) Lysate from wild-type worms untreated (WT) or injected with dsRNA of indicated genes were analyzed by Western blotting with antibody to indicated proteins. To each lane, whole-worm lysates from 20 injected worms was loaded. (C) Embryos expressing indicated protein fused with GFP were dissected from adult hermaphrodites untreated (WT) or injected with spdl-1 dsRNA and analyzed by time-lapse fluorescence microscopy. Images of embryos at metaphase of the first mitosis are shown. For BUB-1, wild-type embryos were dissected from wild-type adult hermaphrodites untreated (WT) or injected with spdl-1 dsRNA, fixed, and stained with anti–BUB-1 antibody. Kinetochore localization of proteins was not affected in SPDL-1–depleted embryos. Bars, 10 μm.
In reciprocal experiments, we observed kinetochore localization of KNL-3 (Cheeseman et al., 2004), a component of the MIS-12 complex; KBP-4 (Cheeseman et al., 2004), a component of the NDC-80 complex; CZW-1; and BUB-1 in SPDL-1–depleted embryos in metaphase (Fig. 2 C). Depletion of KNL-1 also reduced the targeting of CZW-1 on chromosomes (unpublished data). Together, SPDL-1 is positioned downstream of CZW-1 and KNL-1 in a linear hierarchical dependency of kinetochore assembly. In contrast, localizations of SPDL-1 and BUB-1 are independent of each other, even though both proteins are required for SAC activation in C. elegans embryos.
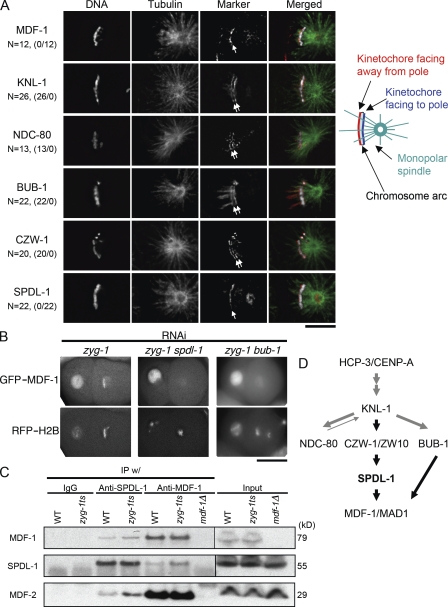
SPDL-1 localizes and targets MDF-1 to the kinetochores that face away from the pole in embryonic cells with monopolar spindle
We next tested whether depletion of SPDL-1 affects MDF-1 localization at unattached kinetochores. MDF-1–kinetochore association was barely detectable during normal mitosis (unpublished data). In two-cell–stage embryos depleted of ZYG-1, condensed chromosomes formed an arc around the single microtubule aster nucleated from the mono centrosome (Fig. 3 A). In these cells, MDF-1 accumulated along the outer side of the chromosome arc that faces away from the centrosome (Fig. 3 A). In contrast, KNL-1 and NDC-80 localized to both the inner and outer sides of the chromosome arc (Fig. 3 A). These results suggest that sister centromeres were resolved and the outer kinetochore was assembled on both sister chromatids, but one assembled on the side facing away from the centrosomes is not captured, or only laterally captured, by microtubules. Monotelic microtubule attachment could cause the lack of tension on both sister kinetochores. Consistent with the previous finding that BUB1 and the RZZ complex localize to tension-free kinetochores (Skoufias et al., 2001; Famulski and Chan, 2007), BUB-1 and CZW-1 localized as KNL-1 and NDC-80 did (Fig. 3 A). In contrast, SPDL-1 asymmetrically localized as MDF-1 did (Fig. 3 A). Furthermore, MDF-1 associating with chromosomes was substantially reduced by depletion of SPDL-1 (Fig. 3 B and Fig. S3 C). These observations strongly suggest that SPDL-1 is specifically required for MDF-1 targeting to unattached kinetochores. Although CZW-1 localized at both sister kinetochores in ZYG-1–depleted embryos (Fig. 3 A), codepletion of CZW-1 and ZYG-1 substantially reduced localization of SPDL-1 and MDF-1 on the kinetochores facing away from the centrosome (Fig. S3 D), suggesting that SPDL-1 requires CZW-1 for its accumulation on unattached kinetochores as it does in normal mitosis. Depletion of BUB-1 did not affect kinetochore localization of SPDL-1 (Fig. 2 A and Fig. S3 A) but reduced MDF-1 on kinetochores in ZYG-1–depleted embryos (Fig. 3 B). Thus, MDF-1 localization depends on two independent molecules, SPDL-1 and BUB-1.
Figure 3.
SPDL-1 localizes to and targets MDF-1 to kinetochores that face away from the mono centrosome. (A) Immunofluorescence images of AB or P1 cells of ZYG-1–depleted embryos in the second mitosis. Indicated proteins (red) were stained with each specific antibody. DNA (white) and tubulin (green) were also stained with DAPI and anti-tubulin antibody, respectively. The numbers before and after the slash in parentheses are, respectively, the numbers of cells in which the indicated protein localized at both sides of kinetochores and those of cells in which the indicated protein localized only at the outer side of the chromosome arc. KNL-1, NDC-80, BUB-1, and CZW-1 localize at both sides of the chromosomes along the longitudinal axis, showing two parallel bars (white arrows). In contrast, SPDL-1 and MDF-1 localize along the side of the chromosome that faces away from the centrosome (white arrows). KNL-1 and BUB-1 also localize on the plus-end terminus region of microtubules that extends beyond chromosomes. Bar, 10 μm. (B) Embryos expressing GFP–MDF-1 and mCherry–histone H2B (RFP–H2B) were depleted of the indicated genes and subjected to time-lapse fluorescence microscopy. Images of embryos at metaphase of the second mitosis in AB cells are shown. The anterior of the embryo is at the right. Kinetochore localization of MDF-1 depends on both SPDL-1 and BUB-1. Bar, 20 μm. (C) Immunoprecipitations were performed on extracts from wild-type (WT), zyg-1(b1) homozygotes (zyg-1ts), and mdf-1(gk2); fzy-1(h1983) homozygotes (mdf-1Δ) using an anti–SPDL-1 antibody, an anti–MDF-1 antibody, or control IgG. Immunoprecipitants were separated on a gel, transferred onto a nitrocellulose membrane, and probed with antibodies to MDF-1, SPDL-1, and MDF-2. SPDL-1 was detected in MDF-1 immunoprecipitants and MDF-1 and MDF-2 in SPDL-1 immunoprecipitants. (D) Schematic model of the position of SPDL-1 in the hierarchical dependency of kinetochore assembly. SPDL-1 is downstream of CZW-1 and required for unattached kinetochore localization of MDF-1. BUB-1 is also required for unattached kinetochore localization of MDF-1 but not for that of SPDL-1. Together, MDF-1 targeting is regulated by two independent pathways, one includes SPDL-1 and the other includes BUB-1.
SPDL-1 physically associates with MDF-1
We next analyzed the physical interaction between SPDL-1 and MDF-1. There was a small but detectable amount of MDF-1 and MDF-2 in SPDL-1 immunoprecipitates from whole-worm lysate of wild-type strain (Fig. 3 C); reciprocally, SPDL-1 and MDF-2 were detected in immunoprecipitates with anti–MDF-1 antibody (Fig. 3 C). Coimmunoprecipitation of MDF-1 and SPDL-1 increased in the lysate from ZYG-1–deficient worms (Fig. 3 C; O'Connell et al., 2001), suggesting that the MDF-1–MDF-2 complex associates more stably with SPDL-1 at unattached kinetochores.
In conclusion, our san-1 synthetic lethal screen identified that SPDL-1 is required for spindle defect–induced activation of the SAC. SPDL-1 localizes at the kinetochore during mitosis in a CZW-1/ZW10–dependent and BUB-1–independent manner (Fig. 3 D). In embryonic cells with the monopolar spindle, SPDL-1 localizes to kinetochores on the side of the chromosome that faces away from the mono centrosome, thereby recruiting MDF-1 via its physical association. MDF-1 localization to unattached kinetochores also depends on BUB-1. We could not show direct interaction of SPDL-1 and MDF-1 by yeast two-hybrid analysis (unpublished data), suggesting that the proteins need additional components for their interaction, or MDF-1 may be modified in a BUB-1 kinase–dependent manner for its interaction with SPDL-1.
Whether SPDL-1 is actively recruited to unattached kinetochores by interacting with other proteins or specifically excluded from attached kinetochores needs study. High amounts of SPDL-1 on metaphase chromosomes (Fig. 1 D) imply that SPDL-1 is removed from kinetochores after microtubule attachment. SPDL-1 is structurally related to Spindly, which plays a role in recruiting dynein to kinetochores and is translocated from kinetochores to microtubules in a dynein-dependent manner (Griffis et al., 2007). Therefore, dynein may remove SPDL-1 from kinetochores attached to microtubules. Although Spindly does not play the same role as SPDL-1 in MAD1 recruitment in other organisms because the property of MAD1 to localize to unattached kinetochores is highly conserved among eukaryotes (including C. elegans), other organisms might also have a similar regulatory mechanism in which other proteins may play the role of SPDL-1 in recruiting MAD1 to unattached kinetochores. Therefore, elucidating how SPDL-1 accumulates specifically at unattached kinetochores should help unravel the molecular mechanisms by which microtubule attachment status of the kinetochore is sensed to initiate SAC function.
Materials and methods
Strains and plasmids
The following C. elegans strains were used in this study: N2 Bristol (wild-type), RB1391 san-1(ok1580) I, XA3501 unc-119(ed3) III; ruIs32[pAZ132; Ppie-1∷GFP∷his-11 unc-119(+)] III; ojIs1[Ppie-1∷GFP∷tbb-2 unc-119(+)], CB270 unc-42(e270) V, AG164 unc-42(e270) mdf-1(av19) V, RQ153 unc-46(e177) V/nT1[let-X(m435)] IV;V, KR3627 unc-46(e177) mdf-1(gk2) V/nT1[let-X(m435)] IV;V, AG170 mdf-2(tm2910) IV, AG169 mat-3(or180) III. VC1049 spdl-1(ok1515)/mln1[mls14 dpy-10(e128)] II, RQ309 spdl-1(ok1515)/mln1[mls14 dpy-10(e128)] II; emb-30(tn377ts) III, OD1 unc-119(ed3) III; itIs1[pIC22; Ppie-1∷GFP∷knl-3 unc-119(+)], OD11 unc-119(ed3) III; itIs7[pIC41; Ppie-1∷GFP∷kbp-4 unc-119(+)], DH1 zyg-1(b1) II, KR3779 fzy-1(h1983) II; unc-46(e177) mdf-1(gk2) V, RQ120 san-1(ok1580) I, RQ283 spdl-1(ok1515) II; unc-119(ed3) III; jzIs41[pRK177; Ppie-1∷GFP∷spdl-1; unc-119(+)]; itIs37[pAA64; Ppie-1∷mCherry∷his-58 unc-119(+)], RQ298 unc-119(ed3) III; jzIs48[pRK226; Ppie-1∷GFP∷czw-1 unc-119(+)], RQ244 unc-119(ed3) III; unc-46(e177) mdf-1(gk2) V; jzIs1[pRK139; Ppie-1∷GFP∷mdf-1 unc-119(+)]; itIs37[pAA64; Ppie-1∷mCherry∷his-58 unc-119(+)]. RQ120 was isolated from RB1391 after outcrossing four times. RQ283, RQ298, and RQ309 were constructed for this study. To generate transgenic strains expressing GFP–SPDL-1 or GFP–CZW-1, we constructed GFP fusions in pIC26 (Cheeseman et al., 2004). The unspliced genomic locus for spdl-1 (C06A8.5) or czw-1 (F20D12.4) was amplified by PCR from N2 genomic DNA and inserted into the pIC26 in frame. The resulting plasmid DNA was integrated into DP38 [unc-119(ed3)] using microparticle bombardment (Praitis et al., 2001) with a PDS-1000/He Biolistic Particle Delivery System (Bio-Rad Laboratories). The expression of GFP proteins in early stage embryos of RQ283 and RQ298 were analyzed by fluorescence microscopy (Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200805185/DC1). The construction of RQ244 has been described previously (Watanabe et al., 2008). KR3779 was described previously (Kitagawa et al., 2002) and provided by A. Rose (University of British Columbia, Vancouver, Canada). The strains carrying itIs37 (McNally et al., 2006) were provided by K. Oegema (Ludwig Institute for Cancer Research, University of California, San Diego, La Jolla, CA) and F.J. McNally (University of California, Davis, Davis, CA). The strains carrying mat-3(or180), mdf-2(tm2910), and mdf-1(av19) were provided by A. Golden (The National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD). All other strains were obtained from the Caenorhabditis Genetics Center.
Ahringer's C. elegans RNAi feeding library (Kamath et al., 2003) was obtained from Geneservice Ltd. dsRNA expression vectors for targeting particular genes except mdf-1 and bub-3 were isolated from the feeding library clones, retransformed, and individually cultured. For targeting mdf-1, C-terminus coding region of mdf-1 cDNA (1,130 −2,040) was cloned into pPD129.36 (Kamath et al., 2001). For targeting bub-3, the cDNA containing the 58th to 1,032nd of bub-3 coding region was cloned into pPD129.36.
RNAi screen for san-1 synthetic genetic interactors
Each RNAi bacterial clone was grown overnight at 37°C in low-salt LB (0.5% NaCl) containing 50 μg/ml ampicillin in each well of 96–deep well plates, induced with 1 mM isopropyl β-d-thiogalactoside at 37°C for 4 h. About 10 synchronized L1 larvae of wild-type or san-1Δ strain in 100 μl of complete S medium containing 50 μg/ml ampicillin and 1 mM isopropyl β-d-thiogalactoside were fed 50 μl of induced bacterial culture in each well of 96-well plates. Wells for wild-type and san-1Δ strains were placed adjacent to each other to allow easy comparison. After 5–8 d of incubation, densities of F1 worms in each well were scored by visual inspection under a microscope. Six-level scores were defined by the densities of F1 worms. If the score level of F1 density in the san-1Δ strain was reduced by more than two levels compared with the wild-type strain, the RNAi clone was considered a positive gene. Positive results were confirmed by repeating the experiments at least three times. Insert DNA sequences of positive clones were confirmed by sequencing. Some of genes positive for synthetic lethality with san-1 were further tested for synthetic genetic interaction with mdf-1 or mdf-2 as described previously (Tarailo et al., 2007b; Hajeri et al., 2008).
RNA-mediated interference
RNAi by the feeding method (Kamath et al., 2001, 2003), by the soaking method (Maeda et al., 2001), and by the microinjection method (Desai et al., 2003) was performed as described previously. For the soaking and microinjection methods, dsRNA was generated as follows: DNA fragments were amplified from the dsRNA expression vector containing a part of the coding region of the targeting gene by PCR, using T7 primer. PCR products were subjected to in vitro RNA transcription using the Megascript T7 kit (Ambion). The final concentration of dsRNA for each gene in the soaking buffer was 2 μg/μL and in the injection buffer was 1 μg/μL. Depletion level of each protein was tested as described previously (Desai et al., 2003).
Live imaging of embryos expressing GFP-fused proteins
Living embryos expressing GFP-fused proteins were dissected, mounted on an agarose pad, covered with mineral oil, and subjected to time-lapse experiments at room temperature (24°C). To follow whole mitosis in one-cell–stage embryos expressing GFP–SPDL-1 or GFP–CZW-1, an image at single focal planes was taken every 30 s. To follow mitosis in one- or two-cell–stage embryos expressing GFP fusion proteins, a set of images at three focal planes at 2-μm intervals was taken at each time point. To analyze centrosome separation, a set of images at five focal planes at 2-μm intervals was taken every 10 s. Images were projected and the distance between centrosomes was measured using Openlab (Improvision).
Live images were taken by a motorized fluorescence microscope (DM IRE2; Leica) equipped with a Plan Apo 63× lens (NA 1.4; Leica) with 1.5× magnification and an ORCA-ER high-resolution digital CCD camera (Hamamatsu Photonics) binning 2× under control of Openlab. Image deconvolution was performed by Volocity (Improvision).
Induction of SAC-dependent mitotic delay by RNAi-mediated depletion of ZYG-1
SAC-dependent mitotic delay was induced by RNAi-mediated depletion of ZYG-1, a protein required for daughter centriole formation (O'Connell et al., 2001), which results in monopolar spindle formation in the second embryonic division. Consequently, sister kinetochores capture only the mono-oriented microtubule attachment, causing SAC-dependent mitotic delay. Embryos expressing GFP–histone H2B and GFP–tubulin were dissected from adult hermaphrodites injected with zyg-1 dsRNA only or a mixture of dsRNAs of zyg-1 and other genes, and subjected to time-lapse differential interference contrast and fluorescent microscopy. The duration of mitosis was defined as an interval between NEBD and chromosome decondensation. Under our experimental conditions, the duration of mitosis of ZYG-1–depleted cells was extended to 600 s, the duration of normal mitosis being 200 s (Fig. 1 A).
Phenotypic analysis of the spdl-1 deletion strain
Chromosome morphology of germ cells in hermaphrodite gonads of wild-type or spdl-1(ok1515) homozygotes were analyzed as described previously. (Kitagawa and Rose, 1999). To analyze the fertility of the strains, more than twenty wild-type–looking L4 larvae segregated from VC1049 [spdl-1(ok1515)/mln1] or RQ309 [spdl-1(ok1515)/mln1; emb-30(tn377ts)] were isolated individually and incubated at 20°C for 12 h, and then the presence of eggs was checked. The homozygosity of spdl-1(ok1515) in fertile worms was confirmed by PCR.
Antibody preparation
For generation of rabbit anti–SPDL-1 antibody and anti–CZW-1 antibody, a full-length cDNA of spdl-1 and a cDNA containing the 1,090th to 2,334th of czw-1 coding region were cloned into pRSETA (Invitrogen). Each purified 6-histidine–tagged recombinant proteins was used to immunize two rabbits. The antiserum generation was performed by Covance ImmunoTechnologies laboratory. For affinity purification, full-length spdl-1 cDNA and a cDNA containing the 1,090th to 2,334th of czw-1 coding region were cloned into pGEX4T1 (GE Healthcare), and purified GST–SPDL-1 or GST–CZW-1 was coupled to an AminoLink column (Thermo Fisher Scientific). Rabbit immunosera were affinity purified on the AminoLink column with coupled GST-fused proteins.
Immunofluorescence microscopic analysis
For immunostaining of proteins except SPDL-1 or CZW-1, embryos dissected or isolated by the alkaline hypochlorite method (Lewis and Fleming, 1995) were transferred onto poly-l-lysine–coated slides, frozen on dry ice, cracked, fixed with −20°C methanol for 10 min, dehydrated with −20°C acetone for 10 min, and rehydrated with PBS, pH 8.0, for 10 min. For SPDL-1 or CZW-1 staining, embryos prepared by the alkaline hypochlorite method were collected in a tube, fixed with −20°C methanol for 10 min, and washed with PBS. Embryos were then blocked with blocking solution (10% skimmed milk in PBS) for 1 h; incubated overnight at 4°C with blocking solution containing rat anti–α-tubulin YL1/2 (1:100; AbD Serotec) and anti–SPDL-1 (1:200), anti–KNL-1 (1 μg/ml; Desai et al., 2003), anti–NDC-80 (1 μg/ml; Desai et al., 2003), anti–BUB-1 (1 μg/ml; Oegema et al., 2001), anti–MDF-1 (1:2,000; Watanabe et al., 2008), or anti–CZW-1 antibodies (1:200); washed with PBST (0.1% Tween-20 in PBS); incubated for 3 h with blocking solution containing Alexa 594–conjugated anti–rabbit IgG (1:2,000; Invitrogen), Alexa 488–conjugated anti–rat IgG (1:2,000; Invitrogen), and 3 μg/ml DAPI; washed with PBST; and mounted on slides. For costaining of SPDL-1 and other kinetochore proteins, embryos were isolated by the alkaline hypochlorite method, fixed with 3% formaldehyde for 30 min, postfixed with −20°C methanol for 5 min, washed with PBST, and then subjected to immunostaining. The anti–SPDL-1 antibody was labeled with DyLight488 or 594 (Thermo Fisher Scientific), and used along with the anti–BUB-1 antibody labeled with Cy3 or the anti–-CZW-1 antibody labeled with DyLight488, respectively. For costaining of SPDL-1 and KNL-1, the specimen was first stained with anti–KNL-1 antibody and Alexa 488–conjugated anti–rabbit IgG (Invitrogen), and then stained with anti–SPDL-1 labeled with DyLight594. Images were taken by the same microscope system as described in a previous section (Live imaging of embryos expressing GFP-fused proteins) except for a Plan Apo 100× lens (NA 1.4; Leica) without auxiliary magnification and binning (1×). Optical section images were collected at focus intervals of 0.5 μm. Antibodies for KNL-1, NDC-80, and BUB-1 were provided by A. Desai (Ludwig Institute for Cancer Research, University of California, San Diego, La Jolla, CA).
Immunoprecipitation/Western blotting analysis
zyg-1(b1) temperature-sensitive strain (O'Connell et al., 2001) was maintained at 16°C and incubated at 25°C for 12 h before protein lysate preparation. Protein lysate from gravid worms was prepared in RIPA buffer (50 mM Tris-Cl, pH 8.0, 200 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, and 0.1% SDS) and immunoprecipitation was performed as described previously (Watanabe et al., 2008). For Western blot analysis, the following primary antibodies were used: anti–MDF-1 (1:20,000; Watanabe et al., 2008), anti–SPDL-1 (1:2,500), and anti–MDF-2 (1:1,000; Kitagawa and Rose, 1999).
Online supplemental material
Fig. S1 shows genetic and cell biological characterization of the physiological function of spdl-1. Fig. S2 shows fluorescence microscopic analysis of molecular dependency of SPDL-1 on other kinetochore-associating proteins for its kinetochore localization. Fig. S3 shows fluorescence microscopic analysis of molecular dependency of MDF-1 and other kinetochore-associating proteins for its kinetochore localization. Video 1 shows the time-lapse imaging analysis of the one-cell–stage embryo expressing GFP–SPDL-1 in spdl-1Δ background. Video 2 shows the time-lapse imaging analysis of the one-cell–stage embryo expressing GFP–CZW-1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200805185/DC1.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetics Center; Arshad Desai for providing antibodies and useful discussion; Andy Golden and David Greenstein for critically reading the manuscript; and Elaine Law, Christen N. Gregory, Brynne Hancock, and Leslie Douglas for technical assistance.
This work was supported by the Cancer Center Support grant CA021765 from the National Cancer Institute and by the American Lebanese Syrian Associated Charities. A. Essex was supported by the University of California, San Diego, Genetics Training grant.
Abbreviations used in this paper: APC/C, anaphase-promoting complex/cyclosome; dsRNA, double-stranded RNA; NEBD, nuclear envelope breakdown; RZZ, ROD–ZW10–ZWILCH; SAC, spindle assembly checkpoint.
References
- Cheeseman, I.M., and A. Desai. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Niessen, S. Anderson, F. Hyndman, J.R. Yates III, K. Oegema, and A. Desai. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni, A., C.G. Pearson, D. Cimini, J.C. Canman, V. Sala, L. Nezi, M. Mapelli, L. Sironi, M. Faretta, E.D. Salmon, and A. Musacchio. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214–225. [DOI] [PubMed] [Google Scholar]
- Desai, A., S. Rybina, T. Muller-Reichert, A. Shevchenko, A. Shevchenko, A. Hyman, and K. Oegema. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada, S.E., J. Willis, R. Lyczak, and B. Bowerman. 2005. A spindle checkpoint functions during mitosis in the early Caenorhabditis elegans embryo. Mol. Biol. Cell. 16:1056–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski, J.K., and G.K. Chan. 2007. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr. Biol. 17:2143–2149. [DOI] [PubMed] [Google Scholar]
- Furuta, T., S. Tuck, J. Kirchner, B. Koch, R. Auty, R. Kitagawa, A.M. Rose, and D. Greenstein. 2000. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell. 11:1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, A., P.L. Sadler, M.R. Wallenfang, J.M. Schumacher, D.R. Hamill, G. Bates, B. Bowerman, G. Seydoux, and D.C. Shakes. 2000. Metaphase to anaphase (mat) transition–defective mutants in Caenorhabditis elegans. J. Cell Biol. 151:1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, E.R., N. Stuurman, and R.D. Vale. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajeri, V.A., A.M. Stewart, L.L. Moore, and P.A. Padilla. 2008. Genetic analysis of the spindle checkpoint genes san-1, mdf-2, bub-3 and the CENP-F homologues hcp-1 and hcp-2 in Caenorhabditis elegans. Cell Div. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M.A., L. Totis, and B.T. Roberts. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517. [DOI] [PubMed] [Google Scholar]
- Iwasaki, K., J. McCarter, R. Francis, and T. Schedl. 1996. emo-1, a Caenorhabditis elegans Sec61p γ homologue, is required for oocyte development and ovulation. J. Cell Biol. 134:699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., M. Martinez-Campos, P. Zipperlen, A.G. Fraser, and J. Ahringer. 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2:RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., A.G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. [DOI] [PubMed] [Google Scholar]
- Karess, R. 2005. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15:386–392. [DOI] [PubMed] [Google Scholar]
- Kitagawa, R., and A.M. Rose. 1999. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1:514–521. [DOI] [PubMed] [Google Scholar]
- Kitagawa, R., E. Law, L. Tang, and A.M. Rose. 2002. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12:2118–2123. [DOI] [PubMed] [Google Scholar]
- Kops, G.J., Y. Kim, B.A. Weaver, Y. Mao, I. McLeod, J.R. Yates III, M. Tagaya, and D.W. Cleveland. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.A., and J.T. Fleming. 1995. Basic culture methods. Methods Cell Biol. 48:3–29. [PubMed] [Google Scholar]
- Li, R., and A.W. Murray. 1991. Feedback control of mitosis in budding yeast. Cell. 66:519–531. [DOI] [PubMed] [Google Scholar]
- Maeda, I., Y. Kohara, M. Yamamoto, and A. Sugimoto. 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11:171–176. [DOI] [PubMed] [Google Scholar]
- May, K.M., and K.G. Hardwick. 2006. The spindle checkpoint. J. Cell Sci. 119:4139–4142. [DOI] [PubMed] [Google Scholar]
- McNally, K., A. Audhya, K. Oegema, and F.J. McNally. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio, A., and E.D. Salmon. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. [DOI] [PubMed] [Google Scholar]
- Nystul, T.G., J.P. Goldmark, P.A. Padilla, and M.B. Roth. 2003. Suspended animation in C. elegans requires the spindle checkpoint. Science. 302:1038–1041. [DOI] [PubMed] [Google Scholar]
- O'Connell, K.F., C. Caron, K.R. Kopish, D.D. Hurd, K.J. Kemphues, Y. Li, and J.G. White. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 105:547–558. [DOI] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar, and J. Austin. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi, L., M. Melixetian, M. Faretta, E. Prosperini, K. Helin, and A. Musacchio. 2001. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 20:6371–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias, D.A., P.R. Andreassen, F.B. Lacroix, L. Wilson, and R.L. Margolis. 2001. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. USA. 98:4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen, B., L.B. Koski, A. Walsh, P. Marschall, B. Neumann, M. Brehm, A.M. Alleaume, J. Artelt, P. Bettencourt, E. Cassin, et al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 434:462–469. [DOI] [PubMed] [Google Scholar]
- Starr, D.A., B.C. Williams, Z. Li, B. Etemad-Moghadam, R.K. Dawe, and M.L. Goldberg. 1997. Conservation of the centromere/kinetochore protein ZW10. J. Cell Biol. 138:1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D.A., B.C. Williams, T.S. Hays, and M.L. Goldberg. 1998. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 142:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, K.K., E.S. Davis, T. Hays, and A. Golden. 2007. Components of the spindle assembly checkpoint regulate the anaphase-promoting complex during meiosis in Caenorhabditis elegans. Genetics. 175:107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo, M., R. Kitagawa, and A.M. Rose. 2007. a. Suppressors of spindle checkpoint defect (such) mutants identify new mdf-1/MAD1 interactors in Caenorhabditis elegans. Genetics. 175:1665–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo, M., S. Tarailo, and A.M. Rose. 2007. b. Synthetic lethal interactions identify phenotypic “interologs” of the spindle assembly checkpoint components. Genetics. 177:2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S., T.G. Yamamoto, and R. Kitagawa. 2008. Spindle assembly checkpoint gene mdf-1 regulates germ cell proliferation in response to nutrition signals in C. elegans. EMBO J. 27:1085–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.