Abstract
Reversible unfolding of helical transmembrane proteins could provide valuable information about the free energy of interaction between transmembrane helices. Thermal unfolding experiments suggest that this process for integral membrane proteins is irreversible. Chemical unfolding has been accomplished with organic acids, but the unfolding or refolding pathways involve irreversible steps. Sodium dodecyl sulfate (SDS) has been used as a perturbant to study reversible unfolding and refolding kinetics. However, the interpretation of these experiments is not straightforward. It is shown that the results could be explained by SDS binding without substantial unfolding. Furthermore, the SDS perturbed state is unlikely to include all of the entropy terms involved in an unfolding process. Alternative directions for future research are suggested: fluorinated alcohols in homogeneous solvent systems; inverse micelles; and fragment association studies.
Genomic sequencing indicates that integral membrane proteins are abundant, and there are numerous examples of disease states which result from membrane protein misfolding (1). These ought to be sufficient incentives to study the folding of integral membrane proteins, but some biochemists have been discouraged by the notorious difficulty of determining membrane protein 3D structures and the restrictions of working with detergent or vesicle systems. However, the outlook in this field has brightened, as demonstrated in several recent reviews. Bowie (2) has provided an excellent summary of the main issues in current membrane protein folding research, and MacKenzie (3) recently comprehensively surveyed the folding and stability of helical membrane proteins. I will restrict my comments here to a critical evaluation of some past and recent attempts to reversibly unfold helical integral membrane proteins, followed by some suggestions for promising future research directions.
At the outset, it may be useful to explain the rationale for studying folding and unfolding of proteins under conditions which may be far removed from native membranes. Most integral membrane proteins are inserted into the membrane either directly from the ribosome through the translocon of the rough endoplasmic reticulum (4), or bacterial translocons (5), or through chaperone/translocase machinery, such as that found in mitochondria (6). In some cases, toxins or other proteins spontaneously insert directly into the membrane. In the case of insertion from ribosomes, the translocon directs peptide segments either into the bilayer or across the membrane into the endoplasmic reticulum lumen, depending on the peptide's hydrophobicity (4). Membrane protein secondary structure appears to form in the translocon, and some tertiary interactions also may form there as well (7). The folding of a helical membrane protein is completed in the bilayer as transmembrane helices interact to form a compact structure (8). A major part of the stability of membrane proteins is likely to be in these interactions between helices which occur after transmembrane segments are released from the translocon. By separating transmembrane helices under equilibrium conditions, it should be possible to obtain detailed quantitative information about the free energy which stabilizes the folded structure. Free energy is a state function, so the free energy change of protein unfolding is independent of the unfolding path. This means that the unfolding free energy of a membrane protein does not necessarily have to be measured in a native-like lipid bilayer, but the folded and unfolded states must be well-characterized.. An understanding of the thermodynamics of membrane protein folding will also help define the starting and ending points for kinetic studies of folding, which have been fruitful in studies of water soluble proteins, and which may provide clues to redirecting misfolding pathways.
Physical unfolding
Thermal denaturation has been reported for bacteriorhodopsin (BR1), based on calorimetric and spectroscopic observations. A calorimetric transition occurs in purple membrane at approximately 100°C (9), and the changes are reported to be irreversible (9, 10), or partly reversible (11). Substantial secondary structure remains (9,12). Thus, only partial unfolding occurs, even at biologically extreme temperatures. The thermal transition shifts to lower temperatures at alkaline pH and for BR solubilized in detergent/lipid mixed micelles (9, 13, 14). The extent of tertiary unfolding above the transition temperature is unknown, but the denatured protein is more susceptible to proteolysis than native BR (9). Irreversible thermal transitions have also been reported for the GLUT 1 transporter (15), rhodopsin (16) and cytochrome oxidase (17). However, from the data presented, it is not possible to assess the extent to which the denatured states of the proteins were unfolded. For GLUT 1 and rhodopsin, transitions occur at higher temperatures in the presence of bound ligands (glucose or 11-cis retinal, respectively), which presumably stabilize the structures. This indicates that parts of the ligand-binding sites may unfold above the transition. It has been noted (15, 18) that the enthalpy changes measured in membrane protein thermal transitions are smaller than for water-soluble proteins, suggesting that the water-exposed surface loops unfold, but not the bilayer-embedded helices. Chymotrypsin cleavage of the second cytoplasmic surface loop of rhodopsin results in two separate thermal transitions in the absence of the retinal chromophore, but it is not clear whether the chymotrypsin fragments dissociate upon heating or after retinal removal (16).
By atomic force microscopy, single molecules of bacteriorhodopsin (19-21) and sodiumhydrogen anitporter (22) have been unfolded and re-inserted into the membrane. The insertion experiments give an estimate of the free energies of membrane protein insertion. However, this process is different from the way a membrane protein is injected into the membrane during biosynthesis via the translocon complex. In the unfolded state of the cantilever-attached proteins, the secondary structure has been disrupted. By contrast, the translocon releases a partially folded protein into the bilayer, presumably with transmembrane helices already formed.
Chemical denaturation
Urea and guanidinium have not been commonly used as denaturants for helical membrane proteins, in contrast to the effectivness of urea in denaturing β-barrel integral membrane proteins (23-25). A typical case is BO, which does not unfold in neutral solutions of urea or guanidinium (14). Acidic solutions of guanidinium chloride were used to unfold BO for iodosobenzoic acid cleavage (26). Acidic urea solutions solubilize diacylglycerol kinase (DGK) in a form which spontaneously inserts into lipid vesicles and refolds into an active state (27, 28). There are a few reports of solubilization of helical membrane proteins by urea (29) or guanidinium (30) under neutral conditions.
Solvent-induced unfolding and spontaneous refolding of a membrane protein was first demonstrated by Huang et al. (31). After solubilization in trifluoroacetic acid, BR completely lost its secondary and tertiary structure. The unfolded protein was dialyzed into sodium dodecylsulfate (SDS) micelles. The detergent-solubilized protein could then be added to bile salt detergent/lipid mixed micelles, in which a folded and active structure spontaneously formed. A similar procedure was reported for unfolding and refolding of the KcsA potassium channel (32). In this case, the unfolded state was the separation of tetramers into four monomeric two-helix subunits, solubilized in 50% trifluoroethanol containing 1% trifluoroacetic acid. Although in principle, an unfolded membrane protein in trifluoroacetic acid might be directly refolded in a strongly buffered micellar system, this has not been done, partly because of the aggressive nature of trifluoroacetic acid, which can cleave acidsensitive peptide bonds and can trifluoroacetylate protein side chains. In formic acid, BR retains some secondary structure (31). Dilution of formic acid-solubilized bacterio-opsin (BO) with ethanol (33), or transfer to CHCl3/methanol (34) stabilizes the protein. In CHCl3/methanol BO retains considerable secondary structure, but tertiary contacts are lost (35). BR can be refolded from formic acid/ethanol or CHCl3/methanol, but it does not follow a thermodynamically reversible path. Dialysis into SDS results in a precipitate (31, 36), which can be resolubilized with a brief alkaline treatment (34). Precipitation and detergent solubilization is sometimes useful as a step in preparative folding methods (37), but aggregation and precipitation interfere with studying thermodynamically useful folding-unfolding equilibria. By carefully controlling the trifluoroethanol concentration, Barrera et al. (38) succeeded in reversibly unfolding KcsA. The unfolded protein shows both a loss of helicity and dissociation of tetramers to monomers. The transition is reversible if the trifluoroethanol concentration does not exceed 35%. Above this concentration, helicity is regained but the active tetrameric structure does not form upon dilution.
Folding mechanisms
Because proteins are synthesized from the N-terminus, it is possible that the N-terminal helical segments of a transmembrane protein are more stably associated, forming a scaffold for folding the C-terminal part of the protein. Some evidence has been reported on the instability of the C-terminal helices of BO, either individually (39) or in a two-helix fragment (40, 41). The three C-terminal helices of 7-transmembrane (TM) helix proteins were found to have rougher surface topologies and longer axial separations than the N-terminal four helices, suggesting weaker association with the other helices (41). A simulated thermal unfolding computation of bovine rhodopsin found that the C-terminal TM helix was least stable (42). The C-terminal helix of BO was found to be least stably associated with the protein core, as measured by acid-induced unfolding (41). Although this might suggest that C-terminal helix instability is evidence for a sequential folding pathway (39), the evidence is all obtained in rhodopsins, which form a Schiff base linkage between retinal and a lysine on the C-terminal helix. The instability of the C-terminal helix may reflect a conformational flexibility necessary for pigment formation (41). Similar studies on non-retinyl proteins would help resolve this question. Compton et al. compared the rates of folding BO from SDS micelles to lipid bilayers, monitored by global tryptophan fluorescence or by a single fluorescent probe attached to the C-terminal helix (43). No differences were found in the rates, which indicates that the folding of BO under these conditions is highly cooperative and not sequential. To resolve questions of folding pathways, it will be valuable to study unfolded states in conditions other than SDS micelles (see below). Also, it is important to compare folding of 7-TM helix proteins with TM proteins having other topologies, such as that found in the aquaporins.
Folding from SDS
The SDS-solubilized state of several membrane proteins has been studied extensively. Active BR forms spontaneously when SDS-solubilized BO is diluted with certain stabilizing amphiphiles to form mixed micelles and the retinal chromophore is added (31, 44). In a series of pioneering experiments, Booth and co-workers (45) measured the rate of formation of the native bacteriorhodopsin structure from the SDS-destabilized form. Bowie and co-workers used similar detergent systems to examine destabilization of diacylglycerol kinase (DGK) (46) and BR (47, 48) by SDS, using a linear free energy approach previously developed for equilibrium unfolding of water-soluble proteins (49). However, there are several critical questions which should be posed about the nature of SDS “unfolding” of membrane proteins.
Does SDS decrease the per cent helix of membrane proteins?
SDS is generally thought of as a protein “denaturant,” since it is widely used for molecular size estimates by polyacrylamide gel electrophoresis. Lau and Bowie (46) identified an SDS-induced “unfolding” transition in DGK as a 15% decrease in ellipticity at 222 nm. Decreased molar ellipticity was also observed in BR solubilized in SDS (44). It was reported that in SDS, BO is 42% (50) or 65% (40) helix, rather than the 74% helix found in the native structure. Under the conditions used for “denaturing” BR in SDS, an NMR spectrum of a two-helix fragment has been reported (35). The structure shows a high helix content (69%). For a similar 2-helix fragment, Luneberg et al. (40) reported 48% helix using CD, compared with the expected 56% (the mostly structureless C-terminal tail was included in this peptide). SDS is known as an agent for stabilizing α-helical structure. About 40 NMR structures of small helical peptides in SDS are in the Protein Data Bank. In several cases, peptide conformations were examined by both CD and NMR under exactly the same conditions (51-55). The CD measurements consistently report lower per cent helix than what was found by NMR (calculated from the atomic coordinates with the DSSP algorithm (56)), ranging from 9 - 31% lower. This indicates that the standard secondary structure analysis based on CD data is not accurate for proteins in SDS. The discrepancy could be explained if SDS binding to proteins changes the extinction coefficient of the peptide carbonyl group, which would affect the differences observed between absorbance of left and right circularly polarized light. Polet and Steinhardt (57) reported that binding of eight SDS molecules decreases the extinction coefficient of bovine serum albumin (BSA) at 220 nm by 9000 M−1cm−1 (although higher amounts of bound SDS showed an increase at 220 nm, perhaps due to light-scattering.) Considering the possible underestimation of per cent helix by CD in SDS, the question of the extent to which SDS unfolds transmembrane helices is not convincingly settled.
Does SDS cause large conformational changes in membrane proteins?
Addition of SDS to BR or DGK folded in detergent micelles causes highly cooperative spectroscopic changes which have been interpreted as unfolding. A well-studied example of SDS-induced soluble protein unfolding is BSA. With addition of SDS, BSA undergoes a succession of spectroscopic changes (e.g. 58), and at high SDS concentrations an increased BSA hydrodynamic radius is observed (59), indicating partial unfolding. Various conformational effects of SDS on membrane proteins have been reported. The effects range from native-like helix-helix interactions (60) or formation of stable oligomers with apparently native-like structures (61-63), to structures apparently lacking helix-helix interactions (35). The lipid-binding protein saposin C shows a mixture of effects, with some helix-helix interactions remaining intact in SDS, and some being completely disrupted (64). The observed conformational change is probably due to the membrane-binding function of saposin C. Specific interactions were observed between SDS and both polar and non-polar side chains of saposin C. The effect of SDS on NMR chemical shifts in FATPase subunit c was interpreted as evidence for either SDS-induced conformational changes or SDS-protein interactions (65).
Upon addition of SDS to BR, the retinal chromophore bleaches (44). With addition of SDS to BO, there are changes in UV absorption (66) and changes in tryptophan fluorescence (14). Addition of SDS to DGK results in changes in UV absorption (46). However, there is no clear evidence that these spectral changes represent a folding/unfolding equilibrium. The detailed SDS binding and structure analysis done for saposin C (64) is not available for BO or DGK. One necessary condition for an unfolded state of a protein is an increase in volume. Recently we prepared fluorescent-labeled bacterio-opsin for unfolding studies using FRET (67, 41), and we examined SDS-solubilized BO. Surprisingly, the distance between sites on helix B and helix F changes by only 2 Å in SDS (68), indicating that SDS-solubilized BO is not significantly unfolded. More detailed FRET, NMR and hydrodynamic studies should be done to obtain a better picture of the state of unfolding of BO and DGK in SDS.
Does SDS “unfolding” provide meaningful thermodynamic information from linear free energy analyses?
The linear free energy method for studying membrane protein “unfolding” in SDS has been described by Lau & Bowie (46), and more recently this method was applied to BR (47, 48) to examine changes due to site specific mutagenesis. The underlying assumption of this method, as developed for water-soluble proteins (49), is that added perturbant (e.g. urea or guanidinium) shifts the equilibrium between folded and unfolded forms of the protein by stabilizing buried parts of the folded structure in a water-exposed state. The application to unfolding of micellar membrane proteins assumes that a perturbing detergent (i.e. SDS) displaces protein-protein interactions with detergent-protein interactions, and that this causes unfolding of the protein. Because the protein is in a separate micelle phase, rather than in bulk solution, concentrations must refer to the micelle phase. In protein association studies, this has been conveniently done by using mole fraction concentration units (69, 70). For protein unfolding studies in micelles, there are complications which are not encountered in protein association equilibria. First, the micelle itself changes as the perturbing detergent (i.e. SDS) is added. Therefore, the mole fraction cannot be easily calculated without knowing how the mixed micelle structure changes with addition of perturbing detergent. Second, when an ionic perturbing detergent is added to a neutral micelle, the mixing is non-ideal (71). For example, SDS monomers will less readily add to SDS-rich micelles than to micelles containing little SDS, due to charge repulsion. Since the micellar SDS concentration is not linear with total SDS concentration, a linear free energy model is not applicable. In their studies of effects of mutagenesis on BR stability, Bowie and co-workers (47, 48) have avoided some of the hazards of these problems by measuring ΔΔG values over a narrow perturbant concentration range, rather than by extrapolating to zero perturbant concentration. However, the validity of this approach has not been established. The experimental ΔΔG values representing mutagenesis-induced changes in BR stability were computed by Lomize et al. (72) using small molecule data, but for unknown reasons, 8 out of 22 mutation sites gave poor fits.
In summary, for the few membrane proteins examined, SDS does not unwind helices, and in some cases SDS does not cause large conformational changes. Nevertheless, SDS clearly perturbs the structure of membrane proteins. The most detailed information on the structural changes of BR in SDS comes from the NMR structure of a two-helix fragment of BR (35). Although the secondary structure is similar to the native structure, no inter-helix NOE interactions were observed. Because NOEs typically occur over distances of a few Ångstroms, the binding of a single layer of SDS molecules between the helices would be sufficient to eliminate the interhelix NOEs. Binding of SDS would also be consistent with the distance increase of about 2 Å observed by FRET for BO transferred from lipid/detergent micelles to SDS micelles (68). From the limited structural information available about SDS binding to proteins (64, 73, 74) it seems plausible that SDS molecules could insert between helices, stabilized by non-polar interactions with aliphatic and aromatic side chains, and by H-bonded interactions and ionic interactions between the sulfate group and protein polar groups. As an alternative to interpreting the SDS-induced transitions as unfolding, we could treat the data as an SDS binding process. SDS binding to a membrane protein in a mixed micelle can be modeled using a Hill-type equation (75):
| (1) |
where Y is the fraction of protein molecules with n SDS binding sites occupied, measured by a spectroscopic method which probes structure at helix-helix interfaces, such as retinal bleaching in BR (47, 48) or UV difference spectra in DGK (46); [D] is the unbound concentration of SDS, in mole fraction units; n is the Hill constant; and K is the apparent association constant:
| (2) |
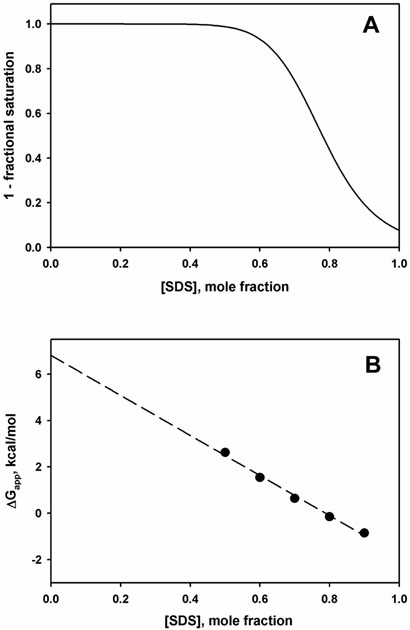
where [PDn] is the concentration of protein with n SDS binding sites occupied and [P] is the concentration of protein with no SDS bound (figure 1A). Since 1-Y is the fraction of protein without SDS bound, the ratio Y/(1-Y) is the same as ratio of “unfolded” to folded protein, using the same data but interpreting it in terms of SDS binding instead of unfolding. Thus, the apparent free energy of “unfolding” calculated from equation 1 is:
| (3) |
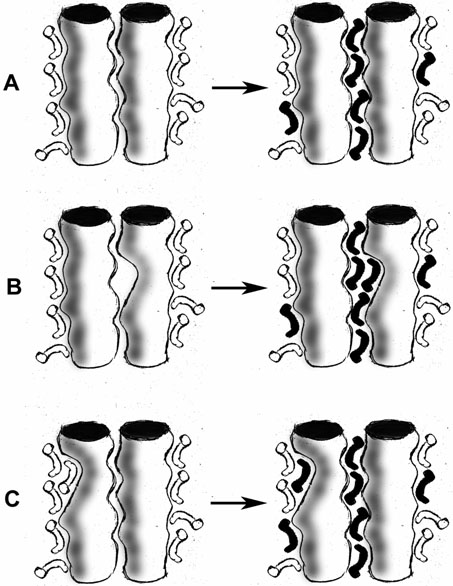
Obviously equation 3 is not linear in SDS concentration, but as a practical matter, the “unfolding” transitions reported (46-48) are relatively sharp and occur above 0.5 mole fraction SDS. When the mole fraction region used for the extrapolation is restricted to the concentration range of the transition, and when this range is at high mole fraction values, there is little change in the slope of the ΔGapp vs. [D] curve (fig. 1B). Thus, it may be difficult to distinguish SDS binding from an unfolding process that follows a linear free energy equation. Other types of binding models which explicitly assign individual association constants to each bound SDS molecule will produce essentially the same result. The measurement of ΔΔG due to mutagenesis is consistent with the SDS binding model in equation 3. Mutations which strengthen SDS binding would be expected to increase K in equation 2, thus shifting the 1-Y vs. [D] curve to the left, in figure 1A. Faham et al. note that the values of ΔΔG of BR mutants are linearly correlated with the amount of buried surface removed from helix-helix interfaces by the mutation, independent of polar interactions (47). This surprising result can be simply explained in terms of an SDS binding model: the removal of buried surface from helix interfaces creates better binding sites for SDS. These sites are not accessible to neutral, activity-supporting detergents (e.g. LM or CHAPSO). By contrast, on the micelle-facing surfaces of the helices, holes created by mutations would be accessible to binding by both the activity-supporting detergents and by the perturbing SDS, so there would be no change in ΔG (fig. 2).
Figure 1. Modeling of SDS-perturbed membrane protein as cooperative SDS binding rather than unfolding.
A. The quantity 1-Y (equation 1) is plotted against SDS concentration, with K = 12 and n = 10. The curve is similar to experimental plots of cooperative spectroscopic changes observed in DGK (46) or bacteriorhodopsin (14, 47, 48) as a function of SDS concentration. B. The SDS binding curve in fig. 1A is interpreted as unfolding, with the equilibrium constant for unfolding equal to Y/(1-Y). The apparent free energy change was calculated from equation 3 for several points. Although equation 3 is not linear with SDS concentration, over the transition region the points fit to a straight line with a 0.99 correlation coefficient.
Figure 2. SDS binding model and mutations.
A. Left: two interacting transmembrane helices are solubilized in a detergent (white) that stabilizes the active structure. Right: after addition of SDS (black), some SDS binds to sites between helices, changing spectroscopic properties. B. Mutation along the interhelix interface creates a cavity which is inaccessible to the activity-supporting detergent (white), but to which SDS (black) can bind. This decreases the concentration of SDS required to cause cooperative spectroscopic changes (fig. 1). C. Mutation on the detergent-facing surface creates a cavity which does not affect the cooperative spectroscopic changes, even if SDS binds to it.
The above discussion might seem to be merely a semantic argument: wouldn't SDS binding between helices actually be a type of unfolding? The free energy changes measured by SDS interactions with membrane proteins may well contain most of the enthalpy of unfolding, since SDS binding would replace helix-helix interactions with helix-SDS interactions. However, because the SDS-protein complex has a compact structure, similar to the native structure, the entropy of unfolding cannot be properly estimated by this method. It has been argued (46) that the entropy of unfolding of a membrane protein (i.e. due to helix separation) will be small compared with the entropy of unfolding of a watersoluble protein, since membrane proteins are spatially constrained within the bilayer, or within a micelle. Nevertheless, the entropy change is unlikely to be 0. A recent estimate, based on model compound studies, is about -4 kcal/mole per helix pair (72). In addition to the entropy of helix separation, changes in side chain entropy may occur upon unfolding, which may not be observed in the SDS-bound state. Therefore, the SDS “unfolding” method gives an incomplete measurement of unfolding free energy because it does not include all the unfolding entropy changes.
While there is not yet any satisfactory procedure for reversibly unfolding helical membrane proteins, I would like to finish with some suggestions about future directions which appear to offer promising new methods.
1) Homogeneous solvent systems
Some of the difficulties with trying to unfold membrane proteins in micelle systems might be avoided by using a single phase solvent system. Several attempts have been published. An ethanol/water system was claimed to permit equilibrium unfolding of BO (67). However, it was later shown that the unfolding in this system was due to acidification as the buffering capacity changed with increasing ethanol concentration; and the acid unfolding of BO was found to be irreversible on a time scale of minutes (41). A fluoroethanol solvent system was found to reversibly dissociate KcsA subunits, but an irreversible unfolding reaction dominates at high fluoroethanol concentrations (38). Popot and co-workers have recently studied fluorocarbon amphiphiles which contain small hydrocarbon regions (76). For example, HF-TAC contains six CF2 groups connected to two CH2 groups. The hydrocarbon appears to interact with the membrane protein lipid-facing surface, leaving the CF2 groups to form an outer lipophobic coating. This seems to stabilize helical transmembrane proteins such as BO and cytochrome bf6 (76), as well as preventing aggregation of hydrophobic surfaces in aqueous solution (77). It may be possible in future experiments to use nonamphiphile versions of mixed hydro- and fluorocarbons to reversibly unfold membrane proteins in a homogeneous solvent.
2) Reverse micelles
Hydrocarbon solvents mixed with sulfonic acid amphiphiles such as sodium bis(2-ethylhexyl)sulfosuccinate (AOT) form well-characterized water-filled reverse micelles (78). Incorporation of integral membrane proteins into this type of micelle system has not yet been systematically studied. Detailed examination of myelin proteolipid in reverse micelles indicates that, at high micelle to protein ratios, one integral membrane protein could bridge two reverse micelles (79). One reason for pursuing reverse micelles for membrane protein unfolding measurements is that the amount of protein surface exposed on unfolding is, in many cases, greater for the lipid-contacting helical segments than for the aqueous-solvated connecting loops. Ordinary micelles constrain the volume changes possible as membrane proteins unfold. By constraining the loops rather than the helices to a fixed internal micelle volume, there may be a higher likelihood of obtaining helix-helix separations in the presence of a perturbant. For example, splitting BR between surface loops exposes loop surfaces that range from 5% to 36% of the corresponding exposed helix surface. However, it is not clear what sort of perturbant could be used to achieve destabilization of interhelix interactions. One possibility is short chain alcohols, which could disrupt interhelix hydrogen bonds. At least in some reverse micelle systems, addition of alcohol has relatively minor effects on the micelle properties (80).
3) Fragment recombination
Currently, some of the best thermodynamic information about interhelix interactions of TM helices comes from studies of oligomerization (70, 81-84). Proteolytic fragments of BR containing various numbers of helices can spontaneously associate to form active protein (34, 36, 67, 85-88), which helped form the basis of the Popot-Engelman two-state model. While not all of the individual BR helices, or groups of BR helices, are stable (39-41), enough combinations are stable and spontaneously associate to suggest that the unfolding free energy might be measured by studying fragment association equilibria. Several other TM proteins with mutliple TM helices have been reconstituted from fragments, including lac permease (89) and various G-protein coupled receptors (90-93). A possible problem with using fragment association to study folding is that some fragments may self-associate (94-96).
If the rate of peptide transfer from one micelle to another is not rate-limiting, it may be possible to also measure folding/unfolding kinetics by fragment recombination/dissociation. Fragment combination kinetics were previously used to study water-soluble protein folding. For example, the rate of association of RNase S-peptide with S-protein has been measured (97, 98). In this system, it was possible to examine the effects of amino acid substitutions on the association rate, permitting analysis by the φ-value method (99). This method can give valuable information about the structure of the transition state and possible folding pathways.
Conclusions
Further work is needed to clarify the nature of the SDS-associated state of helical transmembrane proteins. Regardless of the extent to which SDS unfolds membrane proteins, it would be worthwhile to pursue new strategies for obtaining information about the unfolded state of helical membrane proteins.
Footnotes
Supported by a grant from the National Institutes of Health (GM 08194)
Abbreviations: BR, bacteriorhodopsin; SDS, sodium dodecyl sulfate; BO, bacterio-opsin; TM, transmembrane; DGK, diacylglycerol kinase; CD, circular dichroism; BSA, bovine serum albumin; FRET, fluorescence resonance energy transfer; NOE, nuclear Overhauser effect; LM, lauryl maltoside; CHAPSO, 3-([3-cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate; HF-TAC, C2H5C6F12C2H4-S-poly-Tris-(hydroxymethyl)aminomethane.
REFERENCES
- 1.Sanders CR, Myers JK. Disease-related misassembly of membrane proteins. Annu. Rev. Biophys. Biomol. Struc. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]
- 2.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 3.MacKenzie KR. Folding and stability of α-helical integral membrane proteins. Chem. Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 4.White SH, von Heijne G. Transmembrane helices before, during, and after insertion. Current Opinion in Structural Biology. 2005;15:378–386. doi: 10.1016/j.sbi.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 5.van den Berg B, Clemons WM, Jr., Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 6.Wiedemann N, Frazier AE, Pfanner N. The protein import machinery of mitochondria. J. Biol. Chem. 2004;279:14473–14476. doi: 10.1074/jbc.R400003200. [DOI] [PubMed] [Google Scholar]
- 7.Hermansson M, von Heijne G. Inter-helical hydrogen bond formation during membrane protein integration into the ER membrane. J. Mol. Biol. 2003;334:803–809. doi: 10.1016/j.jmb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Popot JL, Engelman DM. Membrane protein folding and oligomerization: the twostage model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 9.Brouillette CG, Muccio DD, Finney TK. pH dependence of bacteriorhodopsin thermal unfolding. Biochemistry. 1987;26:7431–7438. doi: 10.1021/bi00397a035. [DOI] [PubMed] [Google Scholar]
- 10.Galisteo ML, Sanchez-Ruiz JM. Kinetic study into the irreversible thermal denaturation of bacteriorhodopsin. Eur. Biophys. J. 1993;22:25–30. [Google Scholar]
- 11.Heyes CD, El-Sayed MA. Thermal properties of bacteriorhodopsin. J. Phys. Chem. B. 2003;107:12045–12053. [Google Scholar]
- 12.Taneva SG, Caaveiro JMM, Muga A, Goni FM. A pathway for the thermal destabilization of bacteriorhodopsin. FEBS Lett. 1995;367:297–300. doi: 10.1016/0014-5793(95)00570-y. [DOI] [PubMed] [Google Scholar]
- 13.Brouillette CG, McMichens RB, Stern LJ, Khorana HG. Structure and thermal stability of monomeric bacteriorhodopsin in mixed phospholipid/detergent micelles. Proteins: Struc. Func. Gen. 1989;5:38–46. doi: 10.1002/prot.340050106. [DOI] [PubMed] [Google Scholar]
- 14.Chen GQ, Gouaux E. Probing the folding and unfolding of wild-type and mutant forms of bacteriorhodopsin in micellar solutions: evaluation of reversible unfolding conditions. Biochemistry. 1999;38:15380–15387. doi: 10.1021/bi9909039. [DOI] [PubMed] [Google Scholar]
- 15.Epand RF, Epand RM, Jung CY. Glucose-induced thermal stabilization of the native conformation of GLUT 1. Biochemistry. 1999;38:454–458. doi: 10.1021/bi981893z. [DOI] [PubMed] [Google Scholar]
- 16.Landin JS, Katragadda M, Albert AD. Thermal destabilization of rhodopsin and opsin by proteolytic cleavage in bovine rod outer segment disk membranes. Biochemistry. 2001;40:11176–11183. doi: 10.1021/bi0100539. [DOI] [PubMed] [Google Scholar]
- 17.Morin PE, Diggs D, Freire E. Thermal stability of membrane-reconstituted yeast cytochrome c oxidase7. Biochemistry. 1990;29:781–788. doi: 10.1021/bi00455a028. [DOI] [PubMed] [Google Scholar]
- 18.Haltia T, Freire E. Forces and factors that contribute to the structural stability of membrane proteins. Biochim. Biophys. Acta. 1995;1241:295–322. doi: 10.1016/0304-4157(94)00161-6. [DOI] [PubMed] [Google Scholar]
- 19.Oesterhelt F, Oesterhelt D, Pfeiffer M, Engel A, Gaub HE, Müller DJ. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 20.Müller DJ, Kessler M, Oesterhelt F, Möller C, Oesterhelt D, Gaub H. Stability of bacteriorhodopsin α-helices and loops analyzed by single-molecule force spectroscopy. Biophys. J. 2002;83:3578–3588. doi: 10.1016/S0006-3495(02)75358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler M, Gottschalk KE, Janovjak H, Müller DJ, Gaub HE. Bacteriorhodopsin folds into the membrane against an external force. J. Mol. Biol. 2006;357:644–654. doi: 10.1016/j.jmb.2005.12.065. [DOI] [PubMed] [Google Scholar]
- 22.Kedrov A, Ziegler C, Janovjak H, Kühlbrandt W, Müller DJ. Controlled unfolding and refolding of a single sodium-proton antiporter using atomic force microscopy. J. Mol. Biol. 2004;340:1143–1152. doi: 10.1016/j.jmb.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Surrey T, Jahnig F. Refolding and oriented insertion of a membrane protein into a lipid bilayer. Proc. Nat. Acad. Sci. USA. 1992;89:7457–7461. doi: 10.1073/pnas.89.16.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H, Tamm LK. Elastic coupling of integral membrane protein stability to lipid bilayer forces. Proc. Nat. Acad. Sci. USA. 2004;101:4065–4070. doi: 10.1073/pnas.0400358101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleinschmidt JH. Folding kinetics of the outer membrane proteins OmpA and FomA into phospholipid bilayers. Chem. Phys. Lipids. 2006;141:30–47. doi: 10.1016/j.chemphyslip.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Huang K-S, Radhakrishnan R, Bayley H, Khorana HG. Orientation of retinal in bacteriorhodopsin as studied by cross-linking using a photosensitive analog of retinal. J. Biol. Chem. 1982;257:13616–13623. [PubMed] [Google Scholar]
- 27.Lorch M, Booth PJ. Insertion kinetics of a denatured α helical membrane protein into phospholipid bilayer vesicles. J. Mol. Biol. 2004;344:1109–1121. doi: 10.1016/j.jmb.2004.09.090. [DOI] [PubMed] [Google Scholar]
- 28.Mi D, Kim HJ, Hadziselimovic A, Sanders CR. Irreversible misfolding of diacylglycerol kinase is independent of aggregation and occurs prior to trimerization and membrane association. Biochemistry. 2006;45:10072–10084. doi: 10.1021/bi060887x. [DOI] [PubMed] [Google Scholar]
- 29.Roepe PD, Kaback HR. Characterization and functional reconstitution of a soluble form of the hydrophobic membrane protein lac permease from Escherichia coli. Proc. Nat. Acad. Sci. USA. 1989;86:6087–6091. doi: 10.1073/pnas.86.16.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang C, Horn R, Paulsen H. The light-harvesting chlorophyll a/b complex can be reconstituted in vitro from its completely unfolded apoprotein. Biochemistry. 2003;42:4527–4533. doi: 10.1021/bi0273157. [DOI] [PubMed] [Google Scholar]
- 31.Huang K-S, Bayley H, Liao M-J, London E, Khorana HG. Refolding of an integral membrane protein. Denaturation, renaturation, and reconstitution of intact bacteriorhodopsin and two proteolytic fragments. J. Biol. Chem. 1981;256:3802–3809. [PubMed] [Google Scholar]
- 32.Valiyaveetil FI, Zhou Y, MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 33.Gerber GE, Anderegg RJ, Herlihy WC, Gray CP, Biemann K, Khorana HG. Partial primary structure of bacteriorhodopsin: sequencing methods for membrane proteins. Proc. Nat. Acad. Sci. USA. 1979;76:227–31. doi: 10.1073/pnas.76.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sigrist H, Wenger RH, Kislig E, Wuthrich M. Refolding of bacteriorhodopsin. Protease V8 fragmentation and chromophore reconstitution from proteolytic V8 fragment. Eur. J. Biochem. 1988;177:125–133. doi: 10.1111/j.1432-1033.1988.tb14352.x. [DOI] [PubMed] [Google Scholar]
- 35.Pervushin KV, Orekhov V.Yu., Popov AI, Musina L.Yu., Arseniev AS. Threedimensional structure of (1-71) bacterio-opsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy. Eur. J. Biochem. 1994;219:571–583. doi: 10.1111/j.1432-1033.1994.tb19973.x. [DOI] [PubMed] [Google Scholar]
- 36.Popot J-L, Gerchman S-E, Engelman DM. Refolding of bacteriorhodopsin in lipid bilayers. A thermodynamically controlled two-stage process. J. Mol. Biol. 1987;198:655–676. doi: 10.1016/0022-2836(87)90208-7. [DOI] [PubMed] [Google Scholar]
- 37.Klammt C, Schwarz D, Fendler K, Haase W, Dotsch V, Bernhard F. Evaluation of detergents for the soluble expression of α-helical and β-barrel-type integral membrane proteins by a preparative scale individual cell-free expression system. FEBS J. 2005;272:6024–6038. doi: 10.1111/j.1742-4658.2005.05002.x. [DOI] [PubMed] [Google Scholar]
- 38.Barrera FN, Renart ML, Molina ML, Poveda JA, Encinar JA, Fernández AM, Neira JL, González-Ros JM. Unfolding and refolding in vitro of a tetrameric, α-helical membrane protein: the prokaryotic potassium channel KcsA. Biochemistry. 2005;44:14344–14352. doi: 10.1021/bi050845t. [DOI] [PubMed] [Google Scholar]
- 39.Hunt JF, Earnest TN, Bousche O, Kalghatgi K, Reilly K, Horvath C, Rothschild KJ, Engelman DM. A biophysical study of integral membrane protein folding. Biochemistry. 1997;36:15156–15176. doi: 10.1021/bi970146j. [DOI] [PubMed] [Google Scholar]
- 40.Luneberg J, Widmann M, Dathe M, Marti T. Secondary structure of bacteriorhodopsin fragments. External sequence constraints specify the conformation of transmembrane helices. J. Biol. Chem. 1998;273:28822–28830. doi: 10.1074/jbc.273.44.28822. [DOI] [PubMed] [Google Scholar]
- 41.Valluru N, Silva F, Dhage M, Rodriguez G, Alloor SR, Renthal R. Transmembrane helix-helix association: relative stabilities at low pH. Biochemistry. 2006;45:4371–4377. doi: 10.1021/bi0525268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rader AJ, Anderson G, Isin B, Khorana HG, Bahar I, Klein-Seetharaman J. Identification of core amino acids stabilizing rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7246–7251. doi: 10.1073/pnas.0401429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Compton ELR, Farmer N, Lorch M, Mason JM, Moreton KM, Booth PJ. Kinetics of an individual transmembrane helix during bacteriorhodopsin folding. J. Mol. Biol. 2006;357:325–338. doi: 10.1016/j.jmb.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 44.London E, Khorana HG. Denaturation and renaturation of bacteriorhodopsin in detergents and lipid-detergent mixtures. J. Biol. Chem. 1982;257:7003–7011. [PubMed] [Google Scholar]
- 45.Booth PJ. Unravelling the folding of bacteriorhodopsin. Biochim. Biophys. Acta. 2000;1460:4–14. doi: 10.1016/s0005-2728(00)00125-0. [DOI] [PubMed] [Google Scholar]
- 46.Lau F, Bowie JU. A method for assessing the stability of a membrane protein. Biochemistry. 1997;36:5884–5892. doi: 10.1021/bi963095j. [DOI] [PubMed] [Google Scholar]
- 47.Faham S, Yang D, Bare E, Yohannan S, Whitelegge JP, Bowie JU. Sidechain contributions to membrane protein structure and stability. J. Mol. Biol. 2004;335:297–305. doi: 10.1016/j.jmb.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 48.Yohannan S, Faham S, Yang D, Grosfeld D, Chamberlain AK, Bowie JU. A Ca-H·O hydrogen bond in a membrane protein is not stabilizing. J. Am. Chem. Soc. 2004;126:2284–2285. doi: 10.1021/ja0317574. [DOI] [PubMed] [Google Scholar]
- 49.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 50.Riley ML, Wallace BA, Flitsch SL, Booth PJ. Slow α-helical formation during folding of a membrane protein. Biochemistry. 1997;36:192–196. doi: 10.1021/bi962199r. [DOI] [PubMed] [Google Scholar]
- 51.Rozek A, Buchko GW, Cushley RJ. Conformation of two peptides corresponding to human apolipoprotein C-I residues 7-24 and 35-53 in the presence of sodium dodecyl sulfate by CD and NMR spectroscopy. Biochemistry. 1995;34:7401–7408. [PubMed] [Google Scholar]
- 52.Wang G, Treleaven WD, Cushley RJ. Conformation of human serum apolipoprotein A-I(166-185) in the presence of sodium dodecyl sulfate or dodecylphosphocholine by 1H-NMR and CD. Evidence for specific peptide-SDS interactions. Biochim. Biophys. Acta. 1996;1301:174–184. doi: 10.1016/0005-2760(96)00037-9. [DOI] [PubMed] [Google Scholar]
- 53.Montserret R, McLeish MJ, Böckmann A, Geourjon C, Penin F. Involvement of electrostatic interactions in the mechanism of peptide folding induced by sodium dodecyl sulfate binding. Biochemistry. 2000;39:8362–8373. doi: 10.1021/bi000208x. [DOI] [PubMed] [Google Scholar]
- 54.Storjohann R, Rozek A, Sparrow JT, Cushley RJ. Structure of a biologically active fragment of human serum apolipoprotein C-II in the presence of sodium dodecyl sulfate and dodecylphosphocholine. Biochim. Biophys. Acta. 2000;1486:253–264. doi: 10.1016/s1388-1981(00)00062-7. [DOI] [PubMed] [Google Scholar]
- 55.MacRaild CA, Hatters DM, Howlett GJ, Gooley PR. NMR structure of human apolipoprotein C-II in the presence of sodium dodecyl sulfate. Biochemistry. 2001;40:5414–5421. doi: 10.1021/bi002821m. [DOI] [PubMed] [Google Scholar]
- 56.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 57.Polet J, Steinhardt J. Binding-induced alterations in ultraviolet absorption of native serum albumin. Biochemistry. 1968;7:1348–1356. doi: 10.1021/bi00844a015. [DOI] [PubMed] [Google Scholar]
- 58.Turro NJ, Lei X-G. Spectroscopic probe analysis of protein-surfactant interactions: the BSA/SDS system. Langmuir. 1995;11:2525–2533. [Google Scholar]
- 59.Valstar A, Almgren M, Brown W. The interaction of bovine serum albumin with surfactants studied by light scattering. Langmuir. 2000;16:922–927. [Google Scholar]
- 60.Howell SC, Mesleh MF, Opella SJ. NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry. 2005;44:5196–5206. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- 61.Laubinger W, Dimroth P. Characterization of the ATP synthase of Propionigenium modestum as a primary sodium pump. Biochemistry. 1988;27:7531–7537. doi: 10.1021/bi00419a053. [DOI] [PubMed] [Google Scholar]
- 62.Bormann B-J, Knowles WJ, Marchesi VT. Synthetic peptides mimic the assembly of transmembrane glycoproteins. J. Biol. Chem. 1989;264:4033–4037. [PubMed] [Google Scholar]
- 63.Lemmon MA, Flanagan JM, Hunt JF, Adair BD, Bormann B-J, Dempsey CE, Engelman DM. Glycophorin A dimerization is driven by specific interactions between transmembrane α-helices. J. Biol. Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 64.Hawkins CA, de Alba E, Tjandra N. Solution structure of human saposin C in a detergent environment. J. Mol. Biol. 2005;346:1381–1392. doi: 10.1016/j.jmb.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 65.Krueger-Koplin RD, Sorgen PL, Kruger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. An evaluation of detergents for NMR structural studies of membrane proteins. J. Biomolec. NMR. 2004;17:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 66.Renthal R, Haas P. Effect of transmembrane helix packing on tryptophan and tyrosine environments in detergent-solubilized bacterio-opsin. J. Protein Chem. 1996;15:281–289. doi: 10.1007/BF01887117. [DOI] [PubMed] [Google Scholar]
- 67.Nannepaga SJ, Gawalapu R, Velasquez D, Renthal R. Estimation of helix-helix association free energy from partial unfolding of bacterio-opsin. Biochemistry. 2004;43:550–559. doi: 10.1021/bi034875c. [DOI] [PubMed] [Google Scholar]
- 68.Renthal R, Alloor SR. Partially unfolded membrane protein has a compact conformation. FASEB J. 2006;20 abs. # 348.9 ( http://www.eb2006-online.com/) [Google Scholar]
- 69.Salom D, Hill BR, Lear JD, DeGrado WF. pH-Dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fleming KG. Standardizing the free energy change of transmembrane helix–helix interactions. J. Mol. Biol. 2002;323:563–571. doi: 10.1016/s0022-2836(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 71.Tanford C. The Hydrophobic Effect. 2nd. Wiley; New York: 1980. pp. 81–83. [Google Scholar]
- 72.Lomize AL, Pogozheva ID, Mosberg HI. Quantification of helix–helix binding affinities in micelles and lipid bilayers. Protein Sci. 2004;13:2600–2612. doi: 10.1110/ps.04850804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yonath A, Podjarny A, Honig B, Sielecki A, Traub W. Crystallographic studies of protein denaturation and renaturation. 2. Sodium dodecyl sulfate induced structural changes in triclinic lysozyme. Biochemistry. 1977;16:1418–1424. doi: 10.1021/bi00626a028. [DOI] [PubMed] [Google Scholar]
- 74.Forouhar F, Huang WN, Liu JH, Chien KY, Wu WG, Hsiao CD. Structural basis of membrane-induced cardiotoxin A3 oligomerization. J. Biol. Chem. 2003;278:21980–21988. doi: 10.1074/jbc.M208650200. [DOI] [PubMed] [Google Scholar]
- 75.Edsall JT, Wyman J. Biophysical Chemistry v. 1. Academic Press; New York: 1958. p. 635. [Google Scholar]
- 76.Breyton C, Chabaud E, Chaudier Y, Pucci B, Popot J-L. Hemifluorinated surfactants: a non-dissociating environment for handling membrane proteins in aqueous solutions? FEBS Lett. 2004;564:312–318. doi: 10.1016/S0014-5793(04)00227-3. [DOI] [PubMed] [Google Scholar]
- 77.Palchevskyy SS, Posokhov YO, Olivier B, Popot J-L, Pucci B, Ladokhin AS. Chaperoning of insertion of membrane proteins into lipid bilayers by hemifluorinated surfactants: application to diphtheria toxin. Biochemistry. 2006;45:2629–2635. doi: 10.1021/bi052257l. [DOI] [PubMed] [Google Scholar]
- 78.De TK, Maitra A. Solution behaviour of Aerosol OT in non-polar solvent. Adv. Colloid Interface Sci. 1995;59:95–193. [Google Scholar]
- 79.Merdas A, Gindre M, Le Huerou J-Y, Nicot C, Ober R, Urbach W, Waks M. Bridging of nonionic reverse micelles by a myelin transmembrane protein. J. Phys. Chem. B. 1998;102:528–533. [Google Scholar]
- 80.Nazário LMM, Hatton TA, Crespo JPSG. Nonionic cosurfactants in AOT reversed micelles: effect on percolation, size, and solubilization site. Langmuir. 1996;12:6326–6335. [Google Scholar]
- 81.Fleming KG, Ackerman AL, Engelman DM. The effect of point mutations on the free energy of transmembrane α-helix dimerization. J. Mol. Biol. 1997;272:266–275. doi: 10.1006/jmbi.1997.1236. [DOI] [PubMed] [Google Scholar]
- 82.Kochendoerfer GG, Salom D, Lear JD, Wilk-Orescan R, Kent SB, DeGrado WF. Total chemical synthesis of the integral membrane protein influenza A virus M2:role of its Cterminal domain in tetramer assembly. Biochemistry. 1999;38:11905–11913. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]
- 83.You M, Li E, Wimley WC, Hristova K. Förster resonance energy transfer in liposomes: measurements of transmembrane helix dimerization in the native bilayer environment. Anal. Biochem. 2005;340:154–164. doi: 10.1016/j.ab.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li E, You M, Hristova K. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Förster resonance energy transfer suggest weak interactions between fibroblast growth factor receptor 3 (FGFR3) transmembrane domains in the absence of extracellular domains and ligands. Biochemistry. 2005;44:352–360. doi: 10.1021/bi048480k. [DOI] [PubMed] [Google Scholar]
- 85.Liao MJ, London E, Khorana HG. Regeneration of the native bacteriorhodopsin structure from two chymotryptic fragments. J. Biol. Chem. 1983;258:9949–99455. [PubMed] [Google Scholar]
- 86.Liao MJ, Huang KS, Khorana HG. Regeneration of native bacteriorhodopsin structure from fragments. J. Biol. Chem. 1984;259:4200–4204. [PubMed] [Google Scholar]
- 87.Kahn TW, Engelman DM. Bacteriorhodopsin can be refolded from two independently stable transmembrane helices and the complementary five-helix fragment. Biochemistry. 1992;31:6144–6151. doi: 10.1021/bi00141a027. [DOI] [PubMed] [Google Scholar]
- 88.Marti T. Refolding of bacteriorhodopsin from expressed polypeptide fragments. J. Biol. Chem. 1998;273:9312–9322. doi: 10.1074/jbc.273.15.9312. [DOI] [PubMed] [Google Scholar]
- 89.Sahin-Tóth M, Kaback HR, Friedlnader M. Association between the amino- and carboxyl-terminal halves of lactose permease is specific and mediated by multiple transmembrane domains. Biochemistry. 1996;35:2016–2021. doi: 10.1021/bi952496g. [DOI] [PubMed] [Google Scholar]
- 90.Kobilka BK, Kobilka T-S, Daniel K, Regan JW, Caron MG, Lefkowitz RJ. Chimeric α2-, β2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science. 1988;240:1310–1316. doi: 10.1126/science.2836950. [DOI] [PubMed] [Google Scholar]
- 91.Ridge KD, Lee SS, Abdulaev NG. Examining rhodopsin folding and assembly through expression of polypeptide fragments. J. Biol. Chem. 1996;271:7860–7876. doi: 10.1074/jbc.271.13.7860. [DOI] [PubMed] [Google Scholar]
- 92.Schöneberg T, SandigDagger V, Wess J, Gudermann T, Schultz G. Reconstitution of mutant V2 vasopressin receptors by adenovirus-mediated gene transfer molecular basis and clinical implication. J. Clin. Invest. 1997;100:1547–1556. doi: 10.1172/JCI119678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martin NP, Leavitt LM, Sommers CM, Dumont ME. Assembly of G proteincoupled receptors from fragments: identification of functional receptors with discontinuities in each of the loops connecting transmembrane segments. Biochemistry. 1999;38:682–695. doi: 10.1021/bi982062w. [DOI] [PubMed] [Google Scholar]
- 94.Overton MC, Blumer KJ. The extracellular N-terminal domain and transmembrane domains 1 and 2 mediate oligomerization of a yeast G protein-coupled receptor. J. Biol. Chem. 2002;277:41463–41472. doi: 10.1074/jbc.M205368200. [DOI] [PubMed] [Google Scholar]
- 95.Carrillo JJ, López-Giménez JF, Milligan G. Multiple interactions between transmembrane helices generate the oligomeric α1b-adrenoceptor. Mol. Pharmacol. 2004;66:1123–1137. doi: 10.1124/mol.104.001586. [DOI] [PubMed] [Google Scholar]
- 96.Thévenin D, Lazarova T, Roberts MF, Robinson CR. Oligomerization of the fifth transmembrane domain from the adenosine A2A receptor. Protein Science. 2005;14:2177–2186. doi: 10.1110/ps.051409205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goldberg JM, Baldwin RL. Kinetic mechanism of a partial folding reaction. 1. properties of the reaction and effects of denaturants. Biochemistry. 1998a;37:2546–2555. doi: 10.1021/bi972402y. [DOI] [PubMed] [Google Scholar]
- 98.Goldberg JM, Baldwin RL. Kinetic mechanism of a partial folding reaction. 2. nature of the transition state. Biochemistry. 1998b;37:2556–2563. doi: 10.1021/bi972403q. [DOI] [PubMed] [Google Scholar]
- 99.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. theory of protein engineering analysis of stability and pathway of protein folding. J. Mol. Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]