Abstract
Tautomycin (TTM) is a highly potent and specific protein phosphatase inhibitor isolated from Streptomyces spiroverticillatus. The biological activity of TTM makes it an important lead for drug discovery, whereas its spiroketal-containing polyketide chain and rare dialkylmaleic anhydride moiety draw attention to novel biosynthetic chemistries responsible for its production. To elucidate the biosynthetic machinery associated with these novel molecular features, the ttm biosynthetic gene cluster from S. spiroverticillatus was isolated and characterized, and its involvement in TTM biosynthesis was confirmed by gene inactivation and complementation experiments. The ttm cluster was localized to a 86-kb DNA region, consisting of 20 open reading frames that encode three modular type I polyketide synthases (TtmHIJ), one type II thioesterase (TtmT), five proteins for methoxymalonyl-S-acyl carrier protein biosynthesis (Ttm-ABCDE), eight proteins for dialkylmaleic anhydride biosynthesis and regulation (TtmKLMNOPRS), as well as two additional regulatory proteins (TtmF and TtmQ) and one tailoring enzyme (TtmG). A model for TTM biosynthesis is proposed based on functional assignments from sequence analysis, which agrees well with previous feeding experiments, and has been further supported by in vivo gene inactivation experiments. These findings set the stage to fully investigate TTM biosynthesis and to biosynthetically engineer new TTM analogs.
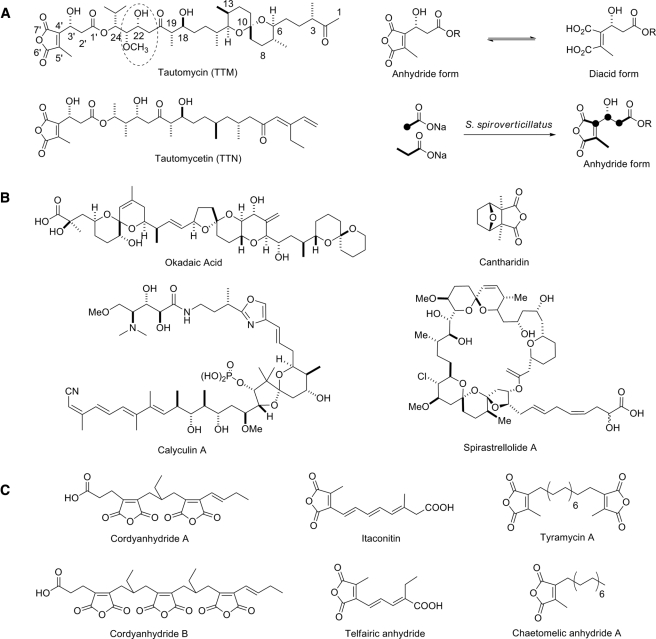
Tautomycin (TTM)2 is a polyketide natural product first isolated in 1987 from Streptomyces spiroverticillatus (1). The structure and stereochemistry of TTM were established on the basis of chemical degradation and spectroscopic evidence (2-4). TTM contains several features not common to polyketide natural products, including a spiroketal group, a methoxymalonate-derived unit, and an acyl chain bearing a dialkylmaleic anhydride moiety. Structurally related to TTM is tautomycetin (TTN), which was first isolated in 1989 from Streptomyces griseochromogenes following the discovery of TTM (5, 6). The structure of TTN was deduced by chemical degradation and spectroscopic analysis (6), and its stereochemistry was established by comparison of spectral data with those of TTN degradation products and synthetic fragments (7). Both TTM and TTN exist as tautomeric mixtures composed of two interconverting anhydride and diacid forms in approximately a 5:4 ratio under neutral conditions (Fig. 1A) (1, 2).
FIGURE 1.
A, structures of TTM and TTN in anhydride or diacid forms, and biosynthetic origin of the dialkylmaleic anhydride by feeding experiments using 13C-labeled acetate and propionate. The methoxymalonate-derived unit in TTM is highlighted by the dotted oval. R, polyketide moiety of TTM or TTN. B, selected natural product inhibitors of PP-1 and PP-2A featuring a spiroketal or dialkylmaleric anhydride moiety. C, selected natural products containing a dialkylmaleic anhydride moiety.
Early studies of TTM revealed its ability to induce morphological changes in leukemia cells (8). However, it was later realized that TTM is a potent and specific inhibitor of protein phosphatases (PPs) PP-1 and PP-2A (9). PP-1 and PP-2A are two of the four major serine/threonine protein phosphatases that regulate diverse cellular events such as cell division, gene expression, muscle contraction, glycogen metabolism, and neuronal signaling in eukaryotic cells (10-12). Many natural product PP-1 and PP-2A inhibitors are known, including okadaic acid (13), calyculin-A (14), phoslactomycin, spirastrellolide, and cantharidin (15) (Fig. 1B), as well as TTM (16, 17), and TTN (18). They have served as useful tools to study PP-involved intracellular events in vivo and as novel leads for drug discovery (10-12). Among these PP inhibitors, TTM and TTN are unique because of their PP-1 selectivity. Despite their structural similarities, TTM exhibits potent specific inhibition of PP-1 and PP-2A with IC50 values of 22-32 nm and only a slight preference for PP-1 (18). Conversely, TTN shows nearly a 40-fold higher binding affinity to PP-1 (IC50 = 1.6 nm) than to PP-2A (IC50 = 62 nm) (18). Because the major structural differences between TTM and TTN reside in the region distal to the dialkylmaleic anhydride moiety (Fig. 1A), it has been proposed that differences in these moieties might be responsible for the PP-1 selectivity (17-19). Finally, TTN also has an impressive immuno-suppressive activity (20, 21), which is apparently devoid for TTM. Clearly, the structural differences between these two polyketides translate into large, exploitable differences in bio-activities, yet an understanding of the biosynthetic origins of these differences remains elusive.
The spiroketal and dialkylmaleic anhydride features of TTM are uncommon for polyketide natural products, as is the methoxymalonate-derived unit (Fig. 1A). Few studies have been carried out for spiroketal biosynthesis, yet it is reasonably common among the phosphatase inhibitors such as calyculin A, okadaic acid, and a few others (Fig. 1B). Less common, but still found in the phosphatase inhibitor cantharidin, as well as TTM and TTN, is the dialkylmaleic anhydride moiety (Fig. 1B); this unit appears in a number of other natural products (Fig. 1C), although the biosynthetic steps leading to this reactive moiety (a protected version of a dicarboxylate) have not been rigorously investigated. Feeding experiments with 13C-labeled precursors indicated that the anhydride of TTM and TTN is assembled from a propionate and an as yet undefined C-5 unit (Fig. 1A), which would require novel chemistry for polyketide biosynthesis (22). TTM differentiates itself from all known PP-1 and PP-2A inhibitors by virtue of its unique combination of both the dialkymaleic anhydride and spiroketal functionalities.
Multiple total syntheses of TTM and a small number of analogs have been reported, confirming the predicted structure and absolute stereochemistry and facilitating structure-activity relationship studies on PP inhibition and apoptosis induction (19, 23-25). These studies revealed that: (i) the C22-C26 carbon chain and the dialkylmaleic anhydride are the minimum requirements for TTM bioactivity; (ii) the C18-C21 carbon chain and 22-hydroxy group are important for PP inhibition; (iii) the spiroketal moiety determines the affinity to specific protein phosphatases; (iv) the active form is most likely the dicarboxylate; and (v) 3′-epi-TTM exhibits 1,000-fold less activity than TTM. However, taken as a whole, none of the analogs had an improved potency or selectivity for PP-1 inhibition than the natural TTM (19, 22-25). As a result, a more specific inhibitor of PP-1 is urgently awaited to differentiate the physiological roles of PP-1 and PP-2A in vivo and to explore PPs as therapeutic targets for drug discovery.
We have undertaken the cloning and characterization of the TTM biosynthetic gene cluster from S. spiroverticillatus as the first step toward engineering TTM biosynthesis for novel analogs (26). We report here: (i) cloning and sequencing of the complete ttm gene cluster, (ii) determination of the ttm gene cluster boundaries, (iii) bioinformatics analysis of the ttm cluster and a proposal for TTM biosynthesis, and (iv) genetic characterization of the TTM pathway to support the proposed pathway. Of particular interest has been the identification of genes possibly related to dialkylmaleic anhydride biosynthesis, the unveiling of the ttm polyketide synthase (PKS) genes predicted to select and incorporate four different starter and extender units for TTM production, and the apparent lack of candidate genes associated with spiroketal formation. These findings now set the stage to engineer TTM analogs for novel PP-1- and PP-2A-specific inhibitors by applying combinatorial biosynthetic methods to the TTM biosynthetic machinery.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids—Escherichia coli DH5α was used as the host for general subcloning. E. coli XL 1-Blue MR (Stratagene, La Jolla, CA) was used as the transduction host for cosmid library construction. E. coli ET12567/pUZ8002 was used as the cosmid donor host for E. coli-Streptomyces conjugation (27). E. coli BW25113/pIJ790 and E. coli DH5α/pIJ773 were provided by John Innes Center (Norwich, UK) as part of the REDIRECT Technology kit (28). The S. spiroverticillatus wild-type strain has been described previously (1, 26). pWHM79 (29) and pBS8004 (30) have been reported before, and SuperKos1, a modified version of SuperCos1 (Stratagene, La Jolla, CA) that lacks the neomycin (neo) resistance marker, was used to construct the S. spiroverticillatus genomic library (26).
Biochemicals, Chemicals, and Media—Common biochemicals and chemicals were from standard commercial sources. E. coli strains carrying plasmids were grown in Luria-Bertani medium with appropriate antibiotics selection. All of the media for Streptomyces growth were prepared according to standard protocols (31). ISP-4 and tryptic soy broth were from Difco Laboratories (Detroit, MI), and modified ISP-4 is ISP-4 supplemented with 0.05% yeast extract and 0.1% tryptone (32).
DNA Isolation and Manipulation—Plasmid extraction and DNA purification were carried out by using commercial kits (Qiagen). Genomic DNAs were isolated according to the literature protocol (31). The digoxigenin-11-dUTP labeling and detection kit (Roche Applied Science) was used for preparation of DNA probes, and detection by colony and Southern hybridization was carried out as per the manufacturer instructions.
Genomic Library Construction and Screening—Genomic library construction and screening were carried out following standard protocols (26, 31). Thus, S. spiroverticillatus genomic DNA was partially digested with Sau3AI to yield a smear of about 45-55 kb, dephosphorylated with shrimp alkaline phosphatase, and ligated into SuperKos1 that was pretreated with XbaI, dephosphorylated, and digested with BamHI. The resulting ligation mixture was packaged with the Gigapack III Gold (Stratagene, La Jolla, CA) and transduced into E. coli XL 1-Blue MR to generate the genomic library. The transduced cells were spread onto LB plates containing 200 μg/ml of ampicillin, and the plates were incubated at 37 °C overnight. The titer of the primary library was ∼105 colony-forming units/μg of DNA. The average size of inserts for the cosmid library was determined to be 40-45 kb by restriction enzyme analysis of 12 randomly selected cosmids.
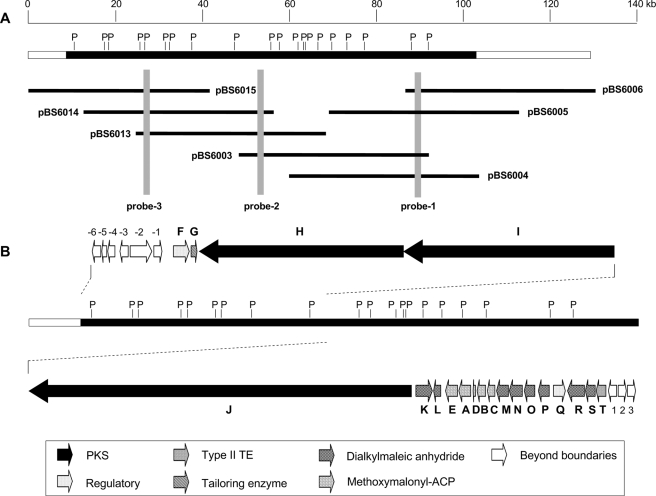
The genomic library was first screened by colony hybridization (31). The positive clones were then rescreened by PCR and confirmed by Southern hybridization. Probe 1 is an acyl-CoA dehydrogenase (ADH) gene fragment (26), and probe 2 (563 bp) and probe 3 (578 bp) were prepared by PCR using the following pairs of primers: P2F, 5′-GGGTAAGAGGTTCGAA-CGCG-3′/P2R, 5′-GCAGGCCTGTGTCCATGG-3′, and P3F, 5′-AAGCGAGCGTCGTCCGG-GCC-3′/P3R, 5′-CCACGGGGACGCTGAGCAC-3′, respectively. A 130-kb DNA region containing the complete ttm biosynthetic gene cluster was localized as represented by overlapping cosmids pBS6003, pBS6004, pBS6005, pBS6006, pBS6013, pBS6014, and pBS6015 (Fig. 2A).
FIGURE 2.
A, restriction map of the 130-kb DNA region from S. spiroverticillatus harboring the entire ttm gene cluster as represented by seven overlapping cosmids. B, genetic organization of the ttm gene cluster. The solid black bar indicates the DNA region sequenced. Proposed functions for individual open reading frames are coded with various patterns and summarized in Table 1. P, PstI.
DNA Sequencing and Analysis—The three cosmids, pBS6003, pBS6013, and pBS6015, were sequenced on both strands by the dideoxynucleotide chain termination method using a shotgun library (Lucigen, Middleton, WI). An additional 10-kb DNA region downstream of pBS6003 was determined by subcloning and primer walking strategies from pBS6004. Sequencing reactions were run using Big Dye Terminator mix (Applied Biosystems, Foster City, CA), purified using CleanSeq magnetic beads (Agencourt Biosciences, Beverly, MA), and sequenced by the University of Wisconsin Biotechnology Center (Madison, WI). Sequence assembly and contiguous alignments were carried out using the Seqman program in the Lasergene software package (DNASTAR Inc. Madison, WI). Assignments of open reading frames (orfs) and their functional predictions were accomplished with the FramePlot 2.3.2 program and Blast program, respectively. The phylogenetic analysis of acyltransferase (AT) domains was performed by using MEGA version 4.0.
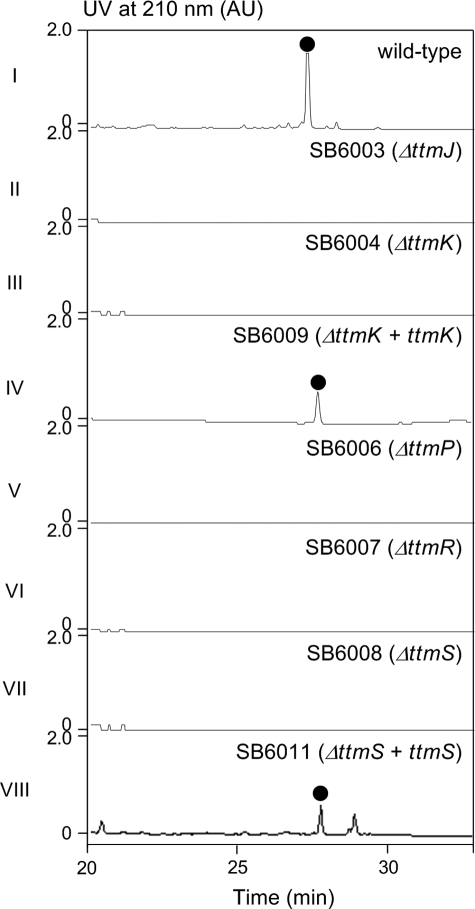
Gene Inactivation and Complementation—Target genes were inactivated by the REDIRECT Technology according to the literature protocol (28). Briefly, an apramycin resistance gene aac(3)IV/oriT cassette was used to replace an internal region of the target gene. To facilitate selection of the double cross-over mutants by resistance phenotype, the neo resistance gene was introduced to pBS6004 and pBS6005 by the REDIRECT Technology using 5.3-kb BspHI fragment from SuperCos1 to afford pBS6016 and pBS6017, respectively. Mutant cosmids pBS6018 (ΔttmJ), pBS6019 (ΔttmK), pBS6021 (ΔttmP), pBS6022 (ΔttmR), and pBS6023 (ΔttmS) were constructed (supplemental Table S1) and introduced into S. spiroverticillatus by conjugation from E. coli ET12567/pUZ8002 according to the literature procedure with the following modifications (31). S. spiroverticillatus spores were suspended in tryptic soy broth medium and heat-shocked at 45 °C for 15 min, followed by incubation at 30 °C for 6 h. Germinated spores (as conjugation recipients) were mixed with E. coli ET12567/pUZ8002 harboring each of the mutant cosmids (as conjugation donor) and spread onto modified ISP-4 plates freshly supplemented with 20 mm MgCl2. After incubation at 28 °C for 16-22 h, each plate was overlaid with 1 ml of sterilized water containing apramycin at a final concentration of 30 μg/ml and nalidixic acid at a final concentration of 50 μg/ml. Incubation continued at 28 °C until exconjugants appeared. The desired double cross-over mutants, selected by the apramycin-resistant and kanamycin-sensitive phenotype, were isolated as SB6003 (ΔttmJ), SB6004 (ΔttmK), SB6006 (ΔttmP), SB6007 (ΔttmR), and SB6008 (ΔttmS), whose genotypes were verified by Southern analysis (supplemental Table S2 and supplemental Figs. S1-S5).
To construct the expression plasmids for mutant complementation, the ErmE* promoter (31) was first amplified from pWHM79, digested with EcoRI, and cloned into the same site of pBS8004 to afford pBS6027. The ttmK and ttmS genes were then amplified from pBS6004, digested with NsiI and XbaI, and cloned into the same sites of pBS6027 to yield pBS6024 (for ttmK expression) and pBS6026 (for ttmS expression). Introduction of pBS6024 and pBS6026 into the mutant strains SB6004 and SB6008 by conjugation finally afford the strains SB6009 and SB6011, in which the ΔttmK and ΔttmS mutations were complemented by the constitutive expression of functional copies of ttmK and ttmS, respectively, under the control of the ErmE* promoter (supplemental Table S3).
Production and Analyses of TTM in S. spiroverticillatus Wild-type and Recombinant Strains—A two-stage fermentation process was adopted (26). Briefly, a spore suspension (5 μl) of the S. spiroverticillatus wild-type or recombinant strains was first inoculated into 50 ml of seed medium in a 250-ml flask and incubated at 28 °C and 250 rpm for 2 days. The resulting seed culture (5 ml) was used to inoculate 50 ml of production medium in a 250-ml flask and incubated at 28 °C and 250 rpm for an additional 3 days. Both seed and production media consisted of 2% glucose (sterile stock solution added after autoclaving), 0.5% soluble starch, 0.05% beef extract, 0.2% dry yeast, 1% soybean flour, 0.2% NaCl, and 0.0025% K2HPO4, pH 7.0. The fermentation broth was adjusted to pH 4.0 with 1.0 n HCl and centrifuged and filtered to remove the mycelia. The resulting supernatant was extracted twice with an equal volume of EtOAc. The combined EtOAc extracts were concentrated in vacuo to afford an oily residue. The latter was dissolved in CH3CN, filtered through a 0.2-μm filter, and subjected to HPLC analysis. The HPLC chromatography system consisted of Varian ProStar 210 pumps and a ProStar 330 photodiode array detector (Varian, Walnut Creek, CA). Analytical HPLC was carried out on a Alltima C18 column (5 μ, 250 × 4.6 mm; Alltech Associates Inc., Deerfield, IL) developed with a linear gradient from 15 to 80% CH3CN/H2O in 20 min followed by an additional 10 min at 100% CH3CN at a flow rate of 1 ml/min and UV detection at 210 nm. Under these conditions, the TTM titer ranges between 10 and 15 mg/liter. The identity of TTM produced by the S. spiroverticillatus wild-type and recombinant strains is confirmed by MS and 1H and 13C NMR analysis. Liquid chromatography-MS was carried out on an Agilent 1100 HPLC-MSD SL quadrupole mass spectrometer equipped with both orthogonal pneumatically assisted electrospray and atmospheric pressure chemical ionization sources (Santa Clara, CA). 1H and 13C NMR data were acquired on a VARIAN Inova-500 spectrometer (Palo Alto, CA). TTM was dissolved in CDCl3 with TMS as an internal standard.
Nucleotide Sequence Accession Number—The nucleotide sequence reported in this paper is available in the GenBank™ data base under accession number EF990140.
RESULTS
Cloning and Sequencing of the ttm Biosynthetic Cluster—Previously, an S. spiroverticillatus cosmid library was constructed and screened for the methoxymalonyl-acyl carrier protein (ACP) biosynthetic locus by using a PCR-amplified ADH gene fragment as a probe (probe 1) (Fig. 2A) (26). Upon end sequencing, all of the positive cosmids have PKS encoding sequences at one end of the inserts, consistent with the polyketide biosynthetic origin of TTM. Additional chromosome walking, carried out with probes 2 and 3, led to the localization of a 130-kb region covered by overlapping cosmids as exemplified by pBS6003, pBS6004, pBS6005, pBS6006, pBS6013, pBS6014, and pBS6015 from which a total of 100-kb region was ultimately sequenced on both strands (Fig. 2A). The nucleotide sequence, possessing an overall G+C content of 72.0%, was deposited in GenBank™ under the accession number EF990140, and the sequence was analyzed for putative orfs with the FramePlot 2.3.2 program. The deduced products for each orf were aligned with homologous sequences using BLAST programs. A total of 29 orfs were identified within the sequenced region, 20 of which were designated as ttm genes (Fig. 2B). The predicted functions for each of the ttm gene products and their homologs are summarized in Table 1. Notably, the deduced gene products include three large type I PKSs with twelve modules (Ttm-HIJ), one type II thioesterase (TE) (TtmT), five proteins responsible for methoxymalonyl-ACP biosynthesis (TtmABCDE), eight enzymes involved in dialkylmaleic anhydride biosynthesis and regulation (TtmKLMNOPRS), two additional regulatory proteins (TtmFQ), and one enzyme catalyzing an additional tailoring step (TtmG) (Fig. 2B and Table 1).
TABLE 1.
Deduced functions of open reading frames in the tautomycin biosynthetic gene cluster
| Gene | Sizea | Proposed function | Homologb | Identity/similarity |
|---|---|---|---|---|
| %/% | ||||
| orf(−3) | 237 | Dehydrogenase | FabG (1Q7C_A) | 47/64 |
| orf(−2) | 589 | Flavohemoprotein | Hmp (CAM78118) | 25/35 |
| orf(−1) | 231 | Unknown | YedU (1ONS_A) | 22/31 |
| Predicted upstream boundary of the ttm cluster | ||||
| ttmF | 429 | Transcriptional activator | StaR (BAC55205) | 12/18 |
| ttmG | 176 | Decarboxylase | Plav_1659 (YP_001412935) | 30/50 |
| ttmH | 5770 | Polyketide synthase (modules 10-12) | ||
| ttmI | 5710 | Polyketide synthase (modules 7-9) | ||
| ttmJ | 10827 | Polyketide synthase (loading module and modules 1-6) | ||
| ttmK | 456 | Esterase | PnbA (1QE3_A) | 29/44 |
| ttmL | 190 | Unknown | Ybhb (1FUX_A) | 23/30 |
| ttmE | 361 | Glyceryl transferase/phosphatase | FkbH (AAF86387) | 67/77 |
| ttmA | 373 | Acyl CoA dehydrogenase | FkbI (1R2J_A) | 62/73 |
| ttmD | 84 | Acyl carrier protein | FkbG (AAF86389) | 57/69 |
| ttmB | 289 | β-Hydroxyacyl CoA dehydrogenase | FkbK (AAF86390) | 56/68 |
| ttmC | 218 | O-Methyl transferase | FkbG (AAF86386) | 61/75 |
| ttmM | 353 | α-Ketoglutarate-dependent hydroxylase | XanA (XP_659714) | 23/34 |
| ttmN | 368 | Unknown | EhpF (AAN40895) | 35/51 |
| ttmO | 314 | Citryl CoA lyase | CitE (1Z6K_A) | 25/36 |
| ttmP | 407 | CoA transferase | CaiB (1XVV_A) | 24/41 |
| ttmQ | 351 | Transcriptional activator | StrR (CAH94333) | 36/48 |
| ttmR | 467 | Dehydratase | PrpD (2HP3_A) | 19/30 |
| ttmS | 274 | Cyclase | PFL_4035 (YP_261132) | 26/40 |
| ttmT | 257 | Thioesterase | PikAV (AAC69333) | 41/52 |
| Predicted downstream boundary of the ttm cluster | ||||
| orf1 | 90 | Unknown | ||
| orf2 | 195 | Unknown | ||
| orf3 | 129 | Unknown |
The numbers are in amino acids.
Given in parentheses are NCBI accession numbers.
Determination of the ttm Gene Cluster Boundaries by Gene Inactivation—The ttm gene cluster boundaries were defined through inactivation of selected genes, orf(-3), orf(-1), and orf3, residing at the distal ends of the sequenced region (Fig. 2B). For the upstream boundary, orf(-3) encodes a putative dehydrogenase containing a conserved short chain dehydrogenase domain, similar to FabG from Brachyspira hyodysenteriae (33). The gene orf(-1) encodes a protein of unknown function that contains a type 1 glutamine amidotransferase-like domain found in proteins similar to the E. coli chaperone Hsp31 (EcHsp31) (34). For the downstream boundary, the deduced gene products of orf1, orf2, and orf3 are all hypothetical proteins. Spot sequencing of regions downstream of orf3 showed genes with deduced products homologous to colossin A and serine/threonine protein kinase, none of which are expected to participate in TTM biosynthesis on the basis of the TTM structure and its biosynthetic origin. Inactivation of orf(-3), orf(-1), and orf3 by gene replacement had no impact on TTM production, confirming that they are not part of the ttm biosynthetic cluster.
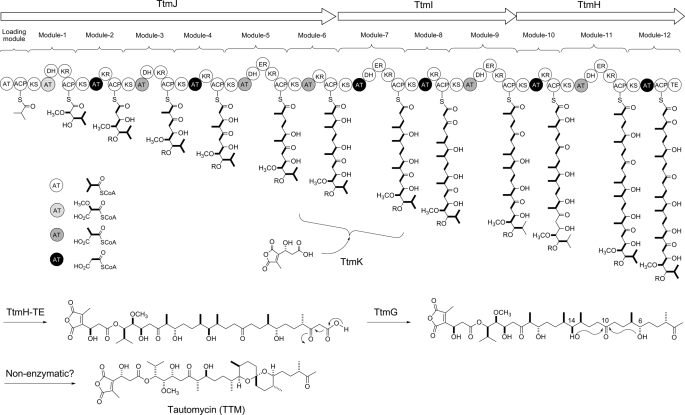
Genes Encoding Modular Polyketide Synthases for the Biosynthesis of the Polyketide Moiety of TTM—Three large open reading frames, designated ttmJ, ttmI, and ttmH, were found to encode type I PKSs responsible for assembly of the TTM polyketide backbone (Table 1 and Figs. 2B and 3). The ttmJ gene encodes the loading module and extension modules 1-6, ttmI encodes the extension modules 7-9, and ttmH encodes extension modules 10-12 and has a C-terminal TE domain. Together, the TTM PKS of TtmJ, TtmH, and TtmI consists of one loading module and 12 extension modules and catalyzes 12 rounds of decarboxylative condensation, using one isobutyryl CoA as a starter unit (loading module) and one methoxymalonyl ACP (module 1), six malonyl CoA (modules 2, 4, 7, 8, 10, 12), and five methyl malonyl CoA (modules 3, 5, 6, 9, 11) as extender units, for initiation, elongation, and termination of the biosynthesis of the polyketide backbone of TTM (Fig. 3).
FIGURE 3.
Deduced module and domain organization of TtmHIJ PKSs and a linear model for the TTM PKS templated assembly of the TTM polyketide backbone featuring four varying starter and extender units as well as key tailoring steps for TTM biosynthesis. R, H or dialkylmeric anhydride unit to emphasize that the precise timing for the TtmK-catalyzed coupling step is unknown. The AT domains are coded with various patterns to highlight their substrate specificity. KS, ketosynthase; ER, enoylreductase.
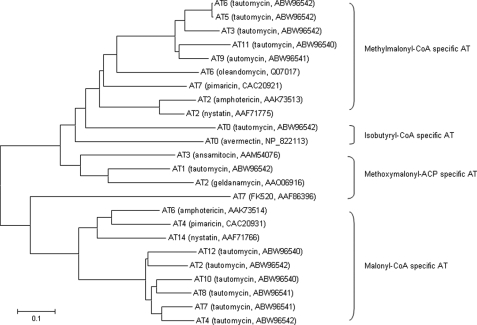
Module and domain organizations within the TtmHIJ PKSs were deduced on the basis of sequence homology comparison with known PKSs (35). The loading module is characterized with AT and ACP domains, and each of the 12 extension modules is minimally characterized by a ketosynthase, AT, and ACP domains. All ketosynthase domains contain the Cys-His-His catalytic triad that is absolutely required for catalyzing the decarboxylative condensation. All the ACP domains feature the highly conserved signature motif of LGGXS. The serine residue of this motif is the site for 4′-phospho-panthetheinylation, a post-translational modification that is essential for polyketide biosynthesis by converting the apo-ACPs into the functional holo-ACPs. The choice of the starter unit for the loading module and the extender unit for each of the extension modules is dictated by the corresponding AT domains, whose specificity is predicted on the basis of sequence comparison with ATs of known substrates (Fig. 4). The 12 extension modules are also characterized with additional domains such as ketoreductase (KR), dehydratase (DH), and enoylreductase, whose presence accounts for the reductive modification of the β-keto group of the growing polyketide intermediate during each cycle of elongation. Functional KR domains containing the conserved consensus sequence GXGXXGXXA associated with NADP(H) binding are predicted for modules 1, 2, 4, 5, 7, 9, 10, and 11 (36). Functional DH domains featuring the conserved consensus sequence HXXXGXXXXP are found for modules 5, 7, and 11, with the exception of the DH domain in module 9 that contains aHXXXEXXXXP motif (37, 38). Functional enoylreductase domains having the conserved sequence GXGXAAXXXA are identified for modules 5, 7, and 9, except for the enoylreductase domain in module 11 with the active site motif GXHX- AAXXXG (36). Intact DH domains are also present in modules 1, 4, 10, and 12, and an intact KR domain is also present in module 3. These activities, however, appear to be unnecessary in their respective modules according to a co-linear model for TTM biosynthesis. Finally, the TE domain at the C terminus of TtmH terminates polyketide biosynthesis by liberating full-length polyketide intermediate from the TTM PKS biosynthetic machinery (Fig. 3).
FIGURE 4.
Phylogenetic analysis of TtmHIJ AT domains and its homologs from type I PKSs that specify malonyl-CoA, methylmalonyl-CoA, methoxymalonyl-ACP, or isobutyryl-CoA. Cluster names and NCBI accession numbers for each of the AT domains are given in parentheses. The scale bar represents 0.1 amino acid substitution per position.
The involvement of TtmHIJ PKS in TTM biosynthesis was confirmed by inactivating the ttmJ gene. To isolate the ΔttmJ mutant, cosmid pBS6018, in which the ketosynthase domain of module 1 encoding sequence of ttmJ was replaced with the aac(3)IV/oriT cassette (supplemental Fig. S1), was introduced into S. spiroverticillatus. Exconjugants that were apramycin-resistant and kanamycin-sensitive were selected as double cross-over mutants, named SB6003, and confirmed by PCR and Southern blot analysis (supplemental Fig. S1). Fermentation of SB6003 followed by extraction and HPLC analysis of the fermentation extract showed that inactivation of ttmJ completely abolished TTM production, consistent with the essential role proposed for TtmJ in TTM biosynthesis (Fig. 5, panel II).
FIGURE 5.
Inactivation of ttmJKPRS, complementation of the ΔttmK and ΔttmS mutants, and HPLC analysis of TTM production in S. spiroverticillatus wild-type (panel I), recombinant strains SB6003 (panel II), SB6004 (panel III), SB6006 (panel V), SB6007 (panel VI), SB6008 (panel VII), and complementation strains SB6009 (panel IV), SB6011 (panel VIII). •, TTM.
Commensurate with methoxymalonyl-ACP serving as an extender unit (module 1), the ttm gene cluster was found to contain a subcluster of five genes ttmABCDE. These newly identified genes are homologous to previously known genes encoding an ADH, β-hydroxyacyl-CoA dehydrogenase, O-methyltransferase, ACP, and a bifunctional phosphatase/glyceryl transferase, respectively (Table 1) (26). The details of this pathway have recently been elucidated (39). Finally, in addition to the chain-terminating TE domain found within TtmH, there was found to be a discrete type II TE, TtmT, within the ttm gene cluster, which may serve as an “editing” enzyme for the TtmHIJ PKSs during polyketide chain elongation (40).
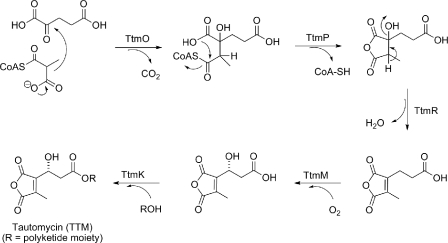
Genes Encoding the Pathway for Dialkylmaleic Anhydride Moiety Biosynthesis—Feeding experiments revealed that the dialkylmaleic anhydride moiety is biosynthesized from one propionate and a C-5 unit such as α-ketoglutarate. A potential route to the anhydride moiety, therefore, is through an inter-molecular aldol condensation between methylmalonyl-CoA and the incipient carbonyl of α-ketoglutarate. Subsequent steps would involve CoA transfer, dehydration, hydroxylation of the resulting allylic position at C-3′, and coupling to the polyketide moiety of TTM (Fig. 6) (22).
FIGURE 6.
Proposed pathway for dialkylmaleic anhydride biosynthesis involving minimally TtmO, TtmP, and TtmM and its coupling with the polyketide moiety catalyzed by TtmK. R, polyketide moiety of TTM.
In support of this sequence of chemical events, eight genes, ttmKL and ttmMNOP, as well as ttmRS, flanking the methoxymalonyl-ACP locus were revealed by sequence analysis. Homology searching revealed that TtmO has high homology to the citryl-CoA lyase (CitE), which converts citryl-CoA to acetyl-CoA and oxaloactate in Mycobacterium tuberculosis (41). Thus, TtmO is a candidate to catalyze the aldol type condensation step. TtmP is highly similar to CaiB, which belongs to the type III CoA transferase family and catalyzes the transfer of the CoA moiety between γ-butyrobetaine-CoA and carnitine forming carnityl-CoA and γ-butyrobetaine (42). As such, TtmP is likely responsible for catalyzing ring closure involving the C4′ carboxylic acid. TtmR could also serves as a candidate for dialkylmaleic anhydride biosynthesis. Installation of the C-4′/C-5′ olefin is likely carried out by TtmR, because it is homologous to the MmgE/PrpD family protein, and PrpD is a known 2-methylcitrate dehydratase involved in propionate catabolism (43). TtmK shows homology to para-nitrobenzyl esterase from Bacillus subtilis (44) and is an excellent candidate to catalyze the convergence of an independently synthesized dialkylmaleic anhydride moiety (or precursor thereof) and the polyketide moiety of TTM. TtmM is homologous to the taurine catabolism dioxygenase TauD, a member of the TfdA family proteins, which contain a conserved Fe(II)-α-ketoglutarate oxygenase domain. Thus, TtmM is likely responsible for catalyzing the C-3′ hydroxylation (Fig. 6) (45).
Of the remaining three open reading frames, TtmL has 30% identity to a Ybhb (1FUX_A) from E. coli containing a conserved bacterial/archael phosphatidyl ethanolamine-binding protein domain in addition to a kinase inhibitory domain (46); TtmN has 51% identity to EhpF (AAN40895) from Pantoea agglomerans, which has a conserved AMP-binding domain and thus is proposed to act as a sensor of cellular energy levels (47); TtmS is a putative cyclase with a conserved HXGTHXDXPXH motif found in the cyclase family (48). Although it is difficult to assign a function to the three gene products on sequence analysis alone, TtmL and TtmN could serve as candidates to perform regulatory functions, probably modulating the biosynthesis of the dialkymaleic anhydride unit, but no apparent role could be speculated for TtmS in TTM biosynthesis.
To gain additional insights into dialkymaleic anhydride biosynthesis, in vivo gene inactivation experiments for ttmKPRS were carried out. The target genes were replaced by the aac(3)IV/oriT cassette in the same direction of the transcription of the genes, yielding mutated cosmids pBS6019, pBS6021, pBS6022, and pBS6023, which was then introduced into Wild-type S. spiroverticillatus (supplemental Table S1). Double cross-over mutants that were apramycin-resistant and kanamycin-sensitive were selected, confirmed by PCR and Southern blot analyses (supplemental Table S2), and named SB6004 (ΔttmK), SB6006 (ΔttmP), SB6007 (ΔttmR), and SB6008 (ΔttmS), respectively (supplemental Table S1). Furthermore, genetic complementation was carried out to eliminate the possibility of polar effects. The vectors pBS6024 and pBS6026, containing the functional copies of ttmK and ttmS whose expressions are under the control of the ErmE* promoter, were introduced into SB6004 and SB6008 to afford SB6009 and SB6011, respectively (supplemental Table S3). All of these strains were tested for TTM production with the wild-type strain as a positive control. No TTM was detected in the fermentation cultures of all the gene inactivation mutants, firmly implicating the involvement of these genes in TTM biosynthesis (Fig. 5, panels III, V, VI, and VII). TTM production was partially restored upon expression of a functional copy of the target gene in trans, as exemplified by ttmK (pBS6024) and ttmS (pBS6026) to SB6004 (ΔttmK) and SB6008 (ΔttmS), respectively, to ∼20 and 30% of the levels seen for the wild-type strain (Fig. 5, panels IV and VIII). The identity of TTM produced by the S. spiroverticillatus wild-type and recombinant strains was finally confirmed by MS and 1H and 13C NMR analysis, all spectra were identical to those of authentic TTM.
Tailoring, Regulatory, and Resistance Genes—Feeding experiments have shown that the terminal methyl group (C-1) of TTM originates from C-2 of acetate (22), suggesting that the nascent polyketide intermediate undergoes decarboxylation upon release from the TTM PKS machinery by the TtmH TE domain. Of the remaining orfs, the deduced gene product of ttmG exhibits 50% identity to the carboxymuconolactone decarboxylase Plav_1659 (YP_001412935) from Parvibaculum lavamentivorans DS-1, supporting the assignment of TtmG as a carboxylase responsible for the decarboxylation step (Fig. 3) (49).
In addition to the ttmLN noted above, two additional genes, ttmF and ttmQ, within the ttm gene cluster could also encode regulatory proteins. A BlastP search revealed the presence of a typical LuxR type helix-turn-helix motif at the C terminus of TtmF (amino acids 363-420). LuxR family regulatory proteins bind to DNA utilizing the helix-turn-helix motif when activated by the binding of an autoinducer molecule such as N-(3-oxo-hexanoyl)-l-homoserine lactone (50). However, unlike typical response regulators, no autoinducer domain was identified at the N terminus of TtmF. TtmQ was found to have 48% identity to StrR (CAA07385), a well characterized pathway-specific transcriptional activator in streptomycin biosynthesis (51). The sequence of TtmQ has the following two notable characteristics: (i) it codes for a protein with a helix-turn-helix motif near the central region, which is typical for a family of bacterial and phage DNA-binding proteins, and (ii) it contains the rare TTA leucine codon at position 49, indicative of its dependence on bldA, the structural gene of tRNAUUA (52, 53).
No orfs encoding apparent resistance proteins were found within the ttm gene cluster boundaries. This is consistent with the fact that TTM exhibits antimicrobial activity against various fungi and yeasts but only limited species of Gram-negative bacteria (1).
DISCUSSION
A pathway featuring novel chemistry for dialkylmaleic anhydride biosynthesis and a type I PKS selecting and incorporating four different starter and extender units for TTM production has been proposed (Figs. 3 and 6). This proposal is based on the findings from cloning and sequencing of the ttm gene cluster from S. spiroverticillatus, precise determination of the ttm gene cluster boundaries, deduced function of the orfs within the ttm cluster, and in vivo analysis of selected genes from the ttm cluster by targeted gene inactivation and mutant complementation. It agrees well with previous feeding experiments using isotope-labeled precursors (22).
Biosynthesis of the TTM Polyketide Moiety—The identification of a subcluster of five genes, as exemplified by ttmABCDE, present in all polyketide biosynthetic gene clusters known to contain a methoxymalonate-derived unit was crucial to the cloning of the ttm cluster (26). These genes encode the enzymes that catalyze the biosynthesis of the methoxymalonyl-ACP extender unit (26). With ttmABCDE as a cluster beacon, it was determined that the polyketide moiety of TTM is biosynthesized by three large modular type I PKSs (TtmHIJ), whose module and domain organization is co-linear with the TTM structure excluding predicted superfluous or nonfunctional DH or KR domains, a common occurrence in known modular type I PKSs. Module and domain organization for the TTM PKSs were deduced by sequence homology to known PKSs (35-38). The TTM PKS catalyzes the biosynthesis of the TTM polyketide backbone by carrying out 12 rounds of decarboxylative condensation using one isobutyryl CoA as the starter unit (loading module) and one methoxymalonyl ACP (module 1), six malonyl CoA (modules 2, 4, 7, 8, 10, and 12), and five methyl malonyl CoA (modules 3, 5, 6, 9, and 11) as extender units. The TTM PKS is characterized with multiple AT domains for the four varying starter and extender units, a striking feature that could be exploited to engineer novel TTM analogs by combinatorial biosynthesis methods (Fig. 3).
Following the assembly of the polyketide intermediate by the TTM PKS, a minimum of three steps are needed to complete the biosynthesis of TTM. They are (i) esterification to couple the dialkylmaleic anhydride moiety with the polyketide chain; (ii) decarboxylation of a polyketide intermediate to afford the C-1 methyl group; and (iii) cyclization, dehydration, and cyclization of a polyketide intermediate to form the spiroketal moiety. The precise timing of each step could now be experimentally investigated.
Biosynthesis of the Dialkylmaleic Anhydride Moiety—TTM is unique among natural products because it contains both dialkylmaleic anhydride and spiroketal moieties (Fig. 1). Particularly intriguing are the steps involved in dialkylmaleic anhydride biosynthesis, which, to our knowledge, have yet to be reported. Functional assignment of the genes within the ttm gene cluster has allowed us to postulate an efficient route to the dialkylmaleic anhydride consistent with the results of feeding experiments (22). We propose that TtmO initiates the pathway by catalyzing aldol condensation of methylmalonyl-CoA and the α-carbonyl group of α-ketoglutarate. TtmP, a putative CoA transferase, and TtmR, a putative dehydratase, are the best candidates to catalyze the subsequent release of CoA and dehydration, respectively. The final reactions involve C-3′ hydroxylation, which is catalyzed by TtmM, and coupling to the polyketide chain, likely catalyzed by esterase homolog TtmK (Fig. 6). Finally, TtmN may act, like its homolog EhpF from P. agglomerans, as a sensor of cellular energy levels to modulate the biosynthesis of the diakylmaleic anhydride moiety.
Spiroketal Formation—It is not difficult to imagine that the mechanism of TTM spiroketal formation involves dual attack of the carbonyl group (C-10) by the hydroxyl moieties at C-6 and C-14 (Fig. 3). Such a proposal would parallel the formation of the 6, 6-spiroketal moiety found in the avermectins and spirangienes, of which sequence analysis of the biosynthetic gene cluster has so far failed to identify any candidate responsible for spiroketal formation (54, 55). In an organizational analogy to the avermectins and spirangenes biosynthetic gene clusters, no candidates could be identified within the ttm gene cluster for installing the TTM spiroketal moiety (Fig. 2 and Table 1). As such, we now favor the hypothesis that spiroketal formation could proceed in a spontaneous fashion, although the possibility of enzymes residing outside of the sequenced ttm cluster to catalyze this step could not be excluded (Fig. 3).
Tailoring Events—Decarboxylation of the nascent polyketide intermediate upon release from the TTM PKS machinery to afford the methyl ketone group and esterification to couple the dialkylmaleic anhydride moiety with C-24 OH of the polyketide intermediate are two key tailoring events envisioned in TTM biosynthesis. As noted previously, TtmK is an excellent candidate for performing the esterification based on its homology to the esterase EstA1 (CAB93516) from Bacillus sp. BP-7 (44). Formation of the methyl ketone via decarboxylation of a polyketide intermediate is attributed to the putative decarboxylase TtmG because of its homology to the carboxymuconolactone decarboxylase Plav_1659 from P. lavamentivorans DS-1 (49). It is unclear at present the timing of the TtmG- and TtmK-catalyzed steps, although the precise orchestration and ordering of both tailoring reactions must be determined for future biosynthetic engineering efforts to afford TTM analogs predictably (Fig. 3).
Timing of the Coupling Reaction—Although the S. spiroverticillatus genetic system developed herein will ultimately allow precise elucidation of the timing of individual steps for TTM biosynthesis, the gene inactivation and mutant complementation experiments reported here have already started to shed insights into the timing of the steps for the biosynthesis of the dialkylmaleic anhydride unit and its coupling to the polyketide moiety. The proposed gene functions for ttmOPRK suggest that the dialkylmaleic anhydride unit is biosynthesized and then coupled to the full-length (convergent model) or a growing (linear model) polyketide intermediate, after which time additional tailoring steps (i.e. decarboxylation and spiroketal formation) may proceed (Fig. 3).
The convergent model would suggest that inactivation of genes encoding dialkylmaleic anhydride biosynthesis would abolish TTM production but allow the accumulation of the polyketide moiety of TTM or metabolites thereof. The ttmKPRS genes are excellent candidates to test this hypothesis because they are all likely to play a role in dialkylmaleic anhydride biosynthesis and installation in TTM, inactivation of any one of which would be expected to allow accumulation of a biosynthetic intermediate or metabolites thereof devoid of the dialkymaleic anhydride moiety. However, this was not found to be the case. Inactivation of each one of them abolished production of TTM but without the concomitant accumulation of any detectable new intermediates. These results suggest that all of these gene products perform early biosynthetic functions that most likely precede the release of the full length of TTM polyketide intermediate from the TtmHIJ PKSs. It is conceivable that inactivation of any one of them could prohibit the ensuing enzyme-dependent events related to polyketide elongation or release from the TtmHIJ PKSs. It is significant that TTM production in SB6004 (ΔttmK) and SB6008 (ΔttmS) mutants could be partially restored by expressing a functional copy of ttmK (SB6009) and ttmS (SB6011) in trans, respectively, thereby excluding the possibility of polar effects (Fig. 5). Taken together, these results argue against the convergent model, and we now favor the linear model in which coupling of the dialkylmaleic anhydride unit to the TTM polyketide backbone proceeds the completion of the full-length polyketide biosynthesis and its liberation from the TTM PKS machinery (Fig. 3).
In summary, TTM features a rare alkyl chain bearing a dialkylmaleic anhydride moiety and a spiroketal-containing polyketide chain incorporating a methoxymalonate-derived unit. Cloning, sequencing, and characterization of the ttm biosynthetic gene cluster from S. spiroverticillatus unveiled new insights into the molecular basis for TTM biosynthesis. With the ttm gene cluster available, the novel chemistries predicted for TTM biosynthesis can now be studied. These results set an excellent stage for future investigations of TTM biosynthesis and engineering. The application of these data will play a crucial role in generating novel TTM analogs for drug discovery by combinatorial biosynthesis methods.
Supplementary Material
Acknowledgments
We thank the Analytical Instrumentation Center of the School of Pharmacy at the University of Wisconsin-Madison for support in obtaining MS and NMR data.
This work was supported, in whole or in part, by National Institutes of Health Grant CA113297. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1-S3 and Figs. S1-S5.
Footnotes
The abbreviations used are: TTM, tautomycin; ACP, acyl carrier protein; ADH, acyl-CoA dehydrogenase; AT, acyltransferase; DH, dehydratase; KR, ketoreductase; PKS, polyketide synthase; PP, protein phosphatase; TE, thioesterase; TTN, tautomycetin; MS, mass spectrometry; HPLC, high pressure liquid chromatography.
References
- 1.Cheng, X. C., Kihara, T., Kusakabe, H., Magae, J., Kobayashi, Y., Fang, R. P., Ni, Z. F., Shen, Y. C., Ko, K., Yamaguchi, I., and Isono, K. (1987) J. Antibiot. 40 907-909 [DOI] [PubMed] [Google Scholar]
- 2.Cheng, X. C., Ubukata, M., and Isono, K. (1990) J. Antibiot. 43 809-819 [DOI] [PubMed] [Google Scholar]
- 3.Ubukata, M., Cheng, X. C., Isobe, M., and Isono, K. (1993) J. Chem. Soc., Perkin Trans. 1 5 617-624 [Google Scholar]
- 4.Ubukata, M., Cheng, X. C., and Isono, K. (1990) Chem. Comm. 3 244-246 [Google Scholar]
- 5.Cheng, X. C., Kihara, T., Ying, X., Uramoto, M., Osada, H., Kusakabe, H., Wang, B. N., Kobayashi, Y., Ko, K., Yamaguchi, I., Shen, Y. C., and Isono, K. (1989) J. Antibiot. 42 141-144 [PubMed] [Google Scholar]
- 6.Cheng, X. C., Ubukata, M., and Isono, K. (1990) J. Antibiot. 43 890-896 [DOI] [PubMed] [Google Scholar]
- 7.Dai, J.-P., Sodeoka, M., and Shibasaki, M. (1996) Tetrahedron Lett. 37 491-494 [Google Scholar]
- 8.Nagae, J., Watanabe, C., Osada, H., Cheng, X. C., and Isono, K. (1988) J. Antibiot. 41 932-937 [DOI] [PubMed] [Google Scholar]
- 9.Honkanen, R. E., and Golden, T. (2002) Curr. Med. Chem. 9 2055-2075 [DOI] [PubMed] [Google Scholar]
- 10.Bialojan, C., and Takai, A. (1988) Biochem. J. 256 283-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakoff, J. A., and McCluskey, A. (2004) Curr. Pharm. Des. 10 1139-1159 [DOI] [PubMed] [Google Scholar]
- 12.Mitsuhashi, S., Shima, H., Tanuma, N., Matsuura, N., Takekawa, M., Urano, T., Kataoka, T., Ubukata, M., and Kikuchi, K. (2003) J. Biol. Chem. 278 82-88 [DOI] [PubMed] [Google Scholar]
- 13.Ishihara, H., Martin, B. L., Brautigan, D. L., Karaki, H., Ozaki, H., Kato, Y., Fusetani, N., Watabe, S., Hashimoto, K., Uemura, D., and Hartshorne, D. J. (1989) Biochem. Biophys. Res. Commun. 159 871-877 [DOI] [PubMed] [Google Scholar]
- 14.Kita, A., Matsunaga, S., Takai, A., Kataiwa, H., Wakimoto, T., Fusetani, N., Isobe, M., and Miki, K. (2002) Structure, 10 715-724 [DOI] [PubMed] [Google Scholar]
- 15.Li, Y. M., and Casida, J. E. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 11867-11870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacKintosh, C., and Klumpp, S. (1990) FEBS Lett. 277 137-140 [DOI] [PubMed] [Google Scholar]
- 17.Colby, D. A., Liu, W., Sheppeck, J. E., Huang, H. B., Nairn, A. C., and Chamberlin, A. R. (2003) Bioorg. Med. Chem. Lett. 13 1601-1605 [DOI] [PubMed] [Google Scholar]
- 18.Mitsuhashi, S., Matsuura, N., Ubukata, M., Oikawa, H., Shima, H., and Kikuchi, K. (2001) Biochem. Biophys. Res. Commun. 287 328-331 [DOI] [PubMed] [Google Scholar]
- 19.Oikawa, H. (2002) Curr. Med. Chem. 9 2033-2054 [DOI] [PubMed] [Google Scholar]
- 20.Shim, J. H., Lee, H. K., Chang, E. J., Chae, W. J., Han, J. H., Han, D. J., Morio, T., Yang, J. J., Bothwell, A., and Lee, S. K. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 10617-10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, D. J., Jeong, Y. L., Wee, Y. M., Lee, A. Y., Lee, H. K., Ha, J. C., Lee, S. K., and Kim, S. C. (2003) Transplantation Proc. 35 547. [DOI] [PubMed] [Google Scholar]
- 22.Ubukata, M., Cheng, X. C., Uzawa, J., and Isono, K. (1995) J. Chem. Soc., Perkin Trans. 1 19 2399-2404 [Google Scholar]
- 23.Sheppeck, J. E., II, Liu, W., and Chamberlin, A. R. (1887) J. Org. Chem. 62 387-398 [DOI] [PubMed] [Google Scholar]
- 24.Oikawa, H. K., Oikawa, M., Ueno, T., and Ichihara, A. (1994) Tetrahedron Lett. 35 4809-4812 [Google Scholar]
- 25.Oikawa, M., Ueno, T., Oikawa, H., and Ichihara, A. (1995) J. Org. Chem. 60 5048-5068 [Google Scholar]
- 26.Li, W., Ju, J., Osada, H., and Shen, B. (2006) J. Bacteriol. 188 4148-4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paget, M. S., Chamberlin, L., Atrih, A., Foster, S. J., and Buttner, M. J. (1999) J. Bacteriol. 181 204-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gust, B., Challis, G. L., Fowler, K., Kieser, T., and Chater, K. F. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1541-1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen, B., and Hutchinson, C. R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 6600-6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao, M., Wang, L., Wendt-Pienkowski, E., George, N. P., Zhang, G., Coughlin, J. M., and Shen, B. (2007) Mol. Biosyst. 3 60-74 [DOI] [PubMed] [Google Scholar]
- 31.Kieser T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood. D. A. (2000) Practical Streptomyces Genetics, The John Innes Foundation, Norwich, UK
- 32.Liu, W., and Shen, B. (2000) Antimicrob. Agents Chemother. 44 382-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, A. C., Zhang, Y. M., Rock, C. O., and White, S. W. (2004) Structure 12 417-428 [DOI] [PubMed] [Google Scholar]
- 34.Quigley, P. M., Korotkov, K., Baneyx, F., and Hol, W. G. (2004) Protein Sci. 13 269-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staunton, J., and Weissman, K. J. (2001) Nat. Prod. Rep. 18 380-416 [DOI] [PubMed] [Google Scholar]
- 36.Aparicio, J. F., Molnar, I., Schwecke, T., Konig, A., Haydock, S. F., Khaw, L. E., Staunton, J., and Leadlay, P. F. (1996) Gene (Amst.) 169 9-16 [DOI] [PubMed] [Google Scholar]
- 37.Bevitt, D. J., Cortes, J., Haydock, S. F., and Leadlay, P. F. (1992) Eur. J. Biochem. 204 39-49 [DOI] [PubMed] [Google Scholar]
- 38.Donadio, S., and Katz, L. (1992) Gene (Amst.) 111 51-60 [DOI] [PubMed] [Google Scholar]
- 39.Dorrestein, P. C., Van Lanen, S. G., Li, W., Zhao, C., Deng, Z., Shen, B., and Kelleher, N. L. (2006) J. Am. Chem. Soc. 128 10386-10387 [DOI] [PubMed] [Google Scholar]
- 40.Kim, B. S., Cropp, T. A., Beck, B. J., Sherman, D. H., and Reynolds, K. A. (2002) J. Biol. Chem. 277 48028-48034 [DOI] [PubMed] [Google Scholar]
- 41.Goulding, C. W., Bowers, P. M., Segelke, B., Lekin, T., Kim, C. Y., Terwilliger, T. C., and Eisenberg, D. J. (2007) Mol. Biol. 365 275-283 [DOI] [PubMed] [Google Scholar]
- 42.Stenmark, P., Gurmu, D., and Nordlund, P. (2004) Biochemistry 43 13996-14003 [DOI] [PubMed] [Google Scholar]
- 43.Horswill, A. R., and Escalante-Semerena, J. C. (2001) Biochemistry 40 4703-4713 [DOI] [PubMed] [Google Scholar]
- 44.Spiller B., Gershenson, A., Arnold, F. H., and Stevens, R. C. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 12305-12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montero-Moran, G. M., Li, M., Rendon-Huerta, E., Jourdan, F., Lowe, D. J., Stumpff-Kane, A. W., Feig, M., Scazzocchio, C., and Hausinger, R. P. (2007) Biochemistry 46 5293-5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serre, L., de Jesus, K. P., Zelwer, C., Bureaud, N., Schoentgen, F., and Benedetti, H. (2001) J. Mol. Biol. 310 617-634 [DOI] [PubMed] [Google Scholar]
- 47.Giddens, S. R., Feng, Y. J., and Mahanty, H. K. (2002) Mol. Microbiol. 45 769-783 [DOI] [PubMed] [Google Scholar]
- 48.Lomovskaya, N., Doi-Katayama, Y., Filippini, S., Nastro, C., Fonstein, L., Gallo, M., Colombo, A. L., and Hutchinson, C. R. (1998) J. Bacteriol. 180 2379-2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schleheck, D., Knepper, T. P., Eichhorn, P., and Cook, A. M. (2007) Appl. Environ. Microbiol. 73 4725-4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sitnikov, D. M., Schineller, J. B., and Baldwin, T. O. (1995) Mol. Microbiol. 17 801-812 [DOI] [PubMed] [Google Scholar]
- 51.Retzlaff, L., and Distler, J. (1995) Mol. Microbiol. 18 151-162 [DOI] [PubMed] [Google Scholar]
- 52.Pabo, C. O., and Sauer, R. T. (1992) Annu. Rev. Biochem. 61 1053-1095 [DOI] [PubMed] [Google Scholar]
- 53.Leskiw, B. K., Bibb, M. J., and Chater, K. F. (1991) Mol. Microbiol. 5 2861-2867 [DOI] [PubMed] [Google Scholar]
- 54.Ikeda, H., Nonomiya, T., Usami, M., Ohta, T., and Omura, S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9509-9514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank, B., Knauber, J., Steinmetz, H., Scharfe, M., Blocker, H., Beyer, S., and Muller, R. (2007) Chem. Biol. 14 221-233 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.