Abstract
Glutamate transporters, also referred to as excitatory amino acid transporters (EAATs), are membrane proteins that regulate glutamatergic signal transmission by clearing excess glutamate after its release at synapses. A structure-based understanding of their molecular mechanisms of function has been elusive until the recent determination of the x-ray structure of an archaeal transporter, GltPh. GltPh exists as a trimer, with each subunit containing a core region that mediates substrate translocation. In the present study a series of molecular dynamics simulations have been conducted and analyzed in light of new experimental data on substrate binding properties of EAATs. The simulations provide for the first time a full atomic description of the time-resolved events that drive the recognition and binding of substrate. The core region of each subunit exhibits an intrinsic tendency to open the helical hairpin HP2 loop, the extracellular gate, within tens of nanoseconds exposing conserved polar residues that serve as attractors for substrate binding. The NMDGT motif on the partially unwound part of the transmembrane helix TM7 and the residues Asp-390 and Asp-394 on TM8 are also distinguished by their important role in substrate binding and close interaction with mediating water molecules and/or sodium ions. The simulations reveal a Na+ binding site comprised in part of Leu-303 on TM7 and Asp-405 on TM8 and support a role for sodium ions in stabilizing substrate-bound conformers. The functional importance of Leu-303 or its counterpart Leu-391 in human EAAT1 (hEAAT1) is confirmed by site-directed mutagenesis and Na+ dependence assays conducted with hEAAT1 mutants L391C and L391A.
Chemical synapses are central sites for signal transmission within the nervous system. When an action potential triggers the release of neurotransmitters, the extracellular (EC)2 concentration of neurotransmitters may increase by several orders of magnitude. Transporters on neurons and glia clear neurotransmitters and regulate the extent of receptor activation. Among them, glutamate transporters allow cells to maintain a substantial concentration gradient of glutamate (1). In fulfilling this function, each glutamate molecule is co-transported with three sodium ions and one proton followed by the counter-transport of a potassium ion (2). Binding of substrate and Na+ also activates an uncoupled chloride conductance, and thus, the transporter also functions as a ligand-gated anion channel (3, 4). Glutamate transporters have a major impact on developmental plasticity and long term potentiation, and they have been implicated in many pathological conditions including stroke, epilepsy, cerebral ischemia, amyotrophic lateral sclerosis, Alzheimer disease, and schizophrenia (5). However, the molecular interactions and structural mechanisms that mediate the transport of glutamate and co-transported ions have been difficult to resolve in the absence of high resolution structural information for the carrier family.
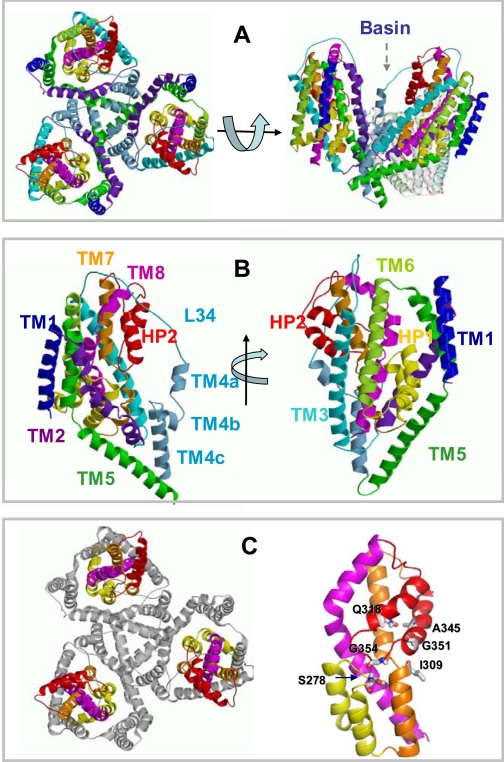
A major breakthrough came with the resolution of the x-ray structure of the archaeal glutamate transporter (GltPh) from Pyrococcus horikoshii under different conditions by Gouaux and co-workers (6, 7). GltPh is a homotrimer (Fig. 1A) with the subunits forming a bowl-shaped concave basin toward the EC side. Each subunit is comprised of eight transmembrane (TM) helices, TM1-TM8, and two helical hairpins, HP1 and HP2 (Fig. 1B), which partially span the lipid bilayer. These structural elements are organized into an N-terminal, outer cylinder region made up of TM helices TM1-TM6 and a C-terminal core comprised of TM7, HP1, HP2, and TM8 (Fig. 1C). The core region is known to encompass the elements of the transport machinery (6).
FIGURE 1.
Structure of an archaeal glutamate transporter (GltPh) resolved by Yernool et al. (6) (PDB code 1xfh). A, top view (left), viewed from the EC region, and side view (right). One of the subunits of the trimer is deleted in the side view for better viewing the interior and concave shape of the basin. B, secondary structural elements (eight helices TM1-TM8 and two hairpins HP1 and HP2, each comprised of a helix-loop-helix motif) shown for one subunit, in two different views. C, N-terminal (gray) and core (colored) domains (left). The N-terminal domain comprises the TM helices 1-6. The core (right) is composed of HP1 (yellow; 259-291), HP2 (red; 338-373), TM7 (orange), and TM8 (magenta). Ser-278 and Gly-354 at the respective tips of hairpins HP1 and HP2 are shown along with a few other resides (Ala-345 and Gly-351 in HP2, Gln-318 in TM7) that are involved in controlling EC gate opening according to the results from simulations (see below).
The crystallographic structural data obtained for GltPh (6, 7) agree, in general, with models based on biochemical and mutagenesis experiments conducted for mammalian excitatory amino acid transporters (EAATs). For example, biochemical studies on human EAAT2 showed that the transporter exists as a trimer (8); cross-linking of pairs of cysteines introduced at the tips of HP1 and HP2 loops, on HP1 tip and TM7, and on HP2 tip and TM7 suggested that these regions are located within close proximity (9, 10), in accord with their spatial distances observed in the resolved GltPh structure. Additionally, residues in the mammalian EAATs equivalent to the GltPh residues Asp-394 and Arg-397, the serine-rich motif (Ser-277-Ser-279), and the conserved NMDGT motif have been proposed to be involved in substrate binding (11-14), consistent with their interaction with the bound aspartate observed in the high resolution structure of GltPh.
These structural data along with the fact that GltPh shares about 36% sequence identity with eukaryotic EAATs suggest that GltPh and its mammalian orthologues share similar structural and biophysical features. The GltPh structure may, therefore, serve as a prototype for gaining new insights into the molecular interactions and mechanisms of glutamate uptake by the Na+/K+-dependent glutamate transporter family (15).
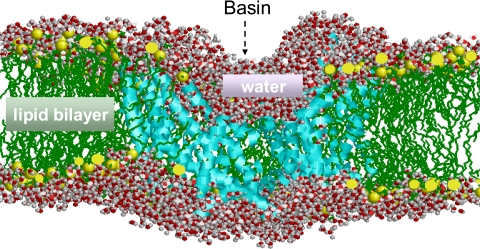
Toward this goal we have performed extensive molecular dynamics (MD) simulations using the available GltPh structure in the presence of explicit solvent, lipid molecules, substrate, and ions. We present here the results from an overall run time of 0.20 μs (see Table 1), which may be viewed as a computational tour de force for this large system of ∼100,000 heavy atoms (see Fig. 2).
TABLE 1.
Description of MD runs conducted in the present study
| Run ID | Duration | Initial structure of the transporter | Total no. of atomsa | Substrates, sodium ions, and their initial position |
|---|---|---|---|---|
| ns | ||||
| MD0 | 40 | GltPh closed formb | 97,748 | None |
| MD1 | 10 | GltPh open formc | 97,631 | Glutamate in aqueous basin (bottom) |
| MD2 | 10 | GltPh open formc | 97,613 | Glutamate in aqueous basin (top) |
| MD3 | 10 | GltPh-Glu complexd | 97,592 | Glutamate + 1 Na+ at binding site |
| MD4 | 30 | GltPh -Glu complexe | 97,577 | Glutamate + 3 Na+ at binding site |
| MD5 | 50 | GltPh closed formb | 98,889 | Aspartatesf + ions at EC and IC regions |
| MD6 | 50 | GltPh closed formb | 99,001 | Glutamatesf + ions at EC and IC regions |
All simulations were performed with explicit water and lipid molecules and with ions at 310 K. The different numbers of atoms refer to differences in the number of ligands, ions, and water molecules.
Closed form refers to the x-ray structure 1xfh (6).
5-ns snapshot from MD0, where subunit A only is open.
4-ns snapshot from MD1.
4-ns snapshot from MD2.
25 substrate molecules included in the EC and IC regions, in accord with the physiological levels of 5-50 and 0.5-1 mm, respectively (30).
FIGURE 2.
A snapshot of simulation environment. The size of the simulated systems is on the order of 105 heavy atoms including the transporter GltPh (cyan), lipid (1-palmitoyl-2-oleoylphosphatidyl ethanolamine) molecules (green), water molecules, substrates, co-transported ions; see Table 1. One of the subunits is deleted in the figure for a clear visualization of the aqueous basin facing the EC region. Water molecules within 5 Å of the protein and 3 Å of lipid molecules are shown in space-filling representation (O, red;H, gray). The hydrophobic tails of the lipid molecules are colored green, and the polar head groups are shown in yellow beads. The basin (∼50 Å in diameter) extends almost halfway (∼30 Å) into the lipid bilayer and is completely filled by water.
Our study demonstrates that the hairpin HP2 loops possess an intrinsic, structure-induced ability to undergo large scale motions even in the absence of substrate. These motions open up the conserved charged or polar residues in the core regions to the central aqueous basin. Exposure of these residues prompts the directed diffusion of substrate toward the putative binding site, provided that the substrate is located within an interaction range in the basin. The intrinsic, structure-induced ability of GltPh to sample the conformational motions that facilitate substrate binding conforms to the functional dynamics observed both experimentally and computationally for many allosteric enzymes and receptors (16-19). Additionally, simulations permit us to visualize the sequence of events that lead to the sequestering of the substrate at the binding site as well as the important role of bound Na+ in controlling EC gate motion and ensuring the tight confinement of the substrate in the core. A well defined pathway is identified where water molecules enter the binding site in single file via a passage between two highly conserved aspartates, Asp-390 and Asp-394, on TM8. Binding of a water molecule to Thr-314 on the NMDGT motif plays a key role in mediating the cooperative rearrangement of amino acids that coordinate the bound substrate. Finally, the residue Leu-303 on TM7 (Leu-391 in hEAAT1) appears to serve as a binding site for Na+. We tested this possibility by examining the Na+ dependence of transport for two human EAAT1 (hEAAT1) mutants (L391A and L391C). The results from these experiments support the significance of this newly defined site for Na+ binding/uptake. Overall, the present structure-based simulations and experiments shed light for the first time on the time-resolved mechanisms of EC gate opening, substrate recognition, and binding and the role of Na+ in modulating the structural dynamics of the transporter.
EXPERIMENTAL PROCEDURES
MD Simulations—We simulated the dynamics of GltPh embedded in a bilayer-solvent environment (Fig. 2) using the GROMACS program 3.2.1 with the Gromacs force field (20). The crystal structure of GltPh (6) was used as the initial conformation (PDB code 1xfh). Missing atoms on the side chains of some residues were constructed using Xplor (20). Constant number of particles (N), pressure (P), and temperature (T) conditions, with a pressure of 1 bar and a coupling constant of τp = 1.0 ps, were adopted. Water, protein, and lipid molecules were coupled separately to a temperature bath at 310 K using a coupling constant of τt = 0.1 ps. The van der Waals interactions were calculated using a cutoff distance of 10 Å, and long range electrostatic interactions were calculated using the particle mesh Ewald method. The single point charge model was adopted for water molecules as a plausible model for lipid simulations (21, 22) along with the lipid parameters used in our previous MD simulations of other membrane proteins (23, 24). The membrane consisted of 353 1-palmitoyl-2-oleoylphosphatidyl ethanolamine molecules, and the system was solvated with ∼23,000 water molecules. Simulations were preceded by energy minimization with steepest descent followed by equilibration periods of 200 ps during which the protein backbone atoms were restrained by harmonic potentials so as to allow for the packing of lipid molecules around the protein. The constraints were then removed for the productive runs.
Table 1 provides a summary of MD runs. MD0 was performed to examine the intrinsic dynamics of the transporter. MD1 and MD2 were generated to examine the substrate binding events using two different initial positions for the substrate (4.5 and 8.8 Å away from the base of the basin, respectively, along the central cylindrical (z) axis). The substrate was seen to drift away to the EC region in the first 1-ns portion of the MD2 trajectory. This was prevented by applying on the substrate backbone nitrogen an acceleration of 10 nm/ps2 along the z direction, toward the basin, similar to the approach used in other applications (25-27). The acceleration had no radial component and was turned off after 1 ns to observe the intermolecular interactions that drive substrate diffusion. The role of sodium ions was examined in the runs MD3 and MD4. MD5 and MD6 were conducted to examine and compare the diffusive properties of aspartate and glutamate at physiological levels in the EC and intracellular (IC) regions. Supplemental Fig. S1 displays the time evolution of the root-mean-square deviation in atomic coordinates relative to starting structure, observed in MD0 (A), MD5 (B), and MD6 (C) for the three subunits. The average deviation in the atomic coordinates of the subunits remains lower than 3.5 Å in MD0 and 4.2 Å in MD5 and MD6, i.e. the overall structure is highly stable in the time frame of simulations. Largest motions occur at the loops and at TM4 (supplemental Fig. S2).
Mutagenesis and Sodium Dependence Assay—Mutants L391A and L391C were made using site-directed mutagenesis method (Stratagene) based on a cysteine-less (CSLS) version of human EAAT1(CSLS) (28). PCR products were subcloned into pCMV5 vector and sequenced by dye terminator cycle sequencing (PerkinElmer Life Sciences). COS-7 cells were passaged and plated into a 24-well plate and transfected using FuGENE 6 reagent (Roche Applied Science). Uptake assays were performed 2 days after transfection. Cells were incubated with 5 μm l-[3H]glutamate (50 nm l-[3H]glutamate and 4.95 μm unlabeled l-glutamate) in different Na+ concentrations (140, 60, 30, 10 mm) for 10 min at room temperature. The uptake buffer contained 140 mm NaCl, 8.1 mm Tris-H3PO4, 2.7 mm KCl, 1.5 mm KH2PO4, 0.1 mm CaCl2, and 1 mm MgCl2 (pH 7.4). Choline chloride was used as the replacement for NaCl in the buffers containing reduced Na+ concentrations. Cells were lysed in 0.1% SDS and counted in a scintillation counter. Cells transfected with pCMV5 vector served as a control for endogenous l-glutamate uptake under all conditions. Km and Vmax values were derived by the Michaelis-Menten equation using Prism 4.0 (Graphpad).
RESULTS
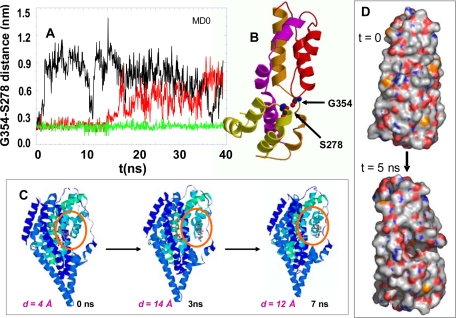
Intrinsic Dynamics of the Transporter in the Absence of Substrate and Spontaneous Opening of HP2 Loop—The first set of simulations (MD0; Table 1) was performed in the absence of substrate in order to investigate the dynamics of the transporter before substrate binding. Fig. 3 illustrates the results. The most striking motions observed in these simulations are the large amplitude fluctuations of the HP2 loop away from the HP1 loop toward the aqueous basin. This type of motion is evidenced by the change in the distance between residues Ser-278 and Gly-354 at the tips of the respective hairpins HP1 and HP2 (panel A). In the closed (starting) conformation the distance between the hydrogen bond-forming backbone nitrogen and oxygen on the respective residues Gly-354 and Ser-278 is 1.90 ± 0.25 Å in all three subunits (panel B). Strikingly, an increase of more than 8 Å was observed in subunit A (black curve in Fig. 3, panel A) succeeding the reconfiguration of the HP2 loop, which literally acts as an “EC gate.” Subunit B (red in Fig. 3A) exhibits a displacement of about 7 Å at 17 ns, whereas subunit C (green) remains closed throughout the entire duration of the run MD0. Once the original tight packing of the two hairpins in the core is disrupted, the HP2 loop tends to remain open and disordered as indicated by the large fluctuations in the HP1-HP2 distance time profile for both subunits A and B.
FIGURE 3.
Opening of EC gate upon disruption of the interaction between the tips of the HP1 and HP2 loops. A, time evolution of the distance between Gly-354 (O) and Ser-278 (N) at the HP2 and HP1 tips, respectively, observed for subunits A (black), B (red), and C (green) in run MD0. Subunit A exhibits a sharp jump at about 1.5 ns, succeeded by large amplitude fluctuations; subunit B exhibits similar, albeit smaller-amplitude fluctuations after ∼16 ns. Whereas subunit C remains closed with minimal fluctuations. B, position of Gly-354 and Ser-278 in the closed form (PDB code 1xfh) of the core domain. The two residues are highlighted in space-filling representation. C, three snapshots from MD0, at t = 0, 3, and 7 ns, displaying the increase in the separation between the hairpins HP1 and HP2 (at the circled region). D, space filling view of the opening of a cavity that binds the substrate, upon the displacement of the HP2 loop away from HP1 loop. Also see supplemental Movie 1 for viewing the opening of the HP2 loop.
The opening of HP2 loop is illustrated in the snapshots of subunit A at three successive times in Fig. 3C and in supplemental Movie 1 and Fig. S3. Largest motions occur at the loop residues Ala-353-Gly-359 between the helical branches HP2a and HP2b of HP2. This loop contains four highly conserved glycines, Gly-351, -354, -357, and -359. Conceivably, these glycines play a dual role of (i) facilitating the sampling of entropically favored (disordered) conformations (open state of the EC gate) by providing access to a broad range of rotameric angles ϕ and ψ and (ii) assisting in the stabilization of the substrate through exposure of their backbone polar groups and opening up a cavity (Fig. 3D) that will bind the substrate.
The propagation of the motion of the HP2 loop to the helical segment HP2a is restrained by its tertiary contacts with TM7. In particular, the interhelical contacts between Gly-354 (HP2 tip) and Ile-309 (TM7) and between Ala-345 (HP2a) and Gln-318 (TM7) limit the motion of HP2 from two ends, thus preventing the wide opening of the EC gate. See Fig. 1C for the position of these residues in the core domain. Supplemental Fig. S3 shows the high stability of the contacts between Gln-318 and the near neighboring residues on HP2a, supporting a role of Gln-318 in controlling the EC gating.
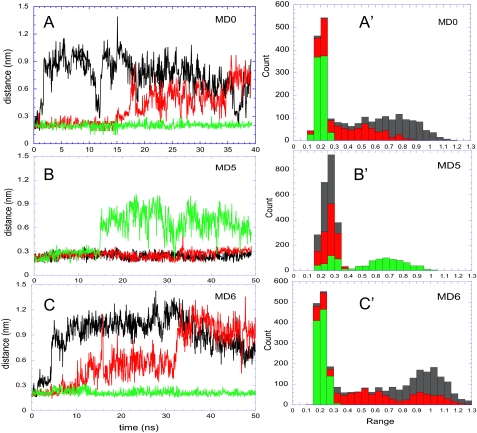
The tendency of the HP2 loop to dissociate from the HP1 loop and interact instead with the aqueous environment is invariably confirmed in three independent runs, MD0, MD5, and MD6. In each case, one or more subunit(s) exhibited this structure-induced tendency to “open up” (Fig. 4), apparently yielding to the competing effect of water molecules that disrupt the interaction between the HP1 and HP2 tips. Note that both helices TM7 and TM8 are partially unwound at this region, and the manifested mobility may partly result from the need to release the internal strain or to satisfy these “frustrated” structural elements including in particular the 310NMDGT314 motif on TM7, as will be further elaborated below. The time profiles in Fig. 4, A-C, show that the transition of HP2 from closed to open form is an abrupt change, suggestive of a conformational “switch.” The onset of the first opening varies from 1.5 ns (MD0, subunit A) to 15 ns (MD5, subunit C). No net correlation was detected between the opening events of the individual subunits. The occurrence of two “states,” open and closed, is also supported by the histograms in panels A′-C′. A bimodal distribution is seen therein for the distance between the tips of HP1 and HP2 loops. The narrow peak around 2 Å refers to the closed form. The open form, on the other hand, samples a broad range of distances, consistent with its disordered state.
FIGURE 4.
EC gate opening events observed in three independent runs. Panels A-C refer to the respective runs MD0, MD5, and MD6 (Table 1). The curves on the left panels describe the time evolution of the distance between Gly-354 and Ser-278 in subunits A (black), B (red), and C (green). Note that panel A is identical to Fig. 3A but is included for completeness. The histograms A′-C′ on the right panels display the distribution of this particular distance in the respective runs, summed over the three subunits (shown in different colors). The peak around 2 Å corresponds to the closed form of the EC gate. The open form, on the other hand, samples a broad range of distances. See also Fig. 10 where the histograms from all runs exhibit a bimodal distribution suggestive of a switch-like transition from closed to open states.
Diffusion of Glutamate toward the Binding Site—The cavity that opens up to the aqueous basin upon EC gating (Fig. 3D) contains charged/polar residues (e.g. Arg-276 and Ser-277-Ser-279 on HP1 and the NMDGT motif on TM7, all conserved among different subtypes of EAATs) with a high affinity for attracting the substrate. HP2, thus, serves not only as an EC gate permitting the entry of the neurotransmitter but also as an attractor driving the diffusion of the substrate toward the binding site.
Next, we explored the possibility of such a directed diffusion of the substrate toward the putative binding site between HP1 and HP2 tips. To address this issue, we performed the runs MD1 and MD2 using as initial conformation an open form of the transporter, e.g. the conformation visited at 5 ns in MD0 where subunit A was open (see Fig. 3). The glutamate was observed to spontaneously move toward the open subunit (subunit A) and slide into the putative binding site in between HP1 and HP2 (Figs. 5 and 6 and supplemental Movie 2) within a nanosecond when it was placed at a position sufficiently distant from the EC region in the aqueous basin. Note that the original radial distance between the glutamate substrate and the closest residues on HP2 was more than 20 Å in both of these simulations and decreases to less than 2 Å within 1 ns (Fig. 5). Moreover, the recognition of the substrate by the open subunit A and the radial diffusion of the substrate toward this particular subunit are achieved without exerting any external forces/biases in that direction but as a result of the long-range attraction exerted by the polar/charged groups exposed to the basin upon HP2 opening.
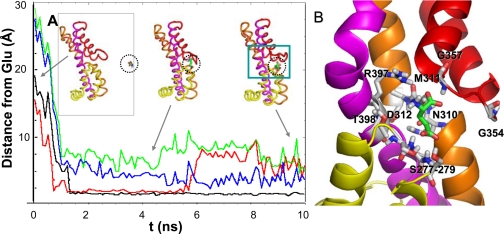
FIGURE 5.
Diffusion of glutamate toward the open EC gate and resulting bound structure. Panel A displays the time evolution of the distance between the glutamate and a series of residues/motifs at the binding site, which interact with the substrate. The distances (ordinate) refer to the closest pair of atoms observed in MD1. The binding event is initiated by the interaction with conserved glycines, Gly-354-Gly-357 (red), on HP2 followed by the interaction with Thr-275-Ser-279 on HP1 loop (black). Approximately 0.5 ns later, the substrate approaches the TM7 residues Asp-312-Thr-314 (green) and TM8 residues Arg-397-Thr-398 (blue). Three snapshots of the core domain and substrate are shown in the inset, corresponding to t = 0 (left), 4.8 ns (middle) and 10 ns (right). The dashed circle highlights the glutamate. Panel B provides an enlarged view of the substrate coordination at the end of the trajectory. See supplemental Movie 2 for viewing the diffusion of glutamate and Fig. 6 for a more detailed description of the successive interactions.
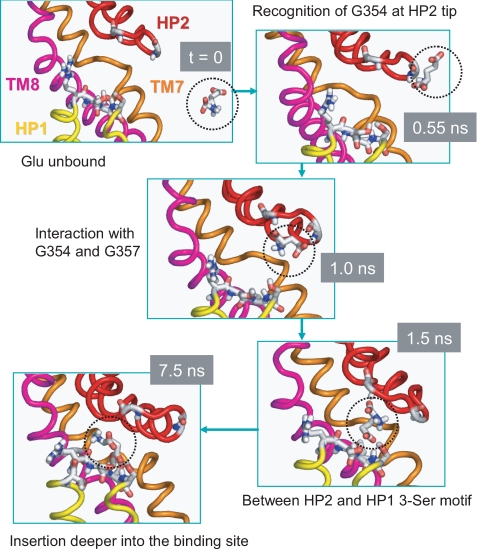
FIGURE 6.
Succession of events leading to substrate binding. Snapshots at different times (as indicated by gray boxes) are displayed to illustrate the time-resolved recognition and binding events. At t = 0, the unbound substrate (encircled) is in the aqueous cavity, whereas HP2 loop of subunit A samples an open conformation that exposes polar and charged residues on HP1 tip. At t = 0.55 ns, the substrate has moved closer toward the HP2 tip to recognize Gly-354. At t = 1.0 ns, it simultaneously interacts with two glycines (Gly-354 and Gly-357) on HP2 tip. At t = 1.5 it occupies an intermediate position between HP1 and HP2 tips, starting to interact with the Ser-277—Ser-278—Ser-279 motif on HP1. At t = 7.5 ns, the substrate dissociates from HP2 and interacts with HP1 tip residues as well as TM7 and TM8 residues (see Fig. 5). The substrate remains sequestered at the binding site for the remaining duration of MD1 run.
The runs MD1 and MD2 demonstrate that the substrate is readily driven toward the open subunit provided that it reaches a relatively deeper position in the basin. Next, we investigated the diffusion of the substrate from the outside (EC region) to the basin. To this aim we conducted two runs, MD5 and MD6, with glutamate and aspartate molecules included at random positions in the EC and IC regions but not in the basin. We also included in the bulk environment sodium, potassium, and chloride ions at physiological levels. No substrate molecule was observed to diffuse to the basin, although the simulations were extended up to 50 ns in each case. These results suggest that the diffusion of the substrate from the EC region to the interior of the basin could be a rate-limiting process in substrate binding. Once the substrate enters the basin, on the other hand, it takes only nanoseconds to “see” the open hairpin HP2 and diffuse to this highly attractive region to reach a low energy state.
We note that the runs MD0 and MD6 differ not only in their concentrations of substrate but also in the ionic strength of the environment. Because MD6 was conducted with near-physiological concentrations of ions (Na+, K+, and Cl-) (Table 1), the comparison with MD0 may provide insights into the effect of ionic strength on conformational dynamics. The corresponding histograms of conformers (closed and open) in panels A′ and C′ of Fig. 4 exhibit little difference, if any, between the two runs, consistent with the concept that the EC gating is predominantly controlled by the intrinsic dynamics of the protein. However, a closer examination (essential dynamics analysis (29)) of all trajectories in the context of the Na+ effect on substrate binding clearly shows how the subunit mobilities tend to be enhanced with higher concentrations of ions in the bulk. The left two panels in supplementary Fig. S4 display the dispersion of the modes obtained from the runs MD0 and MD6 for the hairpin HP1 (dotted) and HP2 (solid) in the three subunits A (black), B (red), and C (green). The ordinate provides a measure of the amplitudes of motions driven by the individual modes (rank-ordered along the abscissa, starting from the lowest frequency mode). Clearly, the HP2 hairpin in the subunit that opens up (A, black) exhibits the largest mobility in the slowest mode compared with the other subunits, and these differences in mobility vanish at higher modes. What is remarkable, however, is the significant enhancement in the mobility of the same subunit in the medium with the higher ionic strength (compare the root mean square amplitude of ∼6.5 Å in MD6 and 3.5 Å in MD0 in mode 1). The right panels refer to the effect of Na+ binding on the HP1 and HP2 mobilities after substrate binds to the transporter, which will be discussed below.
Sequence of Events Leading to Substrate Binding—To gain an understanding of the events that drive the binding of glutamate, we examined the time evolution of the distance between the glutamate and a number of key residues at the binding site. Panel A in Fig. 5 displays the gradual decrease in the distance between these residues and the glutamate molecule, based on their closest interacting atoms, and Fig. 6 displays snapshots at t = 0, 0.55, 1.0, 1.5, and 7.5 ns.
The initiating events are the recognition of the substrate by Gly-354 at the HP2 tip (Fig. 6) followed by the interaction with Gly-354-Gly-357 (red curve in Fig. 5A) and then Thr-275-Ser-279 (on HP1) (black curve), TM7 residues Asp-312-Thr-314 (green), and TM8 residues Arg-397-Thr-398 (blue), as described in the legend to Fig. 5. In particular, the hydrogen bonds formed with the hydroxyl groups on the Ser-277-Ser-278-Ser-279 motif play a major role in stabilizing the substrate, confirmed in both MD1 and MD2, and gradually pulling the substrate deeper into the binding pocket (Fig. 6). The interactions with Ser-277-Ser-278-Ser-279 are closely maintained once formed (black curve in Fig. 5A). We also repeated our simulations by using aspartate and compared the binding pose from simulations with that observed in the high resolution structure (7). Supplemental Fig. S5 shows the similarity between the protein-substrate interactions observed in simulations and experiments.
The Substrate, Once Bound, Does Not Dissociate, but Inserts Deeper into the Binding Pocket—Remarkably, as soon as the glutamate slips into the binding site, the hairpin HP2 moves closer toward HP1 so as to optimize its interaction and maintains a relatively closer interaction with the core residues near the binding site. No escape of glutamate to the aqueous basin was observed in any run. This is consistent with the kinetic data reported by Otis and Jahr (30) where the forward rate constant for glutamate binding is 6 orders of magnitude larger than the backward reaction (dissociation) rate constant. Fig. 5B illustrates the close coordination of the substrate at the binding at the end of MD1. Residues involved in stabilizing the substrate (labeled in the diagram) are in agreement with those coordinating the bound aspartate in the high resolution structure of GltPh (7). As will be shown below, water molecules and Na+ further contribute to stabilizing the substrate.
Mediating Role of Water Molecules and Continual Hydration of Thr-314—In addition to the glutamate, the water molecules in the basin exhibit a tendency to diffuse to the binding site and to move in and out through the putative EC gate before glutamate binding. But once the glutamate blocks the EC gate upon binding, they find another entry through an interstitial region between HP2 and TM8. Two highly conserved aspartic acids on TM8, Asp-390 and Asp-394, are instrumental in facilitating their entry. TM8 has been pointed out to form a portion of the transport pathway (7). Supplemental Fig. S6 and Movie 3 illustrate the trajectories of five water molecules located at distant positions at the beginning of the run MD1, which enter the binding site at around 3.5 ns. Interestingly, the water molecules do not remain at the binding site but diffuse away in less than 0.5 ns and are replaced by other water molecules, entering from the same pathway such that the binding site is continually hydrated.
An interesting phenomenon was the continual presence of a water molecule hydrogen-bonded to Thr-314 on the NMDGT motif. In the absence of glutamate, when the putative EC gate was open, an exchange of water molecules was observed at this site (see supplemental Fig. S7), but once the glutamate was bound, the bound water molecule remained locked at this precise position throughout the rest of the simulations in both MD1 and MD2. The equivalent residue Thr-402 in human EAAT1 is functionally dead when mutated to alanine (data not shown). We also note that two residues located on the adjacent helical turn, Tyr-405 and Glu-406 in hEAAT1, have been proposed to mediate the co-transport of K+ (31, 32).
A Buried Na+ Binding Site on TM7: Leu-303—Experimental data suggest the co-transport of three Na+ with one glutamate (2). To explore the role and the potential binding sites of Na+, the runs MD3 and MD4 were performed (Table 1).
In MD3, one Na+ was placed at the position of the water molecule interacting with Thr-314, postulating that this could be a cation binding site. However, the Na+ moved down deeper into the binding pocket within the first 100 ps, to settle in a highly stable position coordinated by Asn-310 and Asp-312 on the NMGDT motif and Gly-404 on TM8. It remained at this position for the succeeding 10-ns run (see supplemental Fig. S8).
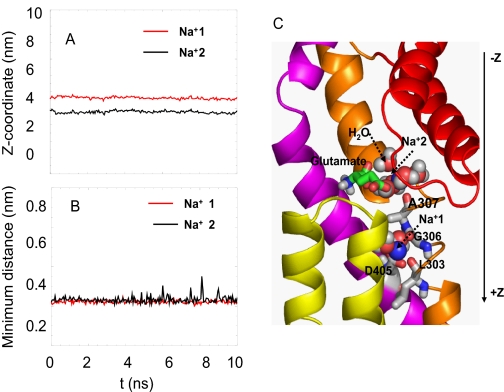
In MD4, three randomly selected water molecules within 5 Å of the bound glutamate were replaced by 3 Na+. One of them escaped to the aqueous region within less than 1 ns. Of the remaining two, one (labeled Na+2 in Fig. 7) settled down, interacting with Ala-307, Asn-310, the bound glutamate, and the water molecule associated with Thr-314, whereas the other (Na+1) moved deeper into the core to interact with Leu-303 and Gly-306 on TM7, Asp-405 on TM8, and a water molecule. These two ions were observed to be remarkably stable throughout the entire duration of MD4. The almost invariant time profiles of their position along the z axis (Fig. 7A) and with respect to the centroid of coordinating residues (Fig. 7B) provide evidence in support of their high stability. Extension of MD4 to 30 ns showed that the profiles remain unchanged.
FIGURE 7.
Two stable binding sites for Na+. Panels A and B shows the high stability of two Na+, labeled Na+1 and Na+2, at their binding sites, and panel C illustrates their coordination in the three-dimensional structure. The ordinate in panel A refers to the positions of the ions along the z axis (cylindrical axis) as a function of time, with z = 0 referring to the EC region and z increasing toward the IC region. Panel B shows the distance between Na+2 and the centroid of its coordinating residues Ala-307, Asn-310, and glutamate (black curve) and the distance between Na+1 and corresponding closest residues Gly-306, Leu-303, and Asp-405 (red curve). The sodium ions are represented as blue spheres, glutamates are in green stick, and the water molecules are red and white spheres. Note that Na+2 is hidden behind the water molecules. Also see supplemental Movie 4.
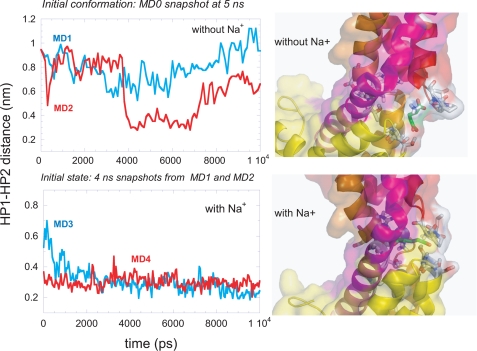
The Presence of Bound Na+ Stabilizes the Substrate and the Closed Form of HP2—The stabilizing role of sodium ions at the binding site is clearly seen in Fig. 8. The top panel displays the results from two runs, MD1 and MD2, in the absence of Na+ at the binding site, and the lower panel displays those from runs MD3 and MD4 in the presence of Na+. The ordinates show the time evolution of the distance between the tips of the hairpins HP1 and HP2 in subunit A. This subunit binds a glutamate molecule within the first 1-2 ns of both runs MD1 and MD2 but continues to either remain open (MD1) or undergo large fluctuations (MD2). Binding of Na+ to the substrate-bound subunit, on the other hand, exhibits a severe suppression in the mobility of the EC gate, in favor of the closed form, as evidenced by the lower panel. These two runs were conducted with two different states of substrate-bound transporter, where the EC gate in subunit A was originally open (MD3) or closed (MD4), and in either case the closed form, evidenced by inter-hairpin distance of about 3 Å, was attained shortly after Na+ binding, and this form remains stable. Comparison of the frequency spectra obtained for MD2 and MD4 in the supplemental Fig. S4, right panels, also confirms the significant effect of Na+ binding on the mobility of the EC gate. The ribbon diagrams in Fig. 8 illustrate how the glutamate penetrates deeper into the binding pocket in the presence of Na+.
FIGURE 8.
Effect of Na+ binding on HP1-HP2 tip distances in the substrate bound simulations. The left panels describe the time evolution of HP1-HP2 tip distances in MD1 (top panel; blue) and MD2 (top panel; red) and in MD3 (bottom panel; blue) and MD4 (bottom panel; red). Note the decrease in HP1-HP2 distance in the presence of Na+ ions (bottom panel). The schematic representations in the right panel focus on the substrate and the residues it interacts with at the binding site, highlighted in stick representation. The lower diagram shows the substrate inserting deeper into the binding site in the presence of sodium ions. The color codes for the secondary structures are same as in Fig. 1C.
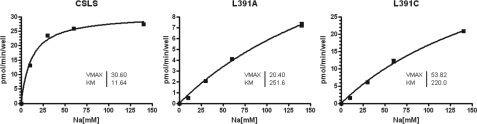
Experimental Verification of the Na+ Binding Role of EAAT1 Leu-391—The first sodium binding site identified here is very similar to that observed in the high resolution GltPh structure. It includes residues Gly-306 and Asp-405 as well as a water molecule trapped at this site, which is also seen in the x-ray structure (7). In addition to these residues, Na+(1) also interacts with Leu-303 (Fig. 7C), which is not seen in the x-ray structure. To test the importance of Leu-303 for Na+ binding, mutants L391A and L391C of the cysteine-less hEAAT1 (Leu-391 is the counterpart of GltPh Leu-303) were constructed, and the Na+ dependence of l-glutamate uptake was examined for each mutant. Both mutants showed an ∼20-fold increase in the Km for Na+ relative to that of the cysteine-less hEAAT1 (Fig. 9). These data suggest that Leu-303 does indeed contribute to one of the Na+ binding sites. Of the two Na+ binding sites detected in the x-ray structure, the second binding site is in the vicinity of that observed in x-ray structure (7) (near Thr-352, Ser-349, Ile-350, and Thr-308).
FIGURE 9.
Impaired sodium binding in L391C and L391A of hEAAT1 (counterpart of Leu-303 in GltPh). The apparent affinity for Na+ was measured as described under “Experimental Procedures.” Data shown are from one of the representative experiments. Sets of 2-4 independent experiments were performed in triplet to confirm the results.
DISCUSSION AND CONCLUSIONS
The present study provides a detailed description of the sequence of events that takes place before glutamate translocation and the effect of Na+ binding on EC gating. Although the time scale of MD simulations is several orders of magnitude shorter than that of transport cycle of glutamate transporters, the substrate binding event is likely to occur within tens of nanoseconds (33-35), and current MD simulations help us gain insights into the molecular events/interactions at that time scale. Table 2 summarizes the residues observed to play a role in our simulations along with their counterparts in hEAAT1 and relevant experimental data available in the literature from several groups using various mammalian orthologues.
TABLE 2.
Comparison of important residues revealed by MD simulations and available experimental observations
Residues in written in boldface are fully or highly conserved among glutamate transporters family members.
| GltPh residue | Structural element | Role observed in MD simulations | EAAT1 counterpart | Role observed in experiments | Reference |
|---|---|---|---|---|---|
| Gly-354, Gly-357 | HP2 | Glu recognition, loop flexibility | Gly-442, Gln-445 | Equivalent residues Ser-440 and Ser-443 in GLT-1 are important for sodium selectivity of the transporter. | Zhang and Kanner (36) |
| Gln-318 | TM7 | Close interaction with HP2a; stabilization of EC gate, confining the bound substrate | Glu-406 | Equivalent residue Glu-373 in EAAC1 has been observed to facilitate glutamate transport when protonated. | Grewer et al. (53) |
| Ser-277-Ser-279 | HP1 | Glu binding | Ser-364-Ser-366 | Mutation and/or sulfhydryl modification of cysteines substituted for these residues significantly disrupts uptake activity. | Seal et al. (2000) (12) and Grunewald and Kanner (41) |
| Asp-312-Thr-314 | TM7 | Glu binding/coordination | Asp-400-Thr-402 | The equivalent residue of Asp-312 (Asp-367 in EAAC1) contributes to sodium binding before substrate binds. | Tao et al. (46) |
| Arg-397-Thr-398 | TM8 | Stabilization of bound glutamate | Arg-479-Thr-480 | The equivalent residue of Arg-397 (Arg-447 in EAAC1) has been proposed to interact with the γ-carboxyl terminal of substrates. | Bendahan et al. (11) |
| Thr-314 | TM7 | Water/Na+ binding | Thr-402 | Mutant T402A is non-functional. Two adjacent residues Tyr-405 and Glu-406 have been implicated in sodium and/or potassium binding. | Kavanaugh et al. (31) and Zhang et al. (s32) |
| Asp-390, Asp-394 | TM8 | Putative pathway for water and Na+ and stabilization of glutamate | Asp-472, Asp-476 | Equivalent residue of Asp-394 (Asp-444 in EAAC1) interacts with the NH2 group of substrates. Equivalent residue Asp-439 in EAAC1 is important for high affinity Na+ binding to glutamate-bound transporter. | Teichman and Kanner (13) and Tao and Grewer (43) |
| Asn-310, Ala-307, Gly-306, Asp-312 | TM7 | Putative Na+ binding site II | Asn-398, Ala-395 Gly-394, Asp-400 | Neutralization of Asp-367 in EAAC1 inhibits Na+ binding. Modification of A395C using sulfhydryl modifying reagents in EAAT1 abolishes transport activity. | Tao et al. (46), Leighton et al. (10), and Seal and Amara (28) |
| Leu-303, Gly-306 | TM7 | Putative Na+ binding site I | Leu-391,Gly-394 | Leu-391 in EAAT1 is Implicated in Na+ binding. | This work |
| Asp-405 | TM8 | Putative Na+ binding site I | Asp-487 | Asp-405 is involved in sodium binding site, and mutant D405N showed reduced aspartate binding affinity. | Boudker et al. (7) |
Structure-encoded Ability of the Subunits to Open the EC Gate—The opening of the HP2 loop in the fast regime sampled by MD simulations supports the view that the transporter possesses an intrinsic, structure-induced ability to open an EC gate for substrate entry. This propensity is supported by a new crystal of GltPh in complex with the non-transportable blocker dl-threo-β-benzyloxyaspartate in which the HP2 loop is dislocated from the substrate binding site compared with the substrate bound structure (7). Gly-354 and Gly-357 (Gly-442 and Gln-445 in hEAAT1) play a dual role in this process: they impart high flexibility to this hairpin, and they assist in recognizing the substrate. Interestingly, the equivalent residues Ser-440 and Ser-443 in GLT-1 have been implicated in Na+ selectivity (36).
Within the time frame of our simulations we have observed the opening of one or two subunits but not all three subunits at the same time. This raises the issue of a possible interdependence between the three subunits which may preclude the simultaneous opening of the three gates. To examine this possibility, we analyzed the statistical distribution of open and closed states. The fraction of time a given subunit resided in the closed form, averaged over all subunits and all runs in Fig. 4, was found to be 0.562. Using this prior probability and assuming the three EC gate opening events to be independent, the expected probabilities of observing 0, 1, 2, or 3 subunits open at a give time are 0.18, 0.42, 0.32, and 0.08, respectively. The fractional residence times of these respective states observed in our simulations are on the other hand, 0.20, 0.44, 0.36, and 0.0. The high correlation (0.98) between the observed distributions and those expected from independent dynamics of the three HP2 loops is in accord with recent studies suggesting that individual subunits function independently for both substrate transport and anion channeling (37-39). The fact that the state with all three HP2 loops open was not observed may be due to inadequate sampling of rare events in MD, although we cannot rule out the possibility of a weak effect induced by intersubunit coupling.
Directed Diffusion and Binding of Substrate within Nanoseconds—The EC gate opening would not be so effective if it were not for the polar groups and, in particular, a sequence of three serines (Ser-277-Ser-278-Ser-279) on HP1 tip that were exposed to the aqueous basin upon opening of the EC gate. These residues serve as efficient attractors directing the diffusion of the glutamate. The substrate remains bound in the core domain for tens of nanoseconds.
The present study underscores the importance of the HP1 serines (Ser-364-Ser-365-Ser-366 in EAAT1). The uptake activities of several EAAT carriers are significantly impaired when residues in this serine stretch are mutated or substituted with cysteine and modified with sulfhydryl modifying reagents (12, 41, 42). Other charged/polar residues that come into play include Asp-390, Asp-394, Arg-397, and Thr-398 on TM8 and Asp-312-Thr-314 on TM7, illustrated in Figs. 5 and 6. Notably, these are all conserved residues among the members of the human EAAT family. Asp-394 and Arg-397 have been proposed to interact with the amino and γ-carboxyl oxygens of the substrate, respectively (7, 11, 13). Asp-312-Thr-314 belongs to the conserved NMDGT motif. The role of Asp-439 (EAAC1), equivalent to Asp-390 (Gltph), has been recently confirmed (43) (Table 2). That of Thr-398 awaits further experimental verification.
Role of Water Molecules—Glutamate binding is preceded by the association of a water molecule with Thr-314, which in turn stabilizes a Na+. These observed interactions presumably compensate for the broken hydrogen bonds at the NMDGT motif of TM7, thus stabilizing a region that is energetically frustrated before substrate/ion binding. In the substrate-bound subunit, water molecules find their way into the binding site through another narrow region, near Asp-390 and Asp-394 on TM8. TM8 forms a kink near Asp-390, centered at Gly-388, and again, similar to the unwound central portion of TM7, this region exhibits a high avidity for forming hydrogen bonds with water molecules and/or salt bridges with the substrate or sodium ions.
Functional Significance of the Unwound Regions of TM Helices—The above results provide evidence for the role of the unwound regions on TM7 and TM8. The marginal (in)stability of these regions predisposes them to engage in intermolecular contacts. Structural models generated by Weinstein and coworkers (44) for neurotransmitter/Na+ symporter family members using the resolved leucine transporter (LeuT) structure (45) as a template also showed that the charged moieties of the substrate interact with the unwound region in the helix TM1. TM1 in LeuT is the structural counterpart of TM7 in GltPh (or EAATs). As noted by Gouaux and co-workers (7), TM1(LeuT)/TM7(GltPh) exhibit similar Na+ binding motifs at their unwound regions despite the low sequence similarity of the families EAATs and neurotransmitter/Na+ symporters. Our study confirms the role of the unwound region on TM7 and also invites attention to that of the unwound region in TM8.
Na+ Binding and Stabilization of the EC Gate—In our simulations one of the sodium ions selects a highly stable conformation coordinated by Asn-310 and Asp-312 on the NMDGT motif and Gly-404 on TM8. It has been proposed that at least one Na+ binds before substrate (35). Interestingly, the equivalent residue of Asp-312 in EAAC1 (D367) has been proposed to be involved in the Na+ binding before substrate binds (46). Therefore, we propose that this could be the Na+ binding site before substrate binding. In MD4, two Na+ binding sites were observed, the first very close to that observed in the x-ray structure, and the second in the vicinity, although not identical. At the second binding site in the x-ray structure, the Na+ interacts with residues on HP2a and Thr-308 on TM7, whereas in the MD simulations, the cation interacts with the bound glutamate, water molecules, and Ala-307 on TM7. During the crystallization of GltPh, thallium was used to enhance the anomalous scattering signaling. Although competition experiments were performed to confirm that the thallium anomalous density peak could be diminished by Na+ competition, it is possible that the actual residues contributing to Na+ binding differ from those obtained in the presence of thallium. Our functional assays with two mutants L391A and L391C (Leu-391 is the equivalent residue in hEAAT1 of Leu-303 in GltPh) showed a substantially increased Na+ Km on both of these mutants, suggesting that Leu-303 could be involved in Na+ binding. Whether Leu-303 directly binds Na+ or whether the experimental data reflect a less direct structural effect remains to be further established.
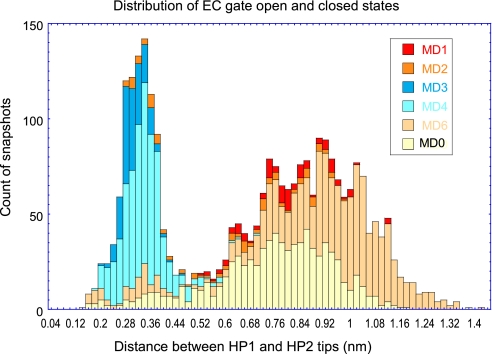
A summary of the results from all runs presented in Fig. 10 permits us to better assess the effect of Na+ binding on EC gate dynamics. The figure shows histograms derived from the essential dynamics analysis of the HP2 motions, which unambiguously display a bimodal distribution of conformations sampled by the EC gate with two peaks corresponding to open and closed states (Fig. 10). In the absence of bound substrate, the subunits exhibit a higher tendency to sample the open state (MD0 and MD6), and this tendency is maintained, albeit at a lesser extent, in the presence of bound substrate (MD1 and MD2). Strikingly, Na+ binding significantly enhances the propensity for closed conformation (MD3 and MD4).
FIGURE 10.
Histogram of HP1-HP2 distance reflecting EC gate dynamics, shown for subunit A in all simulations. The figure displays the bimodal distribution of the HP1-HP2 tip distances in MD0 (yellow), MD1 (red), MD2 (orange), MD3 (blue), MD4(cyan), and MD6 (pink), consistent with two states, open and closed, sampled by the EC gate. The narrow peak around 0.2 nm corresponds to the closed state, whereas the broader peak around 0.9 nm corresponds to the open state. Na+ binding strongly favors the closed state as evidenced by the contributions of MD3 and MD4 to the peak corresponding to the closed state.
Insights into the Structure and Function of the Mammalian Orthologues—On a broader perspective, the intrinsic ability of the transporter to drive the diffusion of the neurotransmitter to its binding site and effectively stabilize it in a constricted region before its release to the IC region is consistent with recent studies pointing to the structure-induced tendency of allosteric enzymes and receptor proteins to undergo conformational changes that are functional (16-19). These observations further suggest that three-dimensional structures have evolved to favor functional motions. GltPh shows a close sequence similarity to its eukaryotic counterparts. Information on the dynamics of GltPh, thus, provides insights into the mechanism of function of mammalian orthologues (Table 2). The similarities in the Na+ coordination geometry (supplemental Fig. S9) and aspartate coordination (supplemental Fig. S5) with the x-ray structure (7) also support mechanisms shared by conserved residues in glutamate transporters.
A widely held view with regard to glutamate transport has been the “alternating access” model (47) in which the binding sites for substrates and co-substrates are alternatively exposed to the EC and IC regions via large scale conformational changes in the transporter. Recent studies, on the other hand, point to small-scale motions that accomplish glutamate uptake in EAATs (40), and a series of small conformational changes have been proposed to propagate across the transporter structure (48). Our simulations also support the importance of local motions for glutamate binding. However, the present study focused on recognition and binding events in substrate transport, and it is conceivable that such events which occur on the nanoseconds regime involve local movements. Further permeation into the protein and release into the cell interior, on the other hand, may require larger scale cooperative motions of the domains and subunits. Coarse-grained normal mode analyses (49, 50) may shed light into allosteric mechanisms that enable the permeation and translocation of substrate, as suggested by applications to other multimeric membrane proteins (51, 52).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R33 GM068400 (to I. B.) and MH080726 (to S. G. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S9 and Movies 1-4.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: EC, extracellular; EAAT, excitatory amino acid transporter; hEAAT, human EAAT; MD, molecular dynamics; TM, transmembrane; HP1, helical hairpin 1; HP2, helical hairpin 2; IC, intracellular; LeuT, leucine transporter.
References
- 1.Amara, S. G., and Fontana, A. C. (2002) Neurochem. Int. 41 313-318 [DOI] [PubMed] [Google Scholar]
- 2.Zerangue, N., and Kavanaugh, M. P. (1996) Nature 383 634-637 [DOI] [PubMed] [Google Scholar]
- 3.Wadiche, J. I., Amara, S. G., and Kavanaugh, M. P. (1995) Neuron 15 721-728 [DOI] [PubMed] [Google Scholar]
- 4.Fairman, W. A., Vandenberg, R. J., Arriza, J. L., Kavanaugh, M. P., and Amara, S. G. (1995) Nature 375 599-603 [DOI] [PubMed] [Google Scholar]
- 5.Danbolt, N. C. (2001) Prog. Neurobiol. 65 1-105 [DOI] [PubMed] [Google Scholar]
- 6.Yernool, D., Boudker, O., Jin, Y., and Gouaux, E. (2004) Nature 431 811-818 [DOI] [PubMed] [Google Scholar]
- 7.Boudker, O., Ryan, R. M., Yernool, D., Shimamoto, K., and Gouaux, E. (2007) Nature 445 387-393 [DOI] [PubMed] [Google Scholar]
- 8.Gendreau, S., Voswinkel, S., Torres-Salazar, D., Lang, N., Heidtmann, H., Detro-Dassen, S., Schmalzing, G., Hidalgo, P., and Fahlke, C. (2004) J. Biol. Chem. 279 39505-39512 [DOI] [PubMed] [Google Scholar]
- 9.Brocke, L., Bendahan, A., Grunewald, M., and Kanner, B. I. (2002) J. Biol. Chem. 277 3985-3992 [DOI] [PubMed] [Google Scholar]
- 10.Leighton, B. H., Seal, R. P., Watts, S. D., Skyba, M. O., and Amara, S. G. (2006) J. Biol. Chem. 281 29788-29796 [DOI] [PubMed] [Google Scholar]
- 11.Bendahan, A., Armon, A., Madani, N., Kavanaugh, M. P., and Kanner, B. I. (2000) J. Biol. Chem. 275 37436-37442 [DOI] [PubMed] [Google Scholar]
- 12.Seal, R. P., Leighton, B. H., and Amara, S. G. (2000) Neuron 25 695-706 [DOI] [PubMed] [Google Scholar]
- 13.Teichman, S., and Kanner, B. I. (2007) J. Gen. Physiol. 129 527-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosental, N., Bendahan, A., and Kanner, B. I. (2006) J. Biol. Chem. 281 27905-27915 [DOI] [PubMed] [Google Scholar]
- 15.Torres, G. E., and Amara, S. G. (2007) Curr. Opin. Neurobiol. 17 304-312 [DOI] [PubMed] [Google Scholar]
- 16.Tobi, D., and Bahar, I. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18908-18913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, L. W., and Bahar, I. (2005) Structure 13 893-904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahar, I., Chennubhotla, C., and Tobi, D. (2007) Curr. Opin. Struct. Biol. 17 633-640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henzler-Wildman, K., and Kern, D. (2007) Nature 450 964-972 [DOI] [PubMed] [Google Scholar]
- 20.Schwieters, C. D., Kuszewski, J. J., Tjandra, N., and Clore, G. M. (2003) J. Magn. Reson. 160 65-73 [DOI] [PubMed] [Google Scholar]
- 21.Berendsen, H., VanderSpoel, D., and VanDrunen, R. (1995) Comput. Phys. Commun. 91 43-56 [Google Scholar]
- 22.Tieleman, D. P., and Berendsen, H. J. C. (1996) J. Chem. Phys. 105 4871-4880 [Google Scholar]
- 23.Shrivastava, I. H., and Sansom, M. S. (2000) Biophys. J. 78 557-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capener, C. E., Shrivastava, I. H., Ranatunga, K. M., Forrest, L. R., Smith, G. R., and Sansom, M. S. (2000) Biophys. J. 78 2929-2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wriggers, W., and Schulten, K. (1999) Proteins 35 262-273 [PubMed] [Google Scholar]
- 26.Jensen, M. O., Park, S., Tajkhorshid, E., and Schulten, K. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6731-6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gullingsrud, J., and Schulten, K. (2003) Biophys. J. 85 2087-2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seal, R. P., and Amara, S. G. (1998) Neuron 28 1487-1498 [DOI] [PubMed] [Google Scholar]
- 29.Amadei, A., Linssen, A. B., and Berendsen, H. J. (1993) Proteins 17 412-425 [DOI] [PubMed] [Google Scholar]
- 30.Otis, T. S., and Jahr, C. E. (1998) J. Neurosci. 18 7099-7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kavanaugh, M. P., Bendahan, A., Zerangue, N., Zhang, Y., and Kanner, B. I. (1997) J. Biol. Chem. 272 1703-1708 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y., Bendahan, A., Zarbiv, R., Kavanaugh, M. P., and Kanner, B. I. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 751-755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grewer, C., Watzke, N., Wiessner, M., and Rauen, T. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 9706-9711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otis, T. S., and Kavanaugh, M. P. (2000) J. Neurosci. 20 2749-2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watzke, N., Bamberg, E., and Grewer, C. (2001) J. Gen. Physiol. 117 547-562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Y., and Kanner, B. I. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 1710-1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grewer, C., Balani, P., Weidenfeller, C., Bartusel, T., Tao, Z., and Rauen, T. (2005) Biochemistry 44 11913-11923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leary, G. P., Stone, E. F., Holley, D. C., and Kavanaugh, M. P. (2007) J. Neurosci. 27 2938-2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch, H. P., Brown, R. L., and Larsson, H. P. (2007) J. Neurosci. 27 2943-2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch, H. P., and Larsson, H. P. (2005) J. Neurosci. 25 1730-1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grunewald, M., and Kanner, B. I. (2000) J. Biol. Chem. 275 9684-9689 [DOI] [PubMed] [Google Scholar]
- 42.Slotboom, D. J., Sobczak, I., Konings, W. N., and Lolkema, J. S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14282-14287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao, Z., and Grewer, C. (2007) J. Gen. Physiol. 129 331-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beuming, T., Shi, L., Javitch, J. A., and Weinstein, H. (2006) Mol. Pharmacol. 70 1630-1642 [DOI] [PubMed] [Google Scholar]
- 45.Yamashita, A., Singh, S. K., Kawate, T., Jin, Y., and Gouaux, E. (2005) Nature 437 215-223 [DOI] [PubMed] [Google Scholar]
- 46.Tao, Z., Zhang, Z., and Grewer, C. (2006) J. Biol. Chem. 281 10263-10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jardetzky, O. (1966) Nature 211 969-970 [DOI] [PubMed] [Google Scholar]
- 48.Koch, H. P., Hubbard, J. M., and Larsson, H. P. (2007) J. Biol. Chem. 282 24547-24553 [DOI] [PubMed] [Google Scholar]
- 49.Ma, J. (2005) Structure 13 373-380 [DOI] [PubMed] [Google Scholar]
- 50.Bahar, I., and Rader, A. J. (2005) Curr. Opin. Struct. Biol. 15 586-592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shrivastava, I. H., and Bahar, I. (2006) Biophys. J. 90 3929-3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taly, A., Corringer, P. J., Grutter, T., Prado de Carvalho, L., Karplus, M., and Changeux, J. P. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16965-16970 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.