Abstract
Formalin-fixed, paraffin-embedded (FFPE) tissue is the most common specimen available for molecular assays on tissue after diagnostic histopathological examination. RNA from FFPE tissue suffers from strand breakage and cross-linking. Despite excellent extraction methods, RNA quality from FFPE material remains variable. To address the RNA quality factors within FFPE tissues, we studied RNA quality, isolating individual elements of the tissue fixation and processing including length of fixation in formalin and the type of buffer incorporated in the fixative. We examined the impact of the length of the tissue processing cycle as well. The optimal fixation period of 12–24 hr in phosphate-buffered formalin resulted in better-quality RNA. Longer tissue processing times were associated with higher quality RNA. We determined that the middle region of gene suffers less damage by these processes as shown by real-time quantitative RT-PCR. These data provide key information for the development of methods of analysis of gene expression in archival FFPE tissues and contribute to the establishment of objective standards for the processing and handling of tissue in surgical pathology. This manuscript contains online supplemental material at http://www.jhc.org. Please visit this article online to view these materials. (J Histochem Cytochem 56:1033–1042, 2008)
Keywords: formalin fixed, paraffin embedded, gene expression, RNA integrity, real-time RT-PCR, tissue fixation and processing
The widespread adoption of transcriptional profiling of tumors has been limited by the inability to isolate high-quality RNA from routine surgical specimens. Most research studies have relied on frozen tissue, which is not practical for most specimens in a routine clinical care environment. RNA can be isolated from formalin-fixed, paraffin-embedded (FFPE) tissue and used in RT-PCR–based assays (Finke et al. 1993; Krafft et al. 1997; Jewell et al. 2002); however, RNA quantity and quality are significant issues in assay development (Hewitt et al. in press). FFPE tissue is anticipated to remain the specimen preservation means of choice for diagnostic histopathology, having been built on over a century of knowledge. Other approaches, including alternative fixatives, have been studied (Sato et al. 1991; Shibutani et al. 2000; Bonin et al. 2005; Lykidis et al. 2007; Hewitt et al. in press); however, none meet the demands of clinical practice as an economic universal fixative, nor do they provide the reproducible artifact of formalin fixation. Individual groups are working on expanding these applications; unfortunately, few data exist comparing different fixation methodologies. Therefore, any data on the impact of tissue handling and processing on RNA quantity and quality is of interest to those developing clinical assays based on RNA analysis.
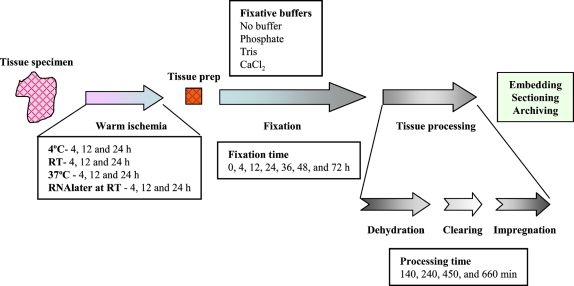
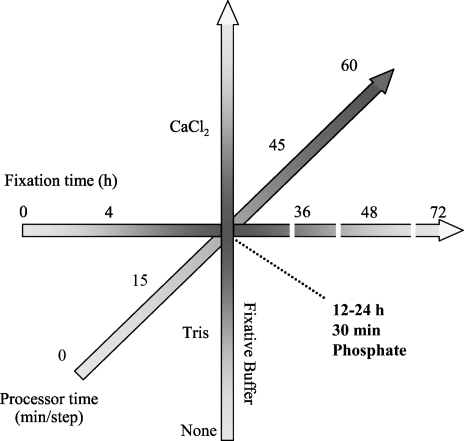
The isolation of RNA from FFPE tissue was first reported by Rupp and Locker (1988), but quality metrics to quantify the quality of RNA isolated from FFPE tissue are limited (Karsten et al. 2002; Auer et al. 2003; Chung et al. 2006; Penland et al. 2007). The quality of RNA obtained from FFPE tissue is widely variable. Methods of isolation no doubt impact the quantity of RNA recovered; however, quality within FFPE specimens is dependent on preanalytical conditions including warm ischemia time and fixation conditions (Hewitt et al. in press). Studies routinely described fixation and processing “per standard protocol,” when in fact there is a lack of standardization of fixation and tissue processing protocols. The process of tissue handling and processing from patient to paraffin block is too frequently invisible to researchers, further limiting the appreciation of the importance of standardization of tissue handling. Namely, there are three major phases (warm ischemia, fixation, and tissue processing) in transformation from patient tissue sample to paraffin block (Figure 1). Warm ischemia refers to the time of transfer from an operation room (or removal of blood supply) to pathology laboratory and may vary from minutes to hours. In most instances, specimens are held at room temperature; however, in some cases, or if long delays are anticipated, they may be refrigerated. Some specimens, more commonly small biopsies, are directly placed in fixative and transported to pathology. Within the pathology laboratory, optimally, the specimen is dissected and sectioned into appropriate size for fixation and processing with minimal delay. However, the length of fixation can vary greatly, largely dependent on specimen size and time of day of receipt of the specimen. Finally, the specimen is placed on a tissue processor, in which the specimen is serially dehydrated in graded alcohols, and the alcohols are replaced with xylene or other clearing agents, followed by impregnation by paraffin (Hewitt et al. in press). The length of this process routinely varies from 4 to 12 hr; however, protocols that are either shorter or longer are regularly encountered. Finally, the tissue is surrounded in a mold with hot paraffin to form a paraffin block, which can be cut with a microtome.
Figure 1.
Diagram of the pathway a tissue specimen takes from removal from patient to an archival tissue block. After devitalization of the specimen, there is a variable length of time before the specimen is placed in fixative. During this period, called “warm ischemia,” the tissue lacks oxygen and anoxic metabolic pathways take over. The specimen undergoes some form of preparation before fixation, depending on the size and complexity of the specimen. The specimen fixes, optimally in 10 volumes of fixative for a duration suitable for complete penetration of the fixative (1 mm/hr). After fixation, the specimen undergoes “tissue processing” on an automated instrument that serially replaces the fluids within a retort to accomplish the replacement of water with paraffin. This three-step process (dehydration, clearing, and impregnation) is accomplished with a variable number of steps (12–14), with more steps devoted to dehydration than clearing or impregnation. Some steps may contain the same reagent, in an effort to complete the exchange of solvents. Most tissue processors operate under vacuum. After tissue processing, the specimen is “embedded” (surrounded with paraffin that acts as a solid support for microtomy). Conditions examined in this study are summarized within the text boxes. For the experiments in this study, the variables were studied in isolation. For experiments on warm ischemia, RNA was assayed before fixation and processing. In all experiments on fixation and tissue processing, warm ischemia was minimized, with a maximum duration of 15 min. For the experiments on fixation, tissue was processed, fixed in phosphate-buffered formalin, and processed at 30 min/step. For experiments on fixative buffer, fixation time was fixed at 24 hr, and tissue was processed for 30 min/step. For experiments on tissue processing, tissue was fixed for 24 hr in phosphate-buffered formalin.
Studies have shown that extended fixation times followed by processing, embedding, and storage of the specimens result in poor quality of RNA (Masuda et al. 1999; Macabeo-Ong et al. 2002; Cronin et al. 2004). However, the preanalytical steps of tissue handling and processing have not been systematically studied for impact on the quality of RNA isolated (Cronin et al. 2004; Hewitt et al. in press). In this study, we dissect the warm ischemia time and its impact on RNA quality. We quantify the effects of fixation time on RNA quality and determine the impact of different buffers added to formalin. Finally, we quantify the effects of time on the tissue processor on RNA.
Materials and Methods
Rats and mice were obtained from the Small Animals Section, Veterinary Resources Branch, National Institutes of Health (NIH), in the United States. Animals were housed and euthanized in accordance with the NIH guidelines for care and use of laboratory animals.
Experimental Methods for Documentation of Effects of Warm Ischemia
We used 8-week-old female Sprague-Dawley rat kidney tissues for quantitative real-time RT-PCR studies. The kidneys were dissected into 40-mg fragments and held as described as an experimental model of warm ischemia. For a time 0 point, RNA extraction was begun within 1 min of death by placement in Trizol and homogenization. Without any treatment, we serially extracted RNA at 4, 25 (room temperature), and 37C in three different time points (4, 12, and 24 hr). In addition, we examined specimens stored in RNAlater (Ambion; Austin, TX) at room temperature and analyzed RNA at the same time points as above.
Experimental Methods for Examination of Effects of Fixation and Tissue Processing
To profile the degradation pattern of RNA depending on fixation time, fixation buffer, and tissue processing time, we used 6-week-old female Balb/c mouse kidneys. A whole kidney was used as the standard tissue specimen for the fixation and processing experiments, and independent experiments were carried out in triplicate. After fixation, ranging from 0 to 72 hr, the fixed tissue was processed according to a defined paraffin-embedding procedure with a Tissue-Tek VIP IV automated tissue processor (Sakura Fineteck USA, Inc.; Torrance, CA) of 30 min per station (see below). For fixative buffer conditions, we performed a routine FFPE procedure after 24-hr fixation in Tris-, phosphate-, and CaCl2-buffered formalins or formalin without buffer. To determine optimal tissue processing time, tissues were fixed for 24 hr in phosphate-buffered 10% formalin. We subsequently processed the tissue based on total processing time ranging from 140 to 660 min in equal 14-step protocols of fixation and dehydration, clearing, and impregnation as follows: two stations of phosphate-buffered 10% formalin, one station of 70% ethanol, two stations of 95% ethanol, three stations of 100% ethanol, two stations of xylene, and four stations of Paraplast Extra paraffin (Tyco Healthcare/Kendall; Mansfield, MA). The time in each station was 5, 15, 30, or 45 min, with additional time required to fill and empty the retort. The instrument was operated with a vacuum and heated to 65C for stations as controlled by the instrument.
Total RNA Isolation From FFPE Sections
RNA extraction from two 10-μm FFPE tissue sections was performed as previously reported (Chung et al. 2006). Briefly, sections were trimmed of excess wax and deparaffinized by three rinses in AutoDewaxer (Open Biosystems; Huntsville, AL) for 15 min at 95C with shaking, followed by three centrifugations at room temperature for 2 min at 10,000 × g. Specimens were briefly rinsed once in 100% ethanol. The sections were resuspended and ground in a solution of 4 M guanidine isothiocyanate, 20 mM sodium acetate, and 25 mM β-mercaptoethanol (pH 5.5), followed by incubation for 72 hr at 65C with mild shaking. After incubation, the solution was mixed with equal volume of phenol-chloroform-isoamyl alcohol (PCI; 25:24:1, pH 4). The phases were separated by centrifugation at 4C for 10 min at 10,000 × g. The aqueous phase was transferred to a fresh tube, and an equal volume of chloroform-isoamyl alcohol (CI; 49:1) was added, followed by centrifugation at 10,000 × g at 4C for 10 min. After extraction, the RNA was precipitated with 100% ethanol. The resulting RNA pellet was washed with ice-cold 75% ethanol and resuspended in diethyl-pyrocarbonate (DEPC)-treated water.
For fresh and frozen tissues, total RNA was obtained using TRIzol (Invitrogen; Carlsbad, CA) at each time point. The RNA was further purified with RNeasy minikits (Qiagen; Valencia, CA) according to the manufacturer's instruction. The amount of RNA was determined by spectrophotometer, and the samples were stored at −80C until use.
Assessment of Quantity and Quality of Total RNA
RNA concentrations were measured with the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies; Wilmington, DE). To assess RNA quality, we used the Agilent 2100 bioanalyzer (Agilent Technologies; Palo Alto, CA) with the RNA 6000 LabChip kit (Agilent Technologies). Rat or mouse kidney total RNA was used as a high-quality reference RNA. To compare electropherograms, we used Agilent 2100 expert software (Agilent Technologies).
Evaluation of RNA Integrity by Branched DNA Assay
RNA integrity was measured by QuantiGene assay (Panomics; Fremont, CA), which is based on branched-DNA signal amplification and detection. To measure RNA degradation from each entity, we used probes against cyclin-dependent kinase 4 (CDK4) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) specific to mouse RNA. The assay was performed as previously described in detail (Kern et al. 1996; Canales et al. 2006). Briefly, 10 μl (200 and 500 ng for GAPDH and CDK4, respectively) of total RNA extracted from FFPE tissue samples was mixed with 40 μl capture buffer, 40 μl lysis mixture, and 10 μl of the target gene probe set (capture extender, label extender, and blocker). The probes were allowed to hybridize to the specific target gene at 53C for 16-20 hr in a 96-well capture plate. Excess probes were removed by wash buffer (Panomics), and 100 μl of branched DNA amplifier was added into an individual well of the capture plates. The plates were incubated at 46C for 1 hr, and 100 μl of label probe working reagent was added. After incubation with substrate dioxetane solution at 46C for 30 min, luminescence was measured with a GloRunnerTM Microplate Luminometer (TunerBiosystems; Sunnyvale, CA). Data are the mean of three independent experiments.
Evaluating RNA Integrity by Real-time Quantitative RT-PCR
To profile RNA integrity in samples prepared from different conditions, we quantified the expression level of the GAPDH gene by three different probes such as 5′-open reading frame (ORF), middle of ORF, and 3′-untranslated regions (UTRs). The amplicon was a 77- to 78- bp portion of the GAPDH gene. The primers and probes were designed with the assistance of Primer Express software (version 1.0; PE Applied Biosystems, Foster City, CA). Details of the primers and probes for GAPDH used in this study are summarized in Supplemental Table 1. Additionally, we also tested the hypoxanthine-guanine phosphoribosyl transferase (HPRT) gene with the same strategy for GAPDH (Supplemental Table 1). Total RNA was treated with DNase (Promega; Madison, WI) to prevent contamination of genomic DNA and resuspended in DEPC water. Reverse transcription was performed with 2 μg total RNA using Superscript II RNA H-Reverse Transcriptase (Invitrogen). For cDNA synthesis, 4 μl 5× first-strand buffer, 2 μl DTT (0.1 M), 2 μl dNTP (10 mM), 1 μl RNAsin (40 U/μl; Promega), 1 μl random hexamers (50 μM; Promega), and 1 μl (200 U) SuperScript II reverse transcriptase were added to 9 μl RNA. The mixture was incubated at 25C for 10 min and 42C for 90 min. The reaction was inactivated by heating to 70C for 15 min. To remove RNA complementary to the cDNA, we added 1 μl RNase H (2 U) to the mixture and further incubated at 37C for 20 min. Five-fold diluted cDNA (5 μl) was used as a template for real-time PCR. The PCR reaction mixture contained 5 μl 5-fold diluted cDNA, 12.5 μl ABI-TaqMan Universal PCR Master Mix (Applied Biosystems), 80 μM of the forward and reverse primers, and 10 μM of the probe. PCR was performed in an ABI Prism 7500 Sequence Detection System (Applied Biosystems). The PCR conditions were 1 cycle at 50C for 2 min, 1 cycle at 95C for 10 min, and 40 cycles of 95C for 15 sec and 60C for 1 min. Serial 10-fold dilutions, from 1 × 103 to 1 × 1010 copies of the amplicon produced by RT-PCR, were used to establish the standard curve. We used the value of 3′-UTR as a control. Quantitative GAPDH data were normalized and expressed as log based on the 3′ UTR value of fresh rat kidney. Each sample was analyzed in triplicate, and the average value of triplicates was used as a quantitative value.
Histology and Photomicroscopy
Blocks generated in the analysis of fixation time, fixative buffers, and processing time were sectioned at 6 μm and stained as a single batch on an automated hematoxylin and eosin (H&E) autostainer (Leica Microsystems, Inc.; Bannockburn, IL). Slides were digitally photographed on a Zeiss Axioplan 2ie with an AxioCam HR camera, with a Plan Apochromat ×20 objective, NA 0.75 and a ×1.6 Optovar for a total magnification of ×320 (Carl Zeiss, Inc.; Thornwood, NY). Illumination and color balance were identical for all the samples.
Results
Simulated Warm Ischemia Times and Degradation Profile of RNA
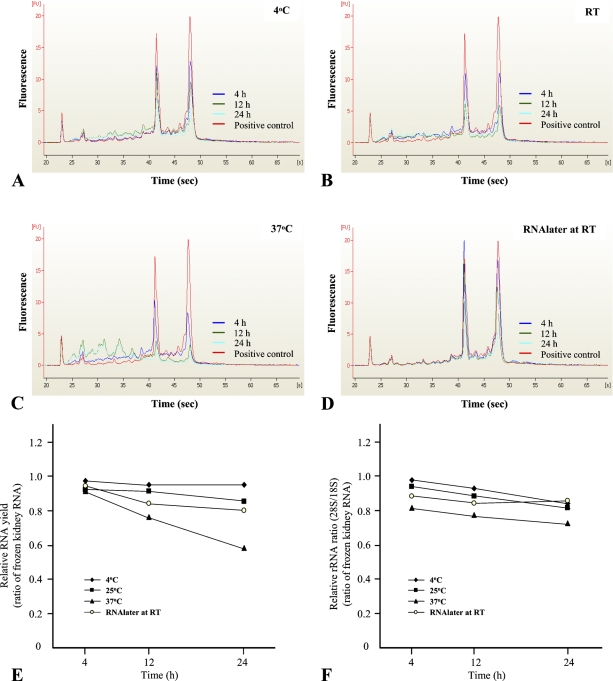
There is significant interest in warm ischemia time and its impact on RNA recovery quantity and quality. To evaluate how storage temperatures and times affect the quantity and quality of isolated RNA before fixation and processing, we analyzed rat kidney RNA extracted from various conditions by spectrophotometer and microcapillary electrophoresis. We used the electropherograms to calculate the ratio of 18S and 28S rRNA as a function of incubation time and temperature. At 4C, the ratio of 28S and 18S rRNA was relatively stable for up to 12 hr, whereas progressive degradation of these two rRNA species was observed after 12 hr of incubation. RNA yield was also stable (95–97%) up to 24-hr incubation (Figures 2A and 2E). With storage at 4C as a reference, the 28S/18S ratio decreased quickly at 25C. The progressive degradation of rRNA was also observed after a 4-hr incubation (Figure 2F). After 24 hr of incubation at 25C, RNA yield was decreased up to 15% compared with the positive control (Figures 2B and 2E). In contrast to results of tissue held at 4C, the representative rRNA pattern had almost completely disappeared after a 12-hr incubation at 37C. The 28S/18S ratio and RNA yield were dramatically decreased in specimens held at 37C (Figures 2C and 2F). Alternatively, RNAlater sustained the stability of RNA up to 24-hr incubation at 25C, but the 28S/18S ratio was marginally decreased compared with the untreated condition (Figures 2D and 2F). RNA yield was also decreased slightly in RNAlater solution compared with the untreated sample, with an ∼5% decrease observed after a 12-hr incubation (Figure 2E).
Figure 2.
The profiling of total RNA extracted from warm ischemia time conditions by microcapillary electrophoresis. We presented electropherograms based on each condition; 4C (A), room temperature (B), 37C (C), and RNAlater at room temperature (D). RNA profiles represented relative RNA yield (E) and relative rRNA ratio (F), respectively. Fresh rat kidney was used as a control, generating an rRNA ratio of 1.42 ± 0.03. Quantities and qualities of RNA recovered are expressed in reference to fresh rat kidney (1.00).
Effect of Fixation Time on RNA Quality
Gene expression profiling in FFPE tissue has been hampered by poor standardization of specimen handling and inefficient RNA extraction methods. To dissect this process, we analyzed RNA quality from each condition by bioanalyzer tracings and QuantiGene (Panomics) assays with the GAPDH and CDK4 genes. The QuantiGene assay uses a branch-DNA technology for direct detection of nucleic acids (Kern et al. 1996; Canales et al. 2006). As formulated for these experiments, total RNA was hybridized directly to the branch-DNA capture probes, with the advantage of no enzyme-mediated activity required for the assay. The dynamic range of QuantiGene assays for GAPDH and CDK4 was determined, and assays were calibrated to within the linear range (Supplemental Figure 1).
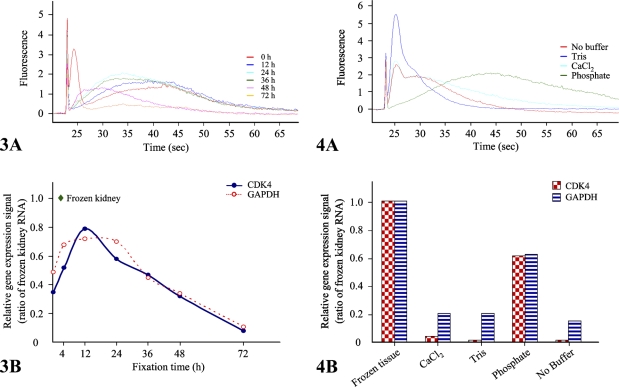
To appreciate the impact of formalin fixation on RNA integrity, we examined the quality of RNA recovered after formalin fixation and paraffin embedding based on a protocol with the only variable being fixation time. Images of H&E histology are shown in Supplemental Figure 2. RNA profiles based on fixation time showed that prolonged fixation results in decreased RNA fragment length after a 48-hr fixation (Figure 3A), supporting the finding that prolonged fixation results in decreased RNA fragment length. These data were confirmed by analysis of GAPDH and CDK4 by QuantiGene assay (Figure 3B), which showed significant differences in detectable RNA dependent only on fixation time. Short times of fixation, 4 hr and less, were also associated with decreased detection of GAPDH and CDK4, which is also reflected in the RNA profile from the bioanalyzer.
Figure 3.
Assessments of RNA profiles according to fixation times. We analyzed RNA quality using the Agilent 2100 bioanalyzer, using 200 ng of total RNA extracted from mouse kidney formalin-fixed, paraffin-embedded (FFPE) tissues after 0, 12, 24, 36, 48, or 72 hr of fixation. Representative data are presented as an electropherogram (A). To measure quantitatively RNA degradation, we performed QuantiGene assays with CDK4 and GAPDH gene-specific probe sets using the QuantiGene reagent system (Panomics). Relative expression signals of both genes are normalized to that of frozen kidney (B). Data are the mean of three independent experiments.
Effect of Different Buffers in Formalin Fixation on RNA Quality
We subsequently examined the effects of different buffering agents used in 10% formalin using the same approach. We examined the differences between phosphate-based buffer, Tris, CaCl2, and no buffer on histology (Supplemental Figure 2) and RNA quality. Although phosphate-buffered formalin is the most common formulation, other chemicals are described and variably encountered in clinical practice (Hewitt et al. in press). As shown in Figure 4, phosphate-buffered formalin showed the best quality of RNA among tested buffers, with ∼60% of that recovered from fresh tissue (Figure 4B). Calcium chloride– and Tris-buffered formalin showed less expressional signal, at 20% and <5% for GAPDH and CDK-4, respectively. Formalin without the addition of a buffering agent performed the worst (Figure 4B).
Figure 4.
RNA quality and gene expression profiles from different fixative buffers. Total RNA was extracted from mouse kidney. Tris-, phosphate-, and CaCl2-buffered formalin and unbuffered formalin-fixed tissues were coupled with paraffin-embedded processing. The total RNA samples (200 ng) were analyzed on an RNA LabChip using the Agilent 2100 bioanalyzer and are shown here as an electropherogram (A). CDK4 and GAPDH gene expressional profiles were measured with 500 and 200 ng of total RNA extracted from those samples, respectively. Relative expressional signal of both genes are normalized to that of frozen kidney (B). Data are the mean of three independent experiments.
Effect of Tissue Processing Time on RNA Quality
The process of converting tissue in an aqueous environment to impregnating in paraffin is termed tissue processing and consists of stepwise replacement of fixative with alcohol (dehydration), the replacement of alcohols with other organic solvents (clearing), and finally impregnation, which is the replacement of the organic solvents with paraffin. This process is typically performed on an automated instrument with between 10 and 14 stations or steps in the process. General parameters that are variable include time, temperature, and the presence of vacuum. There is no standard protocol for the process, with differences in time per step being the most common variable with modern tissue processors. We sought to determine whether tissue processing affected tissue quality. Using tissue processing times obtained from an array of clinical research pathology laboratories, we tested the impact of processor time ranging from 140 to 660 min. Images of the resultant H&E sections are presented in Supplemental Figure 2. As shown by the RNA profiles and the gene-specific measurements, longer tissue processing times resulted in better quality of RNA (Figure 5).
Figure 5.
RNA quality profiles from different tissue processing times. Representative data are presented as an electropherogram (A). Relative expressional signals of CDK4 and GAPDH genes are normalized to that of frozen kidney (B). Frozen mouse kidney tissue RNA was used as a positive control. Data are the mean of three independent experiments.
Optimization of mRNA Targeting by Real-time Quantitative RT-PCR
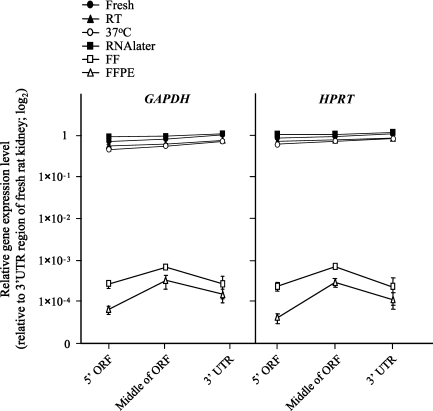
RNA recovered from FFPE tissue is well documented to be short in length; however, little is known about the nature of the residual RNA fragments. To examine this issue, we profiled RNA integrity using real-time quantitative RT-PCR with three specific probes (5′ ORF, middle of ORF, and 3′UTR) for the rat GAPDH gene. The highest expression level was observed in the middle region of the housekeeping gene (Figure 6). Although we performed quantitative real-time RT-PCR with equal amounts of total RNA (2 μg), the expression level of FFPE RNA was very low compared with fresh tissue (∼103–104 fold). To confirm this finding, we performed additional real-time RT-PCR with the HPRT gene using the same strategy with a specific primer and probe set. The expression profile of FFPE RNA was identical to GAPDH (Figure 6). In contrast, the pattern of prefixation samples was slightly decreased toward 5′ upstream (Figure 6). This pattern was identical in all of the RNA samples from each condition, and the highest expression level was shown in the RNAlater-stored specimen at room temperature.
Figure 6.
Gene-target optimization for amplification of GAPDH and HPRT mRNA from fresh, FF, and FFPE tissue RNA isolates. Real-time quantitative RT-PCR was performed in three different target regions [5′ and middle region of open reading frame (ORF) and 3′ untranslational area of target gene] with gene-specific probe sets (see Supplemental Table 1), designed by Primer Express software (Applied Biosystems). The x-axis shows the region that is amplified for the target gene. The y-axis shows expression values for targeted RNA regions (log scale). Fresh rat kidney tissue RNA was used as a high-quality RNA control, and relative expression levels of each sample are normalized to the fresh tissue RNA. Data are from three independent experiments and are expressed as mean ± SD.
Discussion
There is great interest in the use of RNA derived from FFPE tissue in the discovery and validation of biomarkers. The use of these markers is dependent on the quantity and quality of RNA that can be obtained from FFPE tissue. This is a complex challenge that will require improving the collection of tissue, clinical data, and optimized protocols for the isolation of RNA. RNA quantity and quality are closely intertwined. High-quality RNA can be isolated from FFPE tissue. However, there are preanalytical variables that are poorly understood and poorly standardized that impart additional variables in the process (Hewitt et al. in press). The degradation of RNA begins with the removal of oxygenation, and as the cell becomes progressively anoxic, RNA pools are degraded. This is a process that is well appreciated. Slowing the cellular metabolism by cooling the specimen slows the process. RNAlater is able to rapidly enter the cells and protect RNA from degradation.
With fixation in formalin, the same process continues: progressive anoxia and the concomitant degradation of RNA, including hydrolysis of the poly-A tail. Concurrent with this, formalin chemistry results in strand breakage of RNA into short fragments of average length between 100 and 200 nucleotides and protein-RNA cross-links (Godfrey et al. 2000; Cronin et al. 2004; Chung et al. 2006). Damage by this cross-linking is probably predominant in the 5′ and 3′ ends of the RNA, because this is where the signals of translation and degradation are concentrated in mRNA. This model is supported by our RT-PCR data, where the mid-section of the genes are preferentially conserved (Figure 6). Even adjusted for RNA fragment length, efficiency of reverse transcription is markedly reduced and is most likely a function of the damage to nucleosides by cross-linking, predominantly with protein.
It has been well appreciated that fixation time has an impact on RNA quality (Macabeo-Ong et al. 2002). Although shorter fixation times would in theory result in less RNA damage, our data showed that short fixation times result in poorer-quality RNA. We believe the failure to complete fixation leads to further complications downstream in tissue processing, likely related to inadequate dehydration. Formalin is a two-stage fixative, and an element of this process is dehydration by the alcohol phase of the process (Hewitt et al. in press). Fortunately, the differences between 12 and 24 hr of fixation are not overwhelming. Clearly, overfixation, with times in excess of 48 hr, is very detrimental to RNA recovery (Figure 3).
The impact of different buffers in the quality of the RNA recovered has been previously unappreciated. Formalin buffers were originally introduced to remove precipitates and pigments that were common in histological preparations from the formation of formic acid during the fixation process (NCCLS 1999). Numerous buffers are described in the literature (Coolidge and Howard 1979); however, the most commonly used is phosphate-based buffers. Even these buffers exist in multiple commonly encountered formulations (Hewitt et al. in press). Tris-based buffers are rarely used because they have been observed to negatively impact IHC, and subtle differences in buffers can impact histology. The analysis of RNA quality based on processing conditions on the tissue processor has not been previously studied.
The differences in histology as observed with H&E staining (Supplemental Figure 2) are not as pronounced as might be expected, but this is deceiving. Many of the blocks were too brittle to result in 4 or 5-μm sections, and the comparison is limited to 6-μm sections. In particular, the 0-hr formalin fixation time and the 140-min processor time was difficult to section. In general, fixation time and fixative buffers had more direct impact on morphology, whereas processor time had a more noticeable impact on the texture and ability to cut high-quality sections.
The commonly used language is that tissue is processed “per standard protocol.” The protocols are standardized, to a limited extent, within laboratories and routinely categorized as short and long run protocols, which are used for biopsies and excisional specimens, respectively (Hewitt et al. in press). In the histology laboratory setting, the goal is processing and impregnation that results in a tissue that produces a “crisp” histologic appearance (NCCLS 1999). Biopsy specimens are routinely exposed to both protocols, but excisional specimens, because of the volume of the specimen, are excluded from short protocols, because they will not be adequately impregnated, as defined by histological criteria. The diversity of protocols used is extensive and may include differences in time for different steps of dehydration, clearing, and impregnation. Our data suggest that high-quality RNA requires a well-fixed, well-dehydrated, and well-impregnated specimen. In the absence of completion of the fixation process or the dehydration process, residual water is trapped in the tissue, and RNA degrades. It is unclear if the degradation of RNA is purely chemical (hydrolysis) or the action of RNases; however, our data on buffers in formalin suggest it may be chemical. Our experiments showed that far greater attention must be focused on the method of tissue processing and its impact on RNA.
The unanticipated effects of buffers in formalin, as well as time of processing, may account for the poor quality of RNA recovered from archival specimens >10 years old. Although the process of formalin fixation and paraffin has been in use for essentially a century, small elements of the process have been under continuous evolution during this time. During this evolution, the focus has always been histomorphology quality and lack of dramatic change; however, it is clear that the preservation of the biomolecules has changed as the process has evolved.
These experiments were carried out with standardized specimens in an effort to measure the impact of different parameters in tissue handling and processing. In the process, we uncovered additional parameters that require study, including the impact of specimen size and determining which stages of tissue processing mediate the most significant impact. Not addressed are alternative methodologies or reagents that are becoming increasingly common in the histopathology community. Overall, these data suggest that an optimal fixation condition for high-quality RNA could be 16- to 32-hr fixation in phosphate-buffered formalin solution coupled with longer tissue processing (Figure 7).
Figure 7.
The hypothetical basic model of RNA quality depending on FFPE tissue processing. The darker gray color represents conditions associated with better RNA quality from FFPE tissue.
In conclusion, we showed the dramatic effects that preanalytical variables have on the recovery of RNA from FFPE tissue. If robust RNA-based diagnostics are to enter clinical use, true standardized protocols based on hard data are required to balance the demands of histopathology and RNA-based assays. Fortunately, these experiments showed that traditional histology approaches generate high-quality histology and RNA. These experiments also showed that the impact of altering these protocols impacts the biospecimen.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
We acknowledge the technical expertise provided by Kimberly Tuttle and Yvonne Gathright.
References
- Auer H, Lyianarachchi S, Newsom D, Klisovic MI, Marcucci G, Komacker K (2003) Chipping away at the chip bias: RNA degradation in microarray analysis. Nat Genet 35:292–293 [DOI] [PubMed] [Google Scholar]
- Bonin S, Petrera F, Rosai J, Stanta G (2005) DNA and RNA obtained from Bouin's fixed tissues. J Clin Pathol 58:313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, et al. (2006) Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol 24:1115–1122 [DOI] [PubMed] [Google Scholar]
- Chung JY, Braunschweig T, Hewitt SM (2006) Optimization of recovery of RNA from formalin-fixed paraffin-embedded tissue. Diagn Mol Pathol 15:229–236 [DOI] [PubMed] [Google Scholar]
- Coolidge BJ, Howard RM (1979) Animal Histology Procedures (NIH Publication 80–275). Bethesda, National Institutes of Health
- Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, Esteban JM, et al. (2004) Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol 164:35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke J, Fritzen R, Ternes P, Lange W, Dölken G (1993) An improved strategy and a useful housekeeping gene for RNA analysis from formalin-fixed, paraffin-embedded tissue by PCR. Biotechniques 14:448–453 [PubMed] [Google Scholar]
- Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH (2000) Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5′ nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagn 2:84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SM, Lewis F, Cao Y, Conrad RC, Cronin M, Danenberg KD, Goralski TJ, et al. (In Press) Tissue handling and specimen preparation in surgical pathology: issues concerning the recovery of nucleic acids from formalin fixed, paraffin embedded tissue. Arch Pathol Lab Med [DOI] [PubMed]
- Jewell SD, Srinivasan M, McCart LM, Williams N, Grizzle WH, LiVolsi V, MacLennan G, et al. (2002) Analysis of the molecular quality of human tissues: an experience from Cooperative Human Tissue Network. Am J Clin Pathol 118:733–741 [DOI] [PubMed] [Google Scholar]
- Karsten SL, Van Deerlin VM, Sabatti C, Gill LH, Geschwind DH (2002) An evaluation of tyramide signal amplification and archived fixed and frozen tissue in microarray gene expression analysis. Nucleic Acids Res 30:E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin JJ, Sheridan P, et al. (1996) An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 34:3196–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafft AE, Duncan BW, Bijwaard KE, Taubenberger JK, Lichy JH (1997) Optimization of the isolation and amplification of RNA from formalin-fixed, paraffin-embedded tissue: the armed forces institute of pathology experience and literature review. Mol Diagn 2:217–230 [DOI] [PubMed] [Google Scholar]
- Lykidis D, Van Noorden S, Armstrong A, Spencer-Dene B, Li J, Zhuang Z, Stamp GW (2007) Novel zinc-based fixative for high quality DNA, RNA and protein analysis. Nucleic Acids Res 35:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macabeo-Ong M, Ginzinger DG, Dekker N, McMillan A, Regezi JA, Wong DT, Jordan RC (2002) Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analyses. Mod Pathol 15:979–987 [DOI] [PubMed] [Google Scholar]
- Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K (1999) Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res 27:4436–4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCCLS (1999) Quality Assurance for Immunohistochemistry; Approved Guideline (MM4-AC). Wayne, PA, NCCLS
- Penland SK, Keku TO, Torrice C, He X, Krishnamurthy J, Hoadley KA, Woosley JT, et al. (2007) RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab Invest 87:383–391 [DOI] [PubMed] [Google Scholar]
- Rupp GM, Locker J (1988) Purification and analysis of RNA from paraffin embedded tissue. Biotechniques 6:56–60 [PubMed] [Google Scholar]
- Sato Y, Mukai K, Furuya S, Shimosato Y (1991) The AMeX method: a multipurpose tissue-processing and paraffin-embedding method. III. Extraction and purification of RNA and application to slot-blot hybridization analysis. J Pathol 163:81–85 [DOI] [PubMed] [Google Scholar]
- Shibutani M, Uneyama C, Miyazaki K, Toyoda K, Hirose M (2000) Methacarn fixation: a novel tool for analysis of gene expressions in paraffin-embedded tissue specimens. Lab Invest 80:199–208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.