Abstract
Zirconium tetra(tert-butoxide) reacts with surface amide groups of polyamide nylon 6/6 to give (η2-amidate)zirconium complexes in high yield. These surface complexes react to bond the cell-adhesive peptide arginine-glycine-aspartic acid (RGD) to the polymer surface. A surface loading of 0.18 nmol/cm2 of RGD is achieved, which is 20−1000 times higher than previously reported attainable on natural or synthetic polymers by other strategies. Approximately 40% of the nylon surface is covered by the RGD which gives a surface that is both stable to hydrolysis and highly active for cell adhesion and spreading in vitro.
Introduction
Engineered bioactive polymeric scaffolds1 are of increasing importance for use in tissue regeneration in a variety of clinical applications, and interest in them continues to grow because they display significant versatility with wide-ranging physical properties, including biodegradability, compared to metals.2 For these reasons, standard metallic implant technologies may someday be replaced by new, polymer-based ones. Yet, though many polymers do show much promise as biomaterials, the lack of an appropriate interface between the polymer and bodily tissue remains a substantial problem.2 Due in part to their wetting properties,3 polymer surfaces are often prone to nonspecific protein adsorption which can lead to nonspecific cell-type adhesion and fibrous encapsulation.2 Successful strategies to create biocompatible polymeric implant surfaces that support desired cell growth would provide the means to improve device biointegration and would thus significantly impact the biomaterials field.
Metallic implant materials surface-derivatized with high yields of the cell attractive peptide Arg-Gly-Asp (RGD) can foster substantial cell adhesion and growth in vitro.4 But, comparable results have not yet been achieved using therapeutic polymeric devices, as the polymers most often used as biomaterials are not amenable to surface treatments that give high-yield surface coverage.2 Though polymer scaffold materials with improved bioactivity have been prepared by blending,5-8 copolymerization,9-12 or physical treatment,13,14 these methods can alter the bulk properties of the polymer14 and yield only low peptide surface coverages15,16 that do not approach that achieved on metallic substrates.4
We now report a unique, high-yielding strategy for surface activation of pre-cast polymers that we illustrate for nylon polyamides, which are currently used in burn and chronic wound treatment applications.17,18 In our method, a simple zirconium alkoxide complex is allowed to react with surface amide groups of the nylon to give a Zr complex-activated surface that is then easily functionalized with peptides. Fluorescence spectroscopic methods show that approximately 40% of the nylon surface is activated with the RGD. This is the highest yield reported to date for peptide surface attachment by derivatization of a preformed biopolymer, and in vitro studies demonstrate substantially increased fibroblast cell binding and spreading on surfaces functionalized with RGD compared with the untreated polymer. Our activation strategy should be suitable for a range of scaffold materials with acidic N-H moieties to attach any biomolecule containing functionality that is reactive either directly with Zr alkoxides or with their simple derivatives.
Results and discussion
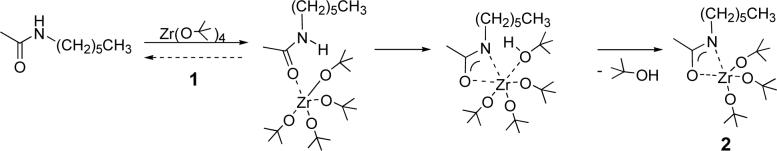
Our novel approach to surface modification enables high surface density derivatization of a preformed polyamide device with RGD under ambient conditions. We hypothesized that, since the surface of nylon 6/6 exposes backbone amide functionality containing acidic N-H bonds, most all of these groups could serve as sites for chemical derivatization if appropriately activated. Coordination of the carbonyl group to an appropriate metallic center would further acidify these N-H bonds and facilitate such activation. Zirconium tetra(tert-butoxide) (1) is an excellent activation reagent because of the high oxyphilicity of Zr and because alkoxide groups remaining in its coordination sphere following reaction with the amide are readily replaceable ligands,19 which accomplishes the desired derivatization of the polymer. This hypothesis was substantiated first in a small-molecule amide model system: N-hexylacetamide, was treated with 1 to yield reactive complex 2 (>95 % by 1H NMR [CDCl3]: δ 0.8 [t, 3H]; 1.3 [m, 35H]; 1.9 [s, 3H]; 3.2 [quartet, 2H]; Scheme 1). The η2-coordination to Zr for the amidate moiety in 2 is indicated by the 8 ppm downfield shift of the acyl carbon vs. the free amide (13C NMR [CDCl3]: δ 170.1 for N-hexylacetamide; δ 178.1 for 2); a similar shift was observed on formation of η2-zirconium carboxylates from carboxylic acids20 while η1-Pd(II) amidates show a smaller downfield shift.21 Zirconium amidate hydroamination catalysts22 show a crystallographic preference for η2-coordination.
Scheme 1.
Reaction of N-hexylacetamide and zirconium tetra(tert-butoxide).
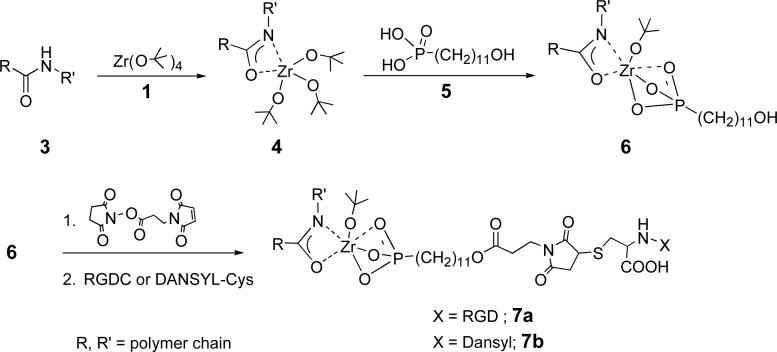
Surface derivatization of solid nylon 6/6 (3) proceeded according to our model system. Films of 3 (R=(CH2)4CO; R’=(CH2)6NH) were cast from formic acid solution on glass microscope slides and were treated with vapor of 1. The IR spectrum of polymer surface-bound Zr complex (4) showed νC-H = 2976 cm−1, indicative of tert-butoxide groups. The 4-coated slide was treated with phosphonoundecanol (5) to yield surface complex 6, which is active for bonding RGDC peptides (Scheme 2). IR analysis of 6 showed peaks in the aliphatic region (νCH2,asym = 2922 cm−1; νCH2,sym = 2851 cm−1) characteristic of disordered alkyl chains.23 Surface 6 was activated for RGDC derivatization by reaction with the N-hydroxysuccinimide ester of 3-maleimidopropionic acid.
Scheme 2.
Reaction of nylon-Zr-amide complex with a phosphonic acid and RGDC or DANSYL-cys coupling.
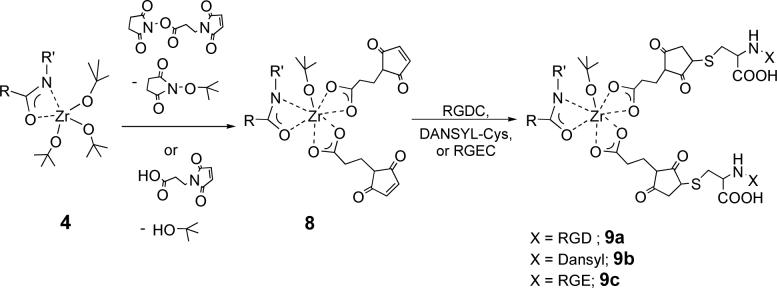
We have also found that an active surface for RGD binding can be prepared by directly reacting surface 4 with the N-hydroxysuccinimide ester of 3-maleimidopropionic acid to produce surface 8. This reaction may proceed by transetherification (Scheme 3); this would give N-(tertbutoxy)succinimide, which was observed as a byproduct (confirmed by LC-MS). It is also possible that adventitious hydrolysis of the succinimidyl ester generates 3-maleimidopropionic acid in situ, which reacts with 4 via ligand exchange. Indeed treating 4 with 3-maleimidopropionic acid (instead of the ester), gave an activated surface that performed identically to 8. IR analysis of 8 showed a characteristic maleimide stretch (νCO = 1705 cm−1). RGDC-modified nylon 7a or 9a was prepared by immersion of 6 or 8 in an aqueous solution of RGDC. A substantial change in surface hydrophilicity was confirmed by a decrease in water contact angle (75° for 3 compared to 50° for 9a).
Scheme 3.
Reaction of nylon-Zr-amide complex 4 with 3-maleimidopropionic acid or its N-hydroxysuccinimidyl ester, and subsequent coupling with RGDC, RGEC or DANSYL-cys.
Hydrolytic stability and surface content of both derivatized nylons 7 and 9 were measured by a fluorescence spectroscopy-based experiment using a DANSYL analog24: DANSYL-Cys was added at the reactive termini instead of RGDC (Schemes 2 and 3). Samples of 7b and 9b were immersed in water (adjusted to pH 7.5 with NaOH) for 7 days. Release of DANSYL groups was measured by fluorescence intensities of supernatants from treated 7b and 9b that were compared to the control sample (3) over this seven-day period. Unreacted DANSYLating reagent desorbed from the nylon surfaces in about 3 hours. No release of surface-bound DANSYL material on 7b or 9b occurred over the next seven days (see, Supplemental Information). Thus, zirconium-amidate surface-bound complexes are stable to hydrolysis under these conditions.
The usefulness of our approach for polymer surface modification is enhanced by the high surface coverage that it can attain: it has been shown25 that cell adhesion and motility both increase as a function of RGD surface density. Surface complex DANSYL contents of 7b and9b were quantified by immersion in water at pH 12 for 3 hrs, which cleaves the Zr complexes from the surface, precipitates ZrO2, and releases fluorophore from 7b and 9b into solution. The amount of DANSYL surface-bound through Zr complexes 7b and 9b was measured to be 0.10 nmol/cm2 and 0.18 nmol/cm2, respectively. These amounts are consistent with the DANSYL : Zr stoichiometries of 1:1 and 2:1 indicated for 7b and 9b, respectively (Schemes 2 and 3). Coverage of the nylon surface by RGD is 20−1000 times higher than has been reported by copolymerization or by purely organic chemical surface modification routes,5-7,9-11,13,15,16,26 and is indeed comparable to that obtained on titanium or other metallic surfaces.4
Nylon 6/6 activated by our procedure and terminated with RGDC peptides is highly active for supporting cell adhesion. NIH3T3 cells attached and spread on the RGD-modified surface 9a, forming membrane extensions that stained with anti-vinculin antibodies (Figure 1A, C) and actin filaments that stained with fluorescent phalloidin (Figure 1C). Significantly more cells attached to the RGDC-modified surface 9a than to untreated nylon (3) (Figure 1D). Similar numbers of cells attached to 9a prepared through treatment of 4 with 3-maleimidopropionic acid compared to treatment with the ester. A one way ANOVA test showed a statistical significance at both 3 and 6 hour time-points (3 hours, p = 1.8×10−5; at 6 hours, p = 7.9×10−6). To determine if the enhanced cell attachment observed for 9a was specific for RGD, we also treated 8 with the non-cell adhesive peptide RGEC, which gives 9c (water wetting contact angle = 50°). Cell adhesion on 9c was less efficient than on 9a (Figure 1B, D); cells on 9c were also less spread, tending to remain round without forming vinculin-positive cellular extensions (Figure 1B, E). One-way ANOVA tests showed statistically significant differences in cell counts (p = 1.9×10−2) and cell spreading (p = 9.0×10−4) on surfaces 9a and 9c. Thus, while cell adhesion on nylon may be somewhat affected by changes in surface composition in general, cell spreading on treated nylon is specifically enhanced by the attachment of RGD.
Figure 1.
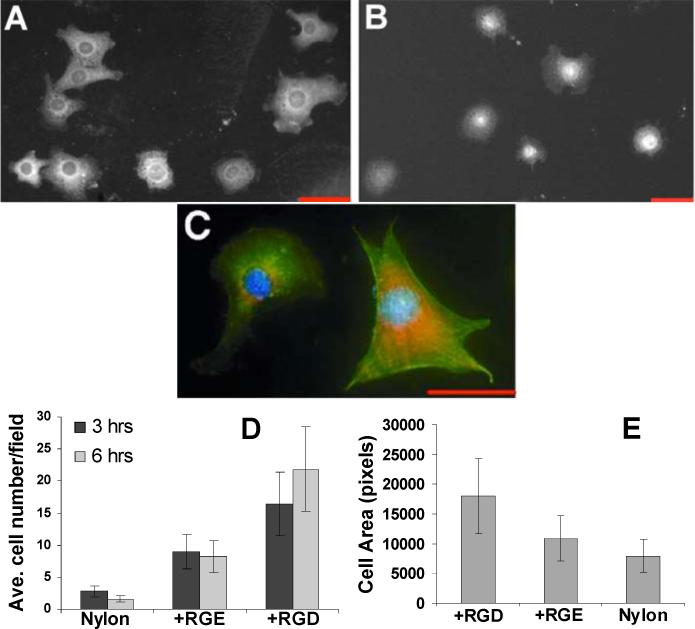
NIH3T3 cell attachment on derivatized nylon. Cells on RGD-modified nylon 6/6 (9a) (A) or RGE-modified nylon 6/6 (9c) (B) surfaces after 3 hours, fixed and stained with anti-vinculin antibodies and rhodamine-conjugated secondary antibodies. (C) Cells on RGD-nylon 6/6 after 3 hours, triple-stained with anti-vinculin antibodies (red), FITC-phalloidin (green), and DAPI (blue). Scale bars are 50 μm. (D) Number of cells per 10X microscope field counted for untreated nylon 6/6, RGD-, and RGE-derivatized nylon 6/6. (E) Average cell area determined for nylon 6/6, RGD-, and RGE-derivatized nylon 6/6 after 3 hours per 20X microscope field with IPLab® software. For (D) and (E), average values for at least 3 fields are shown with error bars representing ± one standard deviation.
Conclusions
We have shown that surface-bound Zr-amidate complexes, which are readily synthesized on the surface of nylon polymer, are effective for activation of that surface for further organic chemical transformation. Our derivatization of nylon 6/6 with RGD peptides, measured to be about 0.1−0.2 nm/cm2 (corresponding to 20−40 % spatial surface coverage for 7a and 9a, respectively) is far higher than has been attained on synthetic and natural polymers by copolymerization or by purely organic chemical surface modification routes,5-7,9-11,13,15,16,26,27 and our RGD-derivatized surfaces are highly cell-attractive. Since our activation process involves simple reaction of amide N-H groups, it should be broadly applicable to other therapeutically important synthetic and natural polymers that contain this functionality such as polyester-co-polyamides,28 polyurethanes,29 polyureas,29 polyimides,30 or even silk.27,31
Experimental Section
General
All reagents were obtained from Aldrich and used as received unless otherwise noted. Tetrahydrofuran was dried over KOH and acetonitrile was dried over CaH2 overnight; both were distilled prior to use. N-Hexylacetamide was synthesized by reaction of acetyl chloride (1.9 g, 24 mmol), hexylamine (2.0 g, 20 mmol) and 0.1 mL triethylamine in CH2Cl2 at 0 °C for 3 hr. The crude product was washed successively with Millipore® water until the pH of the aqueous layer was greater than 6. The CH2Cl2 fraction was dried over Na2SO4, filtered, and evacuated to yield N-hexylacetamide (1H NMR [CDCl3]: δ 0.7 (t, 3H); 1.3 (m, 6H); 1.5 (quintet, 2H); 1.9 (s, 3H); 3.2 (quartet, 2H); 5.5 (s, 1H)). Phosphonoundecanol was synthesized as previously described.24 Surface-modified samples were analyzed using a Midac M2510C Interferometer equipped with a surface optics SOC4000 SH specular reflectance head attachment. Fluorimetry experiments used a Photon Technology International Fluorescence Spectrometer.
(η2-[N-hexyl]amidate)zirconium tri(tert-butoxide), 2
N-Hexylacetamide (0.15 g, 1.0 mmol) was treated with zirconium tetra(tert-butoxide) (Strem), 1 (0.40 g, 1.0 mmol) in dry CH2Cl2 for 1 hr under nitrogen. Solvent and reaction byproducts were removed in vacuo to yield 2 (1H NMR [CDCl3]: δ 0.8 (t, 3H); 1.3 (m, 35H); 1.9 (s, 3H); 3.2 (quartet, 2H)).
Surface reaction of nylon 6/6 with 1
Films of nylon 6/6 (3) were cast from 0.1 mM formic acid solution on glass microscope slides that were then rinsed copiously in Millipore® water and evacuated at 10−2 torr for 3 hours. The coated slides were then placed in a deposition chamber that was equipped with two stopcocks for exposure either to vacuum or to vapor of 1. The chamber was evacuated to 10−3 torr for 30 minutes, and slides of 3 were exposed to vapor of 1 (with external evacuation) for 30 seconds followed by 5 min exposure without external evacuation. This cycle was repeated twice and was then followed by an additional 10 minutes of exposure without external evacuation. The chamber was then evacuated for 16 hours at 10−3 torr to ensure removal of excess 1 to give activated nylon 4.
RGD-modified nylon 6/6 7a
A slide coated with 4 was immersed in a 0.1 mM solution of phosphonoundecanol (5) in dry THF for 15 min to yield Zr phosphonate complex 6. Treatment of 6 in a 0.1 mM solution of 3-maleimidopropionic acid N-hydroxysuccinimide ester for 24 hours under dry N2 followed by copious rinsing successively in acetonitrile and Millipore® water, drying in vacuo, and immersion in a 0.1 mM aqueous solution of RGDC at pH 6.5 for 24 hours produced 7a.
RGD-modified nylon 6/6, 9a
RGD-derivatized surface 9a was prepared by immersing a slide coated with 4 in a 0.1 mM solution of 3-maleimidopropionic acid N-hydroxysuccinimide ester in dry acetonitrile for 16 hours to produce 8. Immersion of 8 in a 0.1 mM aqueous solution of RGDC at pH 6.5 for 24 hours gave 9a.
RGE-modified nylon 6/6, 9c
Immersion of 8 in a 0.1 mM aqueous solution of RGEC (Canadian Peptide) at pH 6.5 for 24 hours gave RGE-derivatized surface 9c.
Determination of nylon 6/6 surface loading using fluorescent molecule-labeled analogues 7b and 9b
These adducts were prepared as described for 7a and 9a, but a 0.1 mM aqueous solution of N-(5-(dimethylamino)-1-naphthylsulfonyl)-cysteine (DANSYL-Cys) was used instead of RGDC (Schemes 2 and 3). To address the issue of solvent-induced polymer swelling, control films of 3 were prepared by soaking in 0.1 mM DANSYL-cys solution for 24 hrs. A calibration curve of fluorescence intensity versus concentration was measured for DANSYL-Cys solutions from 0.16 to 21 μM at pH 7.5 and pH 12. Nylon films (2 cm2) derivatized as 7b and 9b and control films of 3 were immersed in water at pH 7.5 for 7 days at room temperature, and the supernatants were analyzed by fluorescence spectroscopy. The samples were then removed from solution, dried, and immersed in water at pH 12 for 3 hrs, after which the supernatants were again analyzed by fluorescence spectroscopy. The spatial surface coverage of 9a by RGD was calculated from its measured surface loading, 0.2 nmol/cm2; assuming an RGD “footprint” of 40 Å2 (determined using Chem 3D©), this corresponds to coverage of about 0.4 cm2 per cm2 of surface, or 40%.
Fibroblast adhesion and spreading on RGD-derivatized nylon surfaces
Cell response to surfaces 3, 9a and 9c were evaluated in vitro. NIH 3T3 cells maintained in Dulbecco's Modified Eagle's Medium (DMEM) with 10% calf serum (Hyclone) were prepared for cell adhesion experiments as previously described.23 Cells (2.65 × 104 cells/cm2 in serum-free DMEM) were added to tissue culture wells containing untreated or derivatized nylon surfaces which had been pre-blocked for 1 hr in 1% bovine serum albumin in PBS. After 90 minutes, medium with non-adherent cells was removed and replaced with fresh, serum-free DMEM. At 3 and 6 hr cells were fixed, permeabilized, and stained with anti-vinculin antibody (Sigma) followed by rhodamine-IgG secondary antibody (for focal adhesions). In some cases, cells were also stained with FITC-phalloidin (for actin filaments) and DAPI (for DNA). Images were obtained as described previously.32 Brightness and contrast of color levels in Figure 1C were adjusted for the merged image using IPLab software. Cell adhesion was quantified by counting the number of attached cells in at least 3 microscope fields (10X magnification).
Supplementary Material
Acknowledgement
The authors thank the National Science Foundation and the National Institutes of Health for financial support of this research.
Footnotes
Supporting Information Available. Fluorescence intensity vs. time traces showing hydrolytic stability and surface loading of 7b and 9b are available via the internet.
References
- 1.Cima LG, Vacanti JP, Vacanti C, Ingber D, Mooney D, Langer R. Biomech. Eng. 1991;113:143. doi: 10.1115/1.2891228. [DOI] [PubMed] [Google Scholar]
- 2.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 3.Nath N, Hyun J, Ma H, Chilkoi A. Surf. Sci. 2004;570:98. [Google Scholar]
- 4.Gawalt ES, Avaltroni MJ, Danahy MP, Silverman BM, Hanson EL, Midwood KS, Schwarzbauer JE, Schwartz J. Langmuir. 2003;19:200. [Google Scholar]
- 5.Quirk RA, Chan WC, Davies MC, Tendler SBJ, Shakesheff KM. Biomaterials. 2001;22:865. doi: 10.1016/s0142-9612(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 6.Walluscheck KP, Steinhoff G, Kelm S, Haverich A. Eur. J. Vasc. Endovasc. 1996;12:321. doi: 10.1016/s1078-5884(96)80251-6. [DOI] [PubMed] [Google Scholar]
- 7.Yoona JJ, Songa SH, Leeb DS, Park TG. Biomaterials. 2004;25:5613. doi: 10.1016/j.biomaterials.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Dankers P, Harmsen M, Brouwer L, Van Luyn M, Meijer E. Nature Mat. 2005;4:568. doi: 10.1038/nmat1418. [DOI] [PubMed] [Google Scholar]
- 9.Cook AD, Hrkach JS, Gao NS, Johnson IM, Pajvani UB, Cannizzaro SM, Langer RJ. Biomed. Mater. Res. 1997;35:513. doi: 10.1002/(sici)1097-4636(19970615)35:4<513::aid-jbm11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 10.Yamaoka T, Hotta Y, Kobayashi K, Kimura Y. Int. J. Biol. Marcomol. 1999;25:265. doi: 10.1016/s0141-8130(99)00041-0. [DOI] [PubMed] [Google Scholar]
- 11.Smith E, Yang J, McGann L, Sebald W, Uludag H. Biomaterials. 2005;26:7329. doi: 10.1016/j.biomaterials.2005.05.060. [DOI] [PubMed] [Google Scholar]
- 12.Maynard HD, Okada SY, Grubbs RH. J. Am. Chem. Soc. 2006;123:1275. doi: 10.1021/ja003305m. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y, Winn S, Krajbich I, Hollinger J. J. Biomed. Mater. Res. A. 2003;64:583. doi: 10.1002/jbm.a.10438. [DOI] [PubMed] [Google Scholar]
- 14.Sanghvi A, Miller K, Belcher A, Schmidt C. Nature Mat. 2005;4:496. doi: 10.1038/nmat1397. [DOI] [PubMed] [Google Scholar]
- 15.Lin HB, Garcia-Echeverria C, Asakura S, Sun W, Mosher D, Cooper S. J. Biomed. Mater. Res. 1994;28:329. doi: 10.1002/jbm.820280307. [DOI] [PubMed] [Google Scholar]
- 16.Ernsting MJ, Bonina GC, Yang M, Labow RS, Santerre JP. Biomaterials. 2005;26:6536. doi: 10.1016/j.biomaterials.2005.04.047. [DOI] [PubMed] [Google Scholar]
- 17.Shakespeare P. Clin. Dermatol. 2005;23:413. doi: 10.1016/j.clindermatol.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Supp D, Boyce S. Clin. Dermatol. 2005;23:403. doi: 10.1016/j.clindermatol.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 19.Miller JB, Schwartz J. Acta Chem. Scand. 1993;47:292. [Google Scholar]
- 20.Lian B, Lehmann CW, Navarro C, Carpentier JF. Organometallics. 2005;24:2466. [Google Scholar]
- 21.Fujita K, Makoto Y, Puschmann F, Alvarez-Falcon M, Incarvito C, Hartwig J. J. Am. Chem. Soc. 2006;128:9044. doi: 10.1021/ja062333n. [DOI] [PubMed] [Google Scholar]
- 22.Thomson R, Zahariev F, Zhang Z, Patrick B, Wang Y, Schafer L. Inorg. Chem. 2005;44:8680. doi: 10.1021/ic0502980. [DOI] [PubMed] [Google Scholar]
- 23.Gawalt ES, Koch N, Schwartz J. Langmuir. 2001;17:5736. [Google Scholar]
- 24.Danahy MP, Avaltroni MJ, Midwood KS, Schwarzbauer JE, Schwartz J. Langmuir. 2004;20:5333. doi: 10.1021/la036084h. [DOI] [PubMed] [Google Scholar]
- 25.Maheshwari G, Brown G, Lauffenburger D, Wells A, Griffith L. J. Cell Sci. 2000;113:1677. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 26.Kugo K, Okuno M, Masuda K, Nishino J, Masuda H, Iwatsuki M. J. Biomater. Sci. Polym. Ed. 1994;5:325. doi: 10.1163/156856294x00059. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Altman G, Karageorgiou R, Collette A, Volloch V, Colabro T, Kaplan D. J. Biomed. Mater. Res. A. 2003;67:559. doi: 10.1002/jbm.a.10120. [DOI] [PubMed] [Google Scholar]
- 28.Zhiyoung Q, Sai L, Hailian Z, Xiaobo L. Colloid Polym. Sci. 2003;281:869. [Google Scholar]
- 29.Salacinski H, Hamilton G, Seifalian A. J. Biomed. Mater. Res. A. 2002;66:688. doi: 10.1002/jbm.a.10020. [DOI] [PubMed] [Google Scholar]
- 30.Peluso G, Petillo O, Ambrosio L, Nicolais L. J. Mater. Sci. Mater. Med. 1994;4:738. doi: 10.1023/a:1008998604276. [DOI] [PubMed] [Google Scholar]
- 31.Meinel L, Fajardo R, Hofmann S, Langer R, Chen J, Snyder B, Vunjak-Novakovic G, Kaplan D. Bone. 2005;37:688. doi: 10.1016/j.bone.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Midwood KS, Schwarzbauer J. Mol. Biol. Cell. 2002;13:3601. doi: 10.1091/mbc.E02-05-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.