Abstract
Here, we describe the cloning and characterization of RETINOBLASTOMA-RELATED PROTEIN1 (RBR1) from the green alga Volvox carteri. RBR1 expression increases substantially during embryogenesis and in response to the sex-inducer glycoprotein, but it decreases significantly under heat stress. While RBR1 is expressed in gonidia (asexual reproductive cells) and embryos, the largest proportion of RBR1 mRNA is found in parental somatic cells. The presence of 4 splice variants and 15 potential cyclin-dependent kinase phosphorylation sites suggests that RBR1 is subject to control at the posttranscriptional and posttranslational levels. Surprisingly, RBR1 is a gender-specific gene, mapping exclusively to the female mating-type locus. A procedure for stable nuclear transformation of males was established to generate RBR1-expressing males. These transformants exhibit enlarged reproductive cells, altered growth characteristics, and a prolonged embryogenesis. The results suggest that a functionally related analog of RBR1 exists in males. The reason for the divergent evolution of RBRs in females and males appears to be based on sexual development: males and females respond to the same sex-inducer with different cleavage programs and substantial differences in cellular differentiation. Thus, the gender-specific presence of RBR1 provides evidence for an additional, novel role for retinoblastoma family proteins in sexual development.
INTRODUCTION
The human retinoblastoma (RB) protein is the prototype tumor suppressor and has emerged as a key regulator of cell cycle entry; the loss of function of its gene is responsible for predisposition to several malignancies, including retinoblastoma, the cancer of the retina (Friend et al., 1986; Weinberg, 1995; Sherr, 1996; Classon and Harlow, 2002). In animals, RB-related proteins (RBRs) have been demonstrated to be involved not only in the regulation of cell division but also in tumor suppression, DNA repair, DNA damage checkpoint control, differentiation, cellular senescence, and apoptosis (Weinberg, 1995; Lundberg and Weinberg, 1999; Harbour and Dean, 2000; Stevaux and Dyson, 2002; Ben-Porath and Weinberg, 2005). In many mammalian cells, RBRs have been shown to be suppressors that negatively control the cell cycle and thereby play a crucial role in control of the G1- to S-phase transition (Goodrich et al., 1991; Weinberg, 1995; Bartek et al., 1996; Hanahan and Weinberg, 2000; Du and Pogoriler, 2006; Wikenheiser-Brokamp, 2006; Dick, 2007). This control is accomplished in part by the ability of RB family proteins to bind and suppress key transcription factors like E2F, which modulates the successive expression of a battery of genes required for replication initiation, synthesis of G1-/S-phase–specific cyclins, checkpoint control, activation of cyclin-dependent kinases (CDKs), DNA synthesis, and chromosome duplication (Dyson, 1998; Korenjak and Brehm, 2005). E2Fs need to partner with an E2F dimerization partner1 (DP1) subunit to stably interact with RB family proteins and to bind to E2F promoter elements (Chellappan et al., 1991; Helin et al., 1993; Dyson, 1998; Zheng et al., 1999; Stevaux and Dyson, 2002). Genes containing E2F binding sites in their promoter include the v-MYC myelocytomatosis viral oncogene homologous transcription factors c-MYC and N-MYC, the v-MYB myeloblastosis viral oncogene homologous transcription factor c-MYB, thymidine kinase, dihydrofolate reductase, DNA polymerase, cell division cycle2 (CDC2), and RB. The expression of E2F target genes is refined during cell cycle progression due to the acquisition of promoter-specific histone modifications: E2F-associated coactivators create activating histone marks, whereas the recruitment of corepressors associated with E2Fs and RBRs leads to the accumulation of inhibitory histone modifications that affect chromatin compaction. In the latter case, RBRs have been shown to play critical roles in the induction of terminal differentiation and senescence, characterized by irreversible cell cycle arrest (Narita et al., 2003; Ben-Porath and Weinberg, 2005; Blais and Dynlacht, 2007). The activity of RBR proteins is regulated by posttranslational mechanisms, primarily phosphorylation, since transcription factors like E2F bind only the hypophosphorylated form of RB. Phosphorylation of RB is directed by CDKs, including complexes of G1-phase cyclinD/cdk4, G1-/S-phase cyclinE/cdk2, and cyclinA/cdk2 (Hinds et al., 1992; Ewen et al., 1993; Sherr, 2000; Du and Pogoriler, 2006). By virtue of all of the interactions just discussed, RBRs function as central hub proteins in the complex process of cell cycle control, including cell cycle arrest, which requires balancing the activity of many regulators.
RBRs are not restricted to the animal kingdom and seem to play a conserved role in allowing multicellular organisms to develop complex body plans consisting of many different cell types. As in animals, RBRs are involved in controlling entry into S-phase in higher plants, and they have been identified in several dicot and monocot plants (Gutiérrez, 1998; de Jager and Murray, 1999; Lendvai et al., 2007). Despite the fact that key molecular mechanisms of the RB pathway are conserved in plants, some details of their molecular interactions and responses are plant-specific (Mironov et al., 1999; Wildwater et al., 2005). RBRs and components of the RB pathway are not present in yeast or other fungi (Fang et al., 2006), which use unrelated proteins to control the same processes, but they have been identified in the unicellular algae Chlamydomonas reinhardtii (Armbrust et al., 1995; Umen and Goodenough, 2001), Chlamydomonas incerta (GenBank accession number AY781411), and Ostreococcus tauri (Robbens et al., 2005) as well as in the moss Physcomitrella patens (GenBank accession number AJ428952). These findings suggest that the RB pathway has an ancient origin and that fungi have lost it for some reason. It seems to have arisen only once in the evolution of all higher eukaryotes and then to have evolved independently to meet the requirements of the different groups of organisms (Claudio et al., 2002).
Because knowledge of the impact of RBRs is incomplete, particularly in photosynthetic organisms, and in order to learn more about basic, ancient features of the RB pathway in the context of the evolution of multicellularity and cellular differentiation, we sought to characterize an RBR from the multicellular green alga Volvox carteri. This alga represents one of the simplest multicellular organisms, because it is composed of only two cell types, somatic (body) cells and germ (reproductive) cells. V. carteri is capable of both asexual and sexual reproduction. In the asexual mode of reproduction, each adult male and female is a spheroid containing ∼2000 to 4000 biflagellate somatic cells and ∼16 asexual reproductive cells called gonidia; strikingly, the volume of a single gonidium is up to ∼500 times larger than that of a somatic cell. In the sexual mode of reproduction, adult, sexual females contain ∼32 large eggs and ∼2000 to 4000 small somatic cells. By contrast, adult, sexual males have only ∼128 somatic cells and ∼128 sperm packets containing 64 or 128 sperm each (Starr, 1969, 1970; Hallmann et al., 1998; Kirk, 1998). V. carteri is known as an ideal model system for the molecular analysis of basic principles in developmental processes related to cell division, multicellularity, and cellular differentiation (Kirk, 1998, 2000).
Another advantage of RBR characterization in the multicellular alga Volvox is that a RB-related gene, called MATING-TYPE LINKED3 (MAT3), has already been characterized in its unicellular relative C. reinhardtii (Gillham et al., 1987; Armbrust et al., 1995; Umen and Goodenough, 2001). This would allow for comparisons to be made. Null mutations in the MAT3 gene cause supernumerary cell divisions and small cells, suggesting a role for MAT3 in cell size control of C. reinhardtii (Armbrust et al., 1995; Umen and Goodenough, 2001; Fang et al., 2006). Moreover, E2F1/DP1 was shown to act as a positive regulator downstream of MAT3 (Fang et al., 2006). In C. reinhardtii, other key components of the RB pathway have been revealed by genome-wide annotation and expression profiling (Bisova et al., 2005), and a suppressor screen identified novel components of the RB pathway (Fang and Umen, 2008).
Multicellular Volvox evolved from a unicellular Chlamydomonas-like ancestor (Rausch et al., 1989; Kirk, 1998), but what effect volvocine cell differentiation had on the RB pathway is unclear. Since multicellularity evolved in volvocine algae much more recently than it did in the red algae, brown algae, plants, fungi, or animals (Rausch et al., 1989; Sogin, 1991; Wainright et al., 1993), it is thought that the evolutionary pathway leading to volvocine multicellularity may be much less obscured by the passage of time than the pathways leading to the other multicellular groups. The evolution of multicellularity also involved a change in the mode of sexual reproduction from isogamy (as found, for example, in C. reinhardtii), which involves gametes of the same size, to oogamy (as found, for example, in V. carteri), in which the nonmotile female gamete (egg) is significantly larger than the motile male gamete (sperm). In V. carteri, sexual development of both males and females is triggered by a sex-inducer glycoprotein at concentrations as low as 10−16 M (Starr, 1970; Starr and Jaenicke, 1974; Gilles et al., 1984; Tschochner et al., 1987). The consequences of the evolutionary switch to oogamy for the RB pathway are thus far unknown.
Here, we present the cloning and characterization of a RB-related gene, RBR1, from V. carteri. RBR1 mRNA, which exists as four splice variants, is shown to be strongly expressed during embryogenesis of wild-type females and even more strongly in response to the Volvox sex-inducer glycoprotein. Expression decreases significantly under heat stress conditions. RBR1 is expressed in gonidia, embryos, and parent somatic cells; however, parent somatic cells account for the main fraction of the overall expression level of RBR1. Unexpectedly, the RBR1 gene is not present in the genome of wild-type males, but only in females, where it is localized at the mating-type locus. We established a procedure for the stable nuclear transformation of Volvox males and used it to produce transgenic males expressing the female-derived RBR1 gene. The transgenic males show significant differences in growth characteristics compared with wild-type males. These results provide evidence for an additional role for RB family proteins.
RESULTS
The RBR1 cDNA and Gene from V. carteri
Among the mRNAs examined in a large-scale analysis of cell type–specific gene expression in V. carteri (Nematollahi et al., 2006) was an mRNA with sequence similarity to the human RB gene (RB1), which we named RBR1. A 128-bp RBR1 cDNA fragment from this earlier analysis, together with sequence information from Volvox whole-genome shotgun reads at the Joint Genome Institute (JGI) C. reinhardtii genome portal 3.0 (Department of Energy [DOE]; http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), was used to obtain the complete coding sequence of RBR1 (splice variant 1; see below). Successive RT-PCR amplifications were performed using total RNA from wild-type females as a template to yield four overlapping cDNA fragments (see Supplemental Figure 1 online). An additional RBR1 cDNA fragment was identified in an EST library (clone CBOX16325). Clone CBOX16325 was obtained from the DOE JGI. After sequencing of all fragments, the complete RBR1 cDNA (variant 1) was determined to be 3656 bp in length [excluding a poly(A) tail] and contained a 3321-bp open reading frame.
In some of our RT-PCR amplifications, we obtained fragments of unexpected lengths. Sequencing of these fragments showed that RBR1 is subject to alternative splicing. After this finding, we tested a variety of RBR1-specific primers and primer combinations in RT-PCR amplifications. Four different RBR1 splice variants were identified (Figure 1A). Splice variant 1 is the one described above. Splice variant 2 is caused by an alternative 3′ splice site of intron 13, which is located 110 bp downstream of the one used in variant 1. Splice variant 3 is caused by retention of introns 2 and 3 compared with variant 1. Finally, variant 4 is the result of retention of intron 3 compared with variant 1.
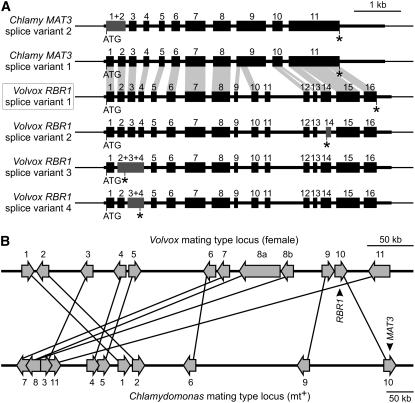
Figure 1.
The Volvox RBR1 Gene: Exon–Intron Structure, Identified Splice Variants, Comparison with the Related Chlamydomonas MAT3 Gene, and Localization within the Female Genome.
(A) Physical map of exon and intron segments in the Volvox RBR1 gene; the Chlamydomonas MAT3 gene is shown for comparison. Four splice variants have been identified for Volvox RBR1 and two are known for Chlamydomonas MAT3 (see Chlre2FinalModels BLAT and Chlre3ACEGS BLAT results for MAT3 at the JGI Chlamydomonas genome portal 3.0; http://genome.jgi-psf.org/Chlre3/Chlre3.home.html). Sequence segments that are similar between Volvox RBR1 (splice variant 1) and Chlamydomonas MAT3 (splice variant 1) are indicated by light gray areas, which connect similar exons or exon fragments. The name of the main splice variant of RBR1, variant 1, is boxed. Exons that are the result of alternative splice events in variants 2 to 4 are highlighted in dark gray. ATG, translation start site; asterisks indicate translation stop sites; thick horizontal bars indicate exons; numbers above exons indicate numbering of exons; thinner horizontal bars represent introns and 5′ and 3′ untranslated regions; and thinnest horizontal bars represent upstream and downstream sequences.
(B) Localization of the Volvox RBR1 gene within the genome of female V. carteri. Comparison of part of the C. reinhardtii mating-type locus (mt+) (scaffold 3/linkage group VI) (Ferris et al., 2002; Merchant et al., 2007) with a genomic fragment of the V. carteri genome (scaffold 43) that contains the RBR1 gene. Putative orthologous genes in this region encode (as numbered): 1, spermine synthase; 2, protein of unknown function; 3, predicted Na+-dependent cotransporter; 4, AMP-activated protein kinase; 5, N-myristoyl transferase; 6, splicing factor 3a, subunit 2; 7, vacuolar ATP synthase, subunit C; 8, cytoplasmic dynein 1b heavy chain; 9, Hyp-rich glycoprotein; 10, retinoblastoma-related protein RBR1/MAT3; 11, predicted sugar kinase. Putative orthologous genes are connected by lines. The sequence comparison of these genes is shown in Supplemental Table 1 online.
In addition to the cDNA variants, the gene of RBR1 was cloned by PCR using genomic DNA of Volvox females as a template. Two RBR1 fragments were produced and sequenced. The fragments show a 185-bp overlap. A unique HpaI restriction site within this overlap was used to join the fragments to give a genomic fragment of 8186 bp, which contains the complete RBR1 gene including upstream and downstream sequences; the sequence between the start ATG and the stop codon is 5744 bp long. The comparison of cDNA and genomic sequences revealed 16 exons and 15 introns within the RBR1 gene (Figure 1A, splice variant 1). The introns have the following lengths (in bp): 69, 66, 64, 146, 206, 198, 129, 84, 286, 134, 702, 73, 74, 107, and 82. Thus, there are seven introns with sizes below 90 bp. Intron 11 is longer than 700 bp. In contrast with the other introns, the 5′ splice site of intron 15 (GC…) differs from the 5′ consensus sequence (GT…); therefore, intron 15 has a nonconsensus splice site.
We intended to determine the position of the RBR1 gene within the Volvox genome. There is a Volvox genome project in progress and genomic sequences have become available (DOE JGI Volvox genome portal 1.0; http://genome.jgi-psf.org/Volca1/Volca1.home.html), but so far the genome is still incomplete and there are no molecular markers that are linked to a specific location on the genome. Fortunately, the genome of its unicellular relative C. reinhardtii has been completed (Merchant et al., 2007) and much is known about its characteristics. Comparison of the genes on a small genomic fragment of the V. carteri genome (scaffold 43), which contains the RBR1 gene, with the complete C. reinhardtii genomic sequence revealed several putative orthologs (see Supplemental Table 1 online), which all map to a distinctive locus: the mating-type locus of Chlamydomonas (Figure 1B). This genomic region of C. reinhardtii also includes the putative ortholog of RBR1, the RB-like gene MAT3 (Gillham et al., 1987; Armbrust et al., 1995; Ferris et al., 2002; Merchant et al., 2007). The synteny is substantial, even if the microcollinearity is disrupted by numerous local rearrangements (Figure 1B). Because shared synteny is a reliable criterion for the orthology of genomic regions in different species (King, 2002; Tang et al., 2008), the comparison discloses that RBR1 is localized at the mating-type locus of the female Volvox.
The Deduced Amino Acid Sequence of the RBR1 Protein
The open reading frame of the cDNA for splice variant 1 encodes a RBR1 polypeptide of 1107 amino acids (Figure 2A). RBR1 (variant 1) displays a domain structure that is characteristic of RB-related proteins (Figure 2B). Sequence similarity to RB-related proteins is not restricted to the so-called A and B domains (Figure 3A) but also includes significant similarity at the N terminus, especially in three areas named N1 to N3 (Figure 2B). These conserved regions at the N terminus may contribute to the subcellular localization of RB family proteins (Durfee et al., 2000). The conserved A and B domains are separated by a spacer region, as in other RB proteins (Lee et al., 1998; Claudio et al., 2002). In human RB proteins, the A and B domains were shown to form a pocket domain, which is responsible for interacting with transcription factors, cyclins, and CDKs, as well as for the activity of the RB protein (Lee et al., 1998; Claudio et al., 2002). The amino acid residues at the A-B contact site, also called the A-B interface, are conserved in RBR1. The A-B contact site is localized at the end of the A domain and at the beginning of the B domain (Figures 2A and 2B). The conserved LxCxE binding site in the second half of the B domain is central to the cellular function of RB proteins in humans; this site is known to interact with a range of proteins containing the RB binding (LxCxE) motif (Lee et al., 1998; Dahiya et al., 2000).
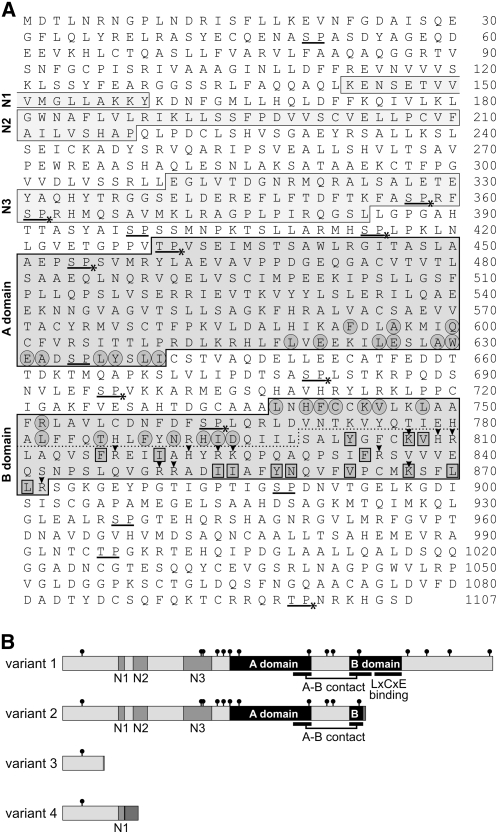
Figure 2.
Amino Acid Sequence of RBR1 (Variant 1) and Schematic Representation of the Four Variants.
(A) Deduced amino acid sequence of the RBR1 protein (variant 1). Boxes indicate conserved A and B domains as well as conserved N-terminal sequence areas (N1 to N3); the dotted line indicates the putative LxCxE binding site within the second half of the B domain; circled amino acid residues are residues that are known to form contacts between the A and B domains in human RB1 and that build the A-B interface (Lee et al., 1998); amino acid residues within rectangles are conserved residues that were shown to be involved in binding of human RB1 to a peptide containing the LxCxE motif (Lee et al., 1998); underlined letters indicate potential CDK phosphorylation sites (S/TP); asterisks indicate potential CDK phosphorylation sites that are conserved between Chlamydomonas MAT3 (Umen and Goodenough, 2001) and Volvox RBR1; arrowheads indicate positively charged amino acid residues within the LxCxE binding site of the B domain.
(B) Schematic representation of the four variants of RBR1 caused by alternative splicing. Black boxes, conserved A and B domains; gray boxes, conserved N-terminal sequence areas (N1 to N3); small black circles, potential CDK phosphorylation sites (S/TP). The A-B contact site and the LxCxE binding site are indicated. Dark gray boxes at the outermost C-terminal end of variants 2, 3, and 4 represent amino acid sequences that differ from variant 1 (variant 2, after residue 773 there is the sequence AGPGFIS; variant 3, after residue 106 there is K; variant 4, after residue 160 there is VSSVVLSSPRLRPAIFRVNLAGQFRHASASVRLL).
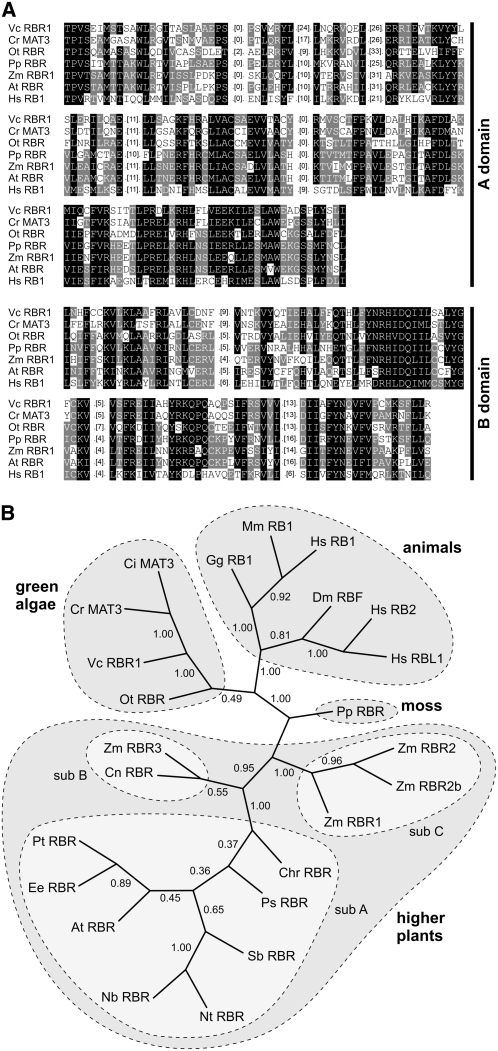
Figure 3.
Protein Sequence Alignment of A and B Domains of RBR1 with Six RB-Related Proteins and Sequence Relationship among 24 RB-Related Proteins.
(A) Protein sequence alignment of A and B domains of RBR1 with six RB-related proteins from green algae, moss, higher plants, and animals. Conserved amino acid residues were shaded using GeneDoc 2.6 (Nicholas et al., 1997) with similarity groups enabled. White letters on black background, conserved in >80% of the sequences at the corresponding position; white letters on gray background, conserved in >40% of the sequences at the corresponding position. For clarity, less-related areas were removed and only the number of removed amino acid residues is given in parentheses. Source organisms are as follows: Vc RBR1, Volvox carteri; Cr MAT3, Chlamydomonas reinhardtii; Ot RBR, Ostreococcus tauri; Pp RBR, Physcomitrella patens; Zm RBR1, Zea mays; At RBR, Arabidopsis thaliana; Hs RB1 (RB1, RB), Homo sapiens.
(B) Relationships among 24 RB-related proteins from green algae, moss, higher plants, and animals. The unrooted tree was calculated using the neighbor-joining method (Saitou and Nei, 1987) of PHYLIP (see Methods). Numbers indicate bootstrap analysis values obtained using 10,000 resampled data sets. The analysis is based on the trimmed alignment given in Supplemental Figure 2 and Supplemental Data Set 1 online. Subfamilies of RB-related proteins within the higher plants are highlighted (Lendvai et al., 2007): subfamily A (sub A) contains dicot RB-related proteins, subfamilies B (sub B) and C (sub C) comprise the monocot RB-related proteins. Source organisms are as follows: Vc RBR1, Volvox carteri; Ci MAT3, Chlamydomonas incerta; Cr MAT3, Chlamydomonas reinhardtii; Ot RBR, Ostreococcus tauri; Pp RBR, Physcomitrella patens; Gg RB1, Gallus gallus; Mm RB1, Mus musculus; Hs RB1 (RB1, RB), Homo sapiens; Hs RB2 (p130), H. sapiens; Hs RBL1 (p107), H. sapiens; Dm RBF, Drosophila melanogaster; Pt RBR, Populus tremula × Populus tremuloides; Ee RBR, Euphorbia esula; At RBR, Arabidopsis thaliana; Nb RBR, Nicotiana benthamiana; Nt RBR, Nicotiana tabacum; Sb RBR, Scutellaria baicalensis; Ps RBR, Pisum sativum; Chr RBR, Chenopodium rubrum; Zm RBR1, Zea mays; Zm RBR2, Z. mays; Zm RBR2b, Z. mays; Zm RBR3, Z. mays; Cn RBR, Cocos nucifera.
Human RB proteins are phosphorylated, and phosphorylation influences their interaction with other proteins (Taya, 1997; Brown and Gallie, 2002). Phosphorylation occurs mainly near, but not within, domains A and B (Taya, 1997; Brown and Gallie, 2002). The Volvox RBR1 sequence reveals 15 potential CDK phosphorylation sites, most of which are located near, but not within, the A and B domains (Figures 2A and 2B). Therefore, the rough localization of (potential) phosphorylation sites is similar in both Volvox RBR1 and human RB proteins. There are even three full CDK consensus phosphorylation sites (Ubersax et al., 2003) in RBR1 (S696, T996, and T1099). CDK phosphorylation sites are often clustered (Moses et al., 2007), and a loose cluster with five such sites is localized in front of the A domain. Nine of the 15 potential phosphorylation sites are conserved between Chlamydomonas MAT3 and RBR1 (Figure 2A), including two full CDK consensus phosphorylation sites and four of the five clustered sites. Transcriptional repression by phosphorylation of RB proteins is dependent on the presence of positively charged amino acid residues within the LxCxE binding site (Brown and Gallie, 2002). Human RB1 contains 11 positively charged residues (eight Ks, two Rs, and one H) in the LxCxE binding site (Brown and Gallie, 2002). Similarly, there are 12 positively charged residues (three Ks, seven Rs, and two Hs) in the putative LxCxE binding site of RBR1 (Figure 2A).
The main difference between RBR1 splice variants 2 to 4 and variant 1 is that variants 2 to 4 result in a shortened RBR1 polypeptide (Figure 2B). The expected molecular masses of the polypeptides encoded by splice variants 1 to 4 are 122, 85, 12, and 21 kD, respectively. The protein product of splice variant 2 includes the A-B interface with a shortened version of the B contact but lacks the LxCxE binding site, which is known to be required for interaction with proteins containing the RB binding (LxCxE) motif and for transcriptional repression by phosphorylation (Dahiya et al., 2000). Therefore, the protein encoded by variant 2 is probably not subject to regulation or repression, in contrast with that of variant 1. Variant 4 includes the N1 region and an 34 additional amino acids at the C terminus of unknown function (Figure 2B).
Phylogenetic Analysis of RBR1
A phylogenetic classification of RBR1 was performed based on trimmed alignments of 24 RB-related proteins including RBR1 (see Supplemental Figure 2 and Supplemental Data Set 1 online), which also includes the exemplary alignments of A and B domain sequences given in Figure 3A. Figure 3B shows an unrooted tree of RB-related proteins calculated using the neighbor-joining method (Saitou and Nei, 1987). In this tree, RBR1 is located within a branch of RB-related proteins derived from green algae. This algal branch is somewhat more closely related to animal RB-related proteins than to those from higher plants. The phylogenetic position of RBR1 is consistent with the phylogenetic position of the Volvox species (Kirk, 2003).
The closest relative of RBR1 is MAT3 from the unicellular C. reinhardtii (Gillham et al., 1987; Armbrust et al., 1995; Umen and Goodenough, 2001). In a BLASTn (nucleotide) search of the National Center for Biotechnology Information databases, the two coding sequences, 3.3 kb (RBR1) and 3.6 kb (MAT3) in length, showed 66% identity in a 962-bp overlap (4% gaps) and similar identities in other, smaller overlaps (see Supplemental Figure 3 online). BLASTp (protein) search revealed 59% identity and 73% similarity between the proteins in a 639–amino acid overlap and a similar match in other, smaller overlaps (see Supplemental Figure 4 online); the polypeptides are 1107 amino acids (RBR1) and 1209 amino acids (MAT3) in length. Despite weak similarities in some sequence regions, both genes can be aligned, and they show a similar intron–exon pattern (Figure 1A). However, RBR1 has 16 exons, whereas MAT3 has only 11. Of the five additional exons in Volvox RBR1, the analogous sequences of four exons can be identified in the MAT3 sequence: the 5′ end of exon 9 in MAT3 corresponds to exons 9 and 10 in RBR1; exon 10 in MAT3 corresponds to exons 12 and 13 in RBR1; and exon 11 in MAT3 corresponds to exons 14, 15, and 16 in RBR1. The 3′ end of exon 9 in MAT3 and of exon 11 in RBR1 cannot be aligned due to the lack of any similarities. Likewise, none of the intron sequences of RBR1 show any significant similarities to the genomic sequence of MAT3.
Like RBR1, MAT3 exhibits splice variants (Figure 1A) (see Chlre2FinalModels BLAT and Chlre3ACEGS BLAT results for MAT3 at the JGI Chlamydomonas genome portal 3.0; http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), but there are only two variants in comparison with the four variants in RBR1. The splice variants of MAT3 result from an optional retention of intron 1, which is not observed in any of the RBR1 variants.
Expression Characteristics of RBR1 in Females
To investigate RBR1 expression at specific developmental stages, Volvox wild-type females were grown synchronously in an 8-h-dark/16-h-light cycle. Under these conditions, the life cycle takes exactly 48 h. Males and females exhibit similar cleavage patterns in the asexual phase, but this changes if the sex-inducer is present, which makes them switch to a gender-specific cleavage program that results in the production of sperm or eggs (Figure 4A) (Starr, 1970).
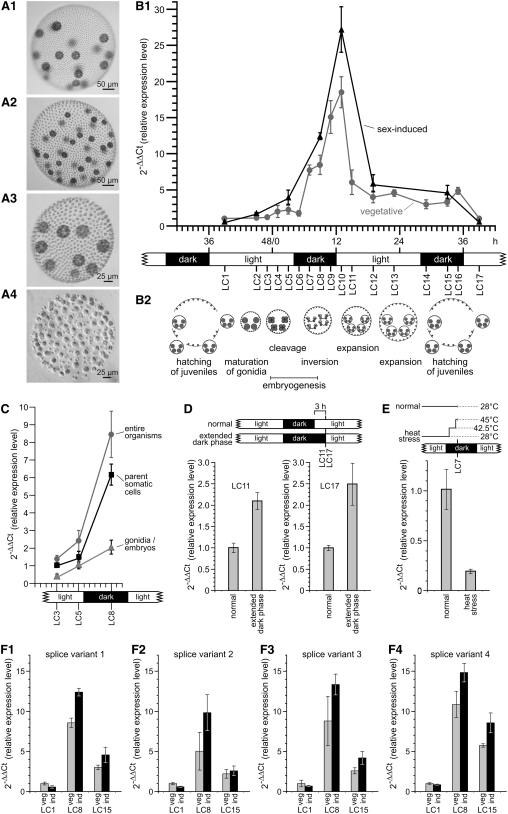
Figure 4.
Expression Characteristics of RBR1 in V. carteri Females.
(A) Phenotypes of vegetative and sexually induced V. carteri females and males. Sexual spheroids were photographed 48 h after sex induction.
(A1) Asexual female containing 18 gonidia.
(A2) Sexual female containing 36 eggs.
(A3) Asexual male containing nine gonidia.
(A4) Sexual male containing ∼128 sperm packets.
(B) to (F) Error bars refer to the sd of the y value. All real-time RT-PCR experiments were performed in triplicate from technical repeats.
(B) Expression kinetics of RBR1 splice variant 1 in females under vegetative conditions and after the addition of sex-inducer.
(B1) Expression of splice variant 1 was analyzed at numerous time points (LC1 to LC17) throughout the life cycle of the female wild-type strain Eve10. Analysis was under vegetative conditions (vegetative) and after addition of the sex-inducer (sex-induced). Under the standard 8-h-dark/16-h-light cycle conditions, the generation time of V. carteri is exactly 48 h; the first cleavage division of the gonidium is defined as 0 h. LC1 of vegetatively grown females was used as the reference point (=1) for calculations of the relative expression level.
(B2) A linear representation of all stages in development is shown in relation to the dark/light cycle.
(C) Expression analysis of RBR1 splice variant 1 in somatic cells of parent spheroids versus their gonidia/embryos at three time points (LC3, LC5, and LC8); expression in entire organisms is shown for comparison. LC3 of parent somatic cells was used as the reference point (=1) for calculations of the relative expression level.
(D) Expression of RBR1 splice variant 1 was analyzed after extension of the dark phase by 3 h and compared with that in organisms grown under normal dark/light conditions; analysis was performed at the time points LC11 and LC17. At both LC11 and LC17, normally grown organisms were used as the reference point (=1) for calculations of the relative expression level.
(E) Expression of RBR1 splice variant 1 in response to heat stress conditions. The culture temperature was elevated from 28 to 42.5°C for 100 min and then to 45°C for 20 min; analysis was performed at the time point LC7. LC7 of females grown at normal growth temperature (28°C) was used as the reference point (=1) for calculations of the relative expression level.
(F) Comparison of the expression levels of the four splice variants of RBR1 at three points in the life cycle of females under vegetative conditions (veg) and after addition of sex-inducer (ind). LC1 of vegetatively grown females was used as the reference point (=1) for calculations of the relative expression level. Expression was analyzed at three different developmental stages in the life cycle of the female wild-type strain Eve10 (LC1, LC8, and LC15). Analysis was performed under vegetative conditions (veg) and after addition of sex inducer (ind).
(F1) Splice variant 1.
(F2) Splice variant 2.
(F3) Splice variant 3.
(F4) Splice variant 4.
Total RNA was isolated at multiple time points throughout the life cycle of vegetative females: after hatching of juveniles (LC1); shortly before the first cleavage division (LC2 and LC3), which defines the beginning of embryogenesis; directly after the first cleavage division (LC4); during the embryonic cleavage divisions (LC5); at the end of cleavage divisions (LC6); during (LC7) and immediately after (LC8) inversion, a morphogenetic process at the end of embryogenesis in which embryos turn inside out; at the beginning of extracellular matrix (ECM) biosynthesis (LC9 and LC10), which starts immediately after inversion; during expansion of the spheroid by ECM biosynthesis (LC11 to LC14); immediately before (LC15) and directly after (LC16) hatching of juveniles; and finally, 5 h after hatching (LC17), which corresponds to the first time point (LC1). The expression levels of Volvox RBR1 (splice variant 1) at the different developmental stages were analyzed by quantitative real-time RT-PCR and calculated using the 2−ΔΔCt method (see Methods) (Bustin, 2000; Pfaffl, 2001), and the results are shown in Figure 4B. RBR1 expression is quite low after hatching of juveniles (LC1) and remains low before cleavage divisions begin (LC2 and LC3). RBR1 expression increases during cell cleavages (LC4 to LC6) and inversion (LC7). At the end of embryogenesis (LC8), which is within the dark phase of the dark/light cycle, RBR1 expression reaches a value that is >700% above the initial one. At LC8, expression of RBR1 is approximately threefold higher than the expression of actin, which indicates that RBR1 mRNA is abundant at this stage. After embryogenesis and at the beginning of ECM biosynthesis, expression increases even further for ∼3 h (LC9 and LC10). Then, RBR1 expression decreases quickly, and this decrease persists as ECM expansion continues (LC11 to LC15); finally, the juveniles hatch (LC16), which correlates with a minimal increase in RBR1 expression.
In an earlier large-scale analysis, RBR1 was shown to be expressed in both cell types, but predominantly in somatic cells (Nematollahi et al., 2006). In this earlier study, the developmental stage LC3 was investigated, but RBR1 expression is quite low at LC3 compared with other stages (Figure 4B). Therefore, we investigated the expression of RBR1 in gonidia/embryos and parent somatic cells during embryogenesis, when expression is higher. Analysis of RBR1 transcripts by quantitative real-time RT-PCR was done shortly before the first cleavage division (LC3), during the embryonic cleavage divisions (LC5), and after the completion of cleavage divisions and inversion (LC8). From LC3 to LC8, RBR1 expression increases in both gonidia/embryos and parent somatic cells (Figure 4C). At all three stages, RBR1 expression is higher in parent somatic cells than in gonidia/embryos.
As mentioned above, expression of RBR1 peaks after the completion of embryogenesis (LC10), which is soon after the end of the dark phase (Figure 4B); rather early within the following light phase, RBR1 expression decreases quickly. To investigate the influence of the dark/light cycle on RBR1 expression, we tested the effects of artificially extending the dark phases. After an extended dark phase, RBR1 expression at LC11 is about twice as high as that measured under normal conditions (Figure 4D) (i.e., under normal conditions, expression decreases by ∼67% between LC10 and LC11 [Figure 4B], whereas after an extended dark phase, it decreases by only ∼32%). The same is true when the experiment is repeated in the following dark phase after hatching at LC17 (Figure 4D). Therefore, it can be concluded that an extended dark phase does not prevent RBR1 expression from decreasing after LC10 and LC16, but it does reduce the amount by which it is decreased.
Sexual development of V. carteri is triggered by a sex-inducer molecule (also called sex-inducing pheromone), a 32-kD glycoprotein (Starr, 1970; Starr and Jaenicke, 1974; Tschochner et al., 1987; Mages et al., 1988). This molecule is one of the most potent biological effector molecules known, triggering sexual development of males and females at concentrations as low as 10−16 M (Starr, 1970; Gilles et al., 1984). It has been shown that both sexes are able to make and release the sex-inducer molecule in a heat stress situation (Kirk and Kirk, 1986). To investigate whether RBR1 expression is influenced by such a heat stress situation, the culture temperature was elevated from 28 to 42.5°C for 100 min and then to 45°C for 20 min, as described previously (Kirk and Kirk, 1986). This heat stress treatment caused RBR1 expression to decline dramatically to ∼20% of the normal expression level (Figure 4E). To analyze RBR1 expression once the sex-inducer is present and sexual development is initiated, the sex-inducer itself was added to a vegetatively grown female population shortly before hatching of juveniles, 2 h before stage LC1. The sex-inducer must be added at this point because, despite its extreme potency, an exposure time of at least 6 h is required before uncleaved gonidia become committed to the sexual cleavage program (Gilles et al., 1984). Quantitative real-time RT-PCR analysis after addition of the sex-inducer was performed as described above for the vegetative development (Figure 4B). The first value (LC1) in the analysis of vegetative females was used as the reference point. Throughout the life cycle, the expression kinetics of RBR1 (splice variant 1; see below) in sex-induced females was similar to that seen in vegetative females; that is, expression was low before cell division (LC1 and LC2), increased over the course of embryogenesis (LC5 to LC8), reached the highest value after embryogenesis at the beginning of ECM biogenesis (LC10), and dropped down during ECM expansion (LC12 to LC15). However, the absolute transcript levels in sex-induced females were much higher than those in the vegetative females. During cell cleavage (LC5), the sex-induced algae show a RBR1 transcript level 78% higher than that of vegetative algae, and it is ∼45% higher by the end of embryogenesis (LC8) and at the beginning of ECM expansion (LC10). The switch to sexual development, therefore, leads to increased RBR1 expression, even though the detectable response to the sex-inducer is delayed: after the sex-inducer has been added, it takes several hours before the RBR1 expression level in sexually induced females is clearly higher than in asexual females (Figure 4B). This also indicates that the 2-h heat stress experiment described above probably was not influenced by the elevated levels of sex-inducer molecule that occur after heat stress.
To investigate whether RBR1 splice variants 2, 3, and 4 show a different expression pattern in the life cycle of Volvox compared with variant 1 described above, primer pairs were established that are specific for each of the four variants. Expression of each splice variant was examined by quantitative real-time RT-PCR at three points in the life cycle of vegetatively grown females: shortly after hatching of juveniles (LC1), at the end of embryogenesis directly after inversion (LC8), and after ECM expansion (LC15). For each splice variant, the first value (LC1) was used as a reference point. In real-time RT-PCR analysis, all four splice variants showed essentially the same expression pattern (Figure 4F). The lowest transcript level of all four variants is observed after hatching of juveniles (LC1), and the highest level is seen at the end of embryogenesis (LC8), when it is 5- to 11-fold higher than LC1.
RBR1 splice variants were also investigated for differential expression after addition of the sex-inducer. Again, all splice variants revealed almost the same expression pattern: after sexual induction, the expression was up to ∼50% higher than at the corresponding vegetative stage in development, with the greatest difference at the end of embryogenesis (LC8) (Figure 4F).
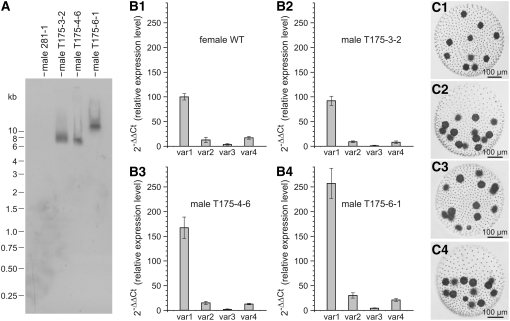
The abundance of the four alternatively spliced transcripts relative to each other was analyzed in vegetative females by quantitative real-time RT-PCR. The amounts of splice variants 2, 3, and 4 were normalized to the level of variant 1, which was set to 100. Variant 1 turned out to be the most abundant, while variants 2, 3, and 4 reached levels of 13, 4, and 17% that of variant 1, respectively (see Figure 6B1).
Figure 6.
Detection of the RBR1 Gene and RBR1 Splice Variants in Transgenic Males Transformed with the Female RBR1 Gene.
(A) DNA gel blot hybridization (standard conditions) of genomic DNA from parent male 281-1 (control) and transgenic males transformed with the female RBR1 gene. Transformants T175-3-2, T175-4-6, and T175-6-1 are descendants of 281-1 that have been transformed with both the nitA gene and the female RBR1 gene. The DNA was digested with AccI. The blot was probed using a 754-bp RBR1 fragment.
(B) Relationship between RBR1 splice variants 1 to 4 (var1 to var4) in wild-type females and in transgenic males expressing the female RBR1 gene. Error bars refer to the sd of the y value. All real-time RT-PCR experiments were performed in triplicate from technical repeats. For calculation of all relative expression levels, the expression level of splice variant 1 in wild-type females was used as the reference point (=100). For isolation of mRNA from each strain, asynchronous, vegetatively grown cultures containing a mixture of the different developmental stages were used.
(B1) Distribution of splice variants in the female wild-type strain Eve10.
(B2) to (B4) Expression of the Volvox RBR1 gene and distribution of splice variants in transgenic males transformed with the female RBR1 gene.
(B2) Male transformant T175-3-2.
(B3) Male transformant T175-4-6.
(B4) Male transformant T175-6-1.
(C) Phenotypes of male TRef1 (control), a transgenic descendant of 281-1 that has been transformed only with the nitA selectable marker gene, and of transgenic males expressing the female RBR1 gene.
(C1) Male TRef1 (control).
(C2) Male transformant T175-3-2.
(C3) Male transformant T175-4-6.
(C4) Male transformant T175-6-1.
Females, but Not Males, Have an RBR1 Gene
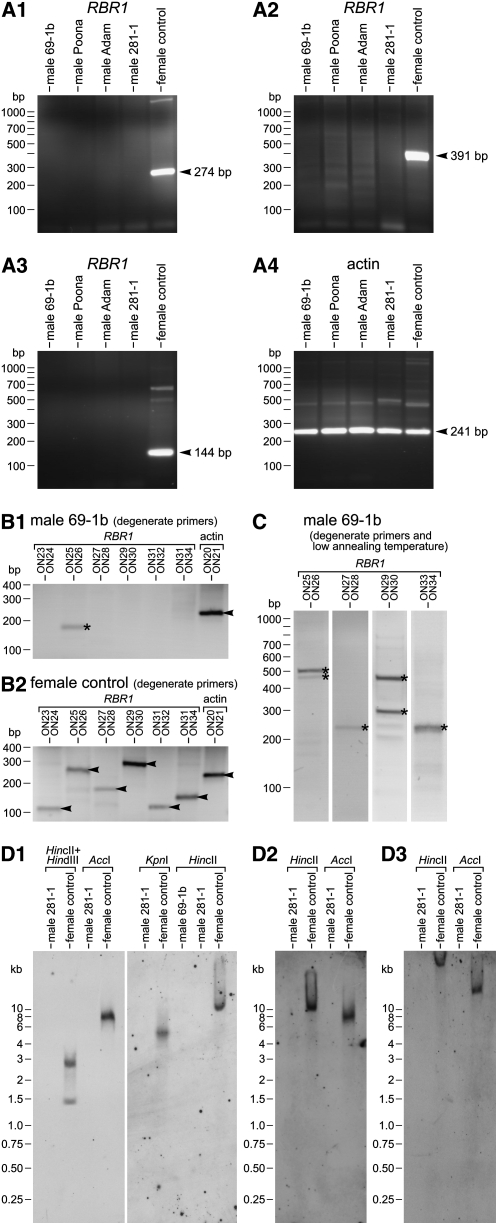
After investigating RBR1 in females, we looked for the RBR1 gene in males. We analyzed the wild-type males 69-1b, Adam, and Poona, as well as the nitA− male 281-1, which was the parent strain for later transformation experiments. Surprisingly, PCR with RBR1-specific primers yielded products only when genomic DNA from female strains was used as a template, but not with genomic DNA from any of the male strains (Figures 5A1 to 5A3). This negative result was obtained not only with the RBR1-specific primer pairs shown in Figures 5A1 to 5A3 but also with several additional RBR1-specific primer pairs (see Supplemental Table 2 online). Control PCRs with actin-specific primers yielded the expected products in all of the strains analyzed (Figure 5A4).
Figure 5.
Absence of RBR1 in Males: PCR Results with RBR1-Specific and Degenerate Primers and Results from DNA Gel Blot Analyses under Standard and Low-Stringency Conditions.
(A) PCR results with specific primers (sequences are presented in Supplemental Table 2 online).
(A1) to (A3) The wild-type V. carteri males 69-1b, Adam, and Poona and the nitA− male 281-1 were analyzed by genomic PCR using RBR1-specific primers. These primers reliably amplified small DNA fragments from different areas of the RBR1 gene when females were used (female control). The expected sizes of RBR1 fragments were as follows: 274 bp (primers ON12 and ON01) (A1); 391 bp (primers ON02 and ON22) (A2); and 144 bp (primers ON13 and ON03) (A3).
(A4) Actin-specific primers were used as a control (expected fragment size, 241 bp; primers ON20 and ON21).
(B) and (C) PCR results with degenerate primers both under standard conditions and at a low annealing temperature. PCR fragments of expected sizes are marked with arrowheads. Fragments marked with asterisks are nonspecific PCR products with unexpected sizes; sequencing showed that they are not related to RBR1 sequences.
(B) Genomic DNA of the male 69-1b (B1) and female control (B2) was analyzed by PCR using degenerate primers that are specific for RBR1. Degenerate primers were designed based on the RBR1 amino acid sequence; each degenerate primer covers all possible base combinations that encode the respective part of the polypeptide sequence. Actin-specific primers were used as a control. The expected sizes of RBR1 fragments were 109, 251, 175, 290, 122, and 155 bp (actin, 241 bp).
(C) The male 69-1b was analyzed by genomic PCR using RBR1-specific, degenerate primers at a low annealing temperature (45°C). This annealing temperature was >10°C lower than the optimal annealing temperature used in (B).
(D) DNA gel blot hybridization of genomic DNA from a male 281-1 and a female control (in the HincII digest of [D1], another male, 69-1b, was also analyzed).
(D1) DNA gel blot hybridization under standard conditions. The blot was probed using probe A, a 754-bp RBR1 fragment.
(D2) DNA gel blot hybridization under low-stringency conditions (hybridization temperature, 42°C; washing with high salt concentration). The blot was probed using probe A, a 754-bp RBR1 fragment.
(D3) DNA gel blot hybridization under low-stringency conditions (hybridization temperature, 42°C; washing with high salt concentration). The blot was probed using probe B, an 802-bp RBR1 fragment.
The localization of PCR primers and DNA gel blot probes is indicated on the map of the RBR1 gene in Supplemental Figure 6 online.
The males were also evaluated by real-time RT-PCR for the presence of any of the RBR1 splice variants. We never observed any RBR1 expression in males, regardless of which splice variant we searched for. The expression level (prior to embryogenesis) of RBR1 splice variants in females was at least ∼4,000-fold higher than the background noise in the real-time PCR experiments with males (see Supplemental Figure 5 online).
The PCR results showed that males have no sequence that is identical to female RBR1. However, recombination is known to be suppressed in the vicinity of the mating-type locus of C. reinhardtii (Gillham, 1969; Ferris and Goodenough, 1994; Ferris, 1995) and probably other Volvocales species. Therefore, there was a possibility that males possess a male-specific RBR1-like allele that encodes a polypeptide that is identical or very similar to the female RBR1, but PCR products of this allele could not be amplified with standard primers due to many synonymous nucleotide interchanges. To investigate this possibility, several degenerate primer pairs were designed based on the RBR1 amino acid sequence (for primer sequences, see Supplemental Table 2 online; for primer localization, see Supplemental Figure 6 online). In PCRs using these degenerate primers, no products were obtained when male DNA was used, except for a single nonspecific band (Figure 5B1). Sequencing of this nonspecific band showed that it is not related to the RBR1 sequence. When female DNA was used, all primer combinations yielded the expected products (Figure 5B2).
We repeated the PCR experiment with male genomic DNA using the degenerate primers at a low annealing temperature, in order to allow the amplification of RBR1-related sequences that are not perfectly complementary to the primers. Under these low-stringency conditions, several primer combinations yielded at least one PCR product (Figure 5C). However, sequencing of these products revealed that all of them are nonspecific products.
In addition to the PCR experiments, DNA gel blot analyses of genomic DNA from males and females were performed using a 754-bp RBR1 fragment as a probe (probe A), which covers the conserved domains N1 and N2 (see Supplemental Figure 6 online). After AccI, KpnI, and HincII restriction digestion, a single band was detectable in the female DNA under standard DNA gel blot hybridization conditions (Figure 5D1); there were two bands after HincII + HindIII digestion, which was expected as HindIII is known to cut within the sequence of the probe. This shows that there is only a single copy of the RBR1 gene in the female genome. Even under low-stringency hybridization conditions, the RBR1 probe detected only a single band in the HincII- and AccI-digested female genome (Figure 5D2). No bands were detected in the male genome after hybridization with the RBR1 probe either under standard hybridization conditions (Figure 5D1) or under low-stringency hybridization conditions (Figure 5D2). In addition, another RBR1 fragment was used as a probe (probe B) for DNA gel blot analysis under low-stringency hybridization conditions (Figure 5D3). Likewise, this 802-bp fragment, which covers the conserved B domain (see Supplemental Figure 6 online), detected bands only in the female genome and not in the male genome.
Moreover, BLASTn analysis of ∼165,000 EST reads from V. carteri males and females (DOE JGI) identified the RBR1 sequence only within the female EST fraction. The ∼72,000 EST reads from males did not exhibit any RBR1-like sequences. Likewise, tBLASTx protein sequence analysis of the EST reads, which were translated into all six possible reading frames, identified only the known RBR1 sequence in the female EST fraction but no other RBR1-like amino acid sequence.
The results of the genomic PCR, DNA gel blot hybridization, RT-PCR, and EST analyses show that RBR1 is a single-copy gene in the female genome. No such gene sequence can be detected in the male genome.
Generation of Transgenic Males Expressing the RBR1 Gene from Females
To learn more about the mode of action of RBR1 in Volvox, we sought to insert the female-specific RBR1 gene into the nuclear genome of Volvox males. At the same time, we wanted to establish a procedure for stably transforming male Volvox. Females are routinely used in transformation experiments, as well as most other kinds of experiments involving Volvox, because they are easier to handle in the laboratory than males. In a vegetatively growing male culture, single males repeatedly switch spontaneously to the sexual pathway and release the sex-inducer, whereas this does not occur in vegetatively growing female cultures (Starr, 1972; Callahan and Huskey, 1980; Weisshaar et al., 1984). Due to the high potency of the sex-inducer glycoprotein, the sexual pathway is then switched on in the entire culture, which upsets the researcher's plans. This problem can be reduced by growing several small cultures in parallel instead of one large-scale culture, by growing the males only up to low densities, and by adding proteases to the culture medium (Tschochner et al., 1987).
Stable nuclear transformation of males was performed using a particle gun as described (Schiedlmeier et al., 1994), but with several modifications according to Hallmann and Wodniok (2006). Gold microprojectiles were coated with a plasmid carrying the RBR1 gene with 1.5-kb upstream and 0.9-kb downstream flanking sequences together with a nitA selectable marker plasmid. Nuclear transformation of Volvox occurs mainly by illegitimate recombination events, resulting in ectopic integration of introduced DNA. In Volvox, it is not possible to obtain the large numbers of transformants seen with bacteria or lower eukaryotic organisms, due to the technical difficulty of treating large numbers of Volvox algae at one time and to the relatively low transformation efficiency (Schiedlmeier et al., 1994). Transformants expressing nitA were evaluated for the presence of the female RBR1 gene by genomic PCR. RBR1-specific primers, which did not yield products on genomic templates from untransformed males (Figures 5A1 to 5A3), did amplify PCR fragments of the correct size from genomic DNA extracted from male transformants (see Supplemental Figure 7 online). In addition to the PCR experiments, DNA gel blot analyses of genomic DNA from transgenic males were performed using the 754-bp RBR1 fragment as a probe (probe A). After AccI restriction enzyme digestion, a single band was detectable in DNA from transgenic males (Figure 6A), which indicates that the RBR1 gene integrated only once into the genome of each transformant.
Expression of the female-specific RBR1 gene in male transformants was analyzed by quantitative real-time RT-PCR. The relative abundance of the four alternatively spliced transcripts was analyzed in asynchronously grown male cultures, and the amount of splice variants 1 to 4 in the male transformants was normalized to the amount of splice variant 1 in asynchronously grown wild-type females, which was defined as 100. All transformants showed a relative abundance of the four alternatively spliced transcripts that was quite similar to that of wild-type females (Figure 6B1). One transformant, male T175-3-2 (Figure 6B2), also showed an expression level that was similar to that of wild-type females. In the male transformant T175-4-6, expression of splice variant 1 was ∼1.7-fold higher than in wild-type females (Figure 6B3), and in male T175-6-1, expression of variant 1 was ∼2.6-fold higher than in females (Figure 6B4). Therefore, Volvox males were successfully transformed with the female-specific RBR1 gene, and they showed similar or higher RBR1 expression levels compared with the females. The fact that the level of transgene expression varies among independent transformants is known from many species; the phenomenon is usually attributed to position effects, chromosomal differences among integration sites that influence gene expression, and gene silencing (Hobbs et al., 1990; Peach and Velten, 1991; Finnegan and McElroy, 1994). Despite the expression of the female RBR1 gene in males, at first sight transgenic males looked and behaved like wild-type males (Figure 6C).
To analyze the stability of the integration into the nuclear genome and sustained expression of the RBR1 transgene, genomic PCR and real-time RT-PCR experiments were repeated after an 8-month propagation period. After this period, which corresponds to ∼120 generations, the transformed males were still positive for the RBR1 transgene, as determined by genomic PCR. Moreover, the expression patterns and expression levels of RBR1 splice variants did not change.
Transgenic Males Expressing the Female-Specific RBR1 Gene Showed Altered Growth Characteristics
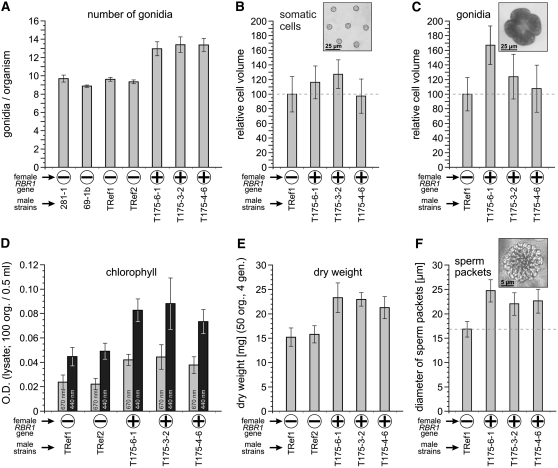
A thorough comparison of wild-type males and transgenic males expressing the RBR1 gene revealed several differences. Initially, we identified an anomaly in the number of gonidia in transformants relative to the strains that lacked RBR1. The average number of gonidia was between 8.9 and 9.7 in strains without the female RBR1 gene, whereas RBR1-expressing transformants showed an average number of gonidia between 12.9 and 13.4 (Figure 7A). Thus, expression of the female-specific RBR1 gene in males led to an increase in the number of gonidia of approximately four, which corresponds to ∼40%. It is difficult to determine the exact number of somatic cells, but because the diameter of the entire spheroids and the number of somatic cells per field of view in the microscope were about the same in transformants and strains that lacked RBR1, expression of RBR1 in transformants does not seem to have a significant effect on the total number of somatic cells.
Figure 7.
Productivity of Transgenic Males Expressing the Female RBR1 Gene.
Characteristics of the male transformants expressing RBR1 were compared with those of male strains that lack the female RBR1 gene. 69-1b is a wild-type male, 281-1 is the parent male strain (nitA−), TRef1 and TRef2 are transgenic male descendants of 281-1 that have been transformed only with the nitA selectable marker gene, and transformants T175-6-1, T175-3-2, and T175-4-6 are descendants of 281-1 that have been transformed with both the nitA gene and the female RBR1 gene. Algae of the same developmental stage were used for each comparative experiment. The presence or absence of the female RBR1 gene is indicated by a plus (+) or a minus (−), respectively. Error bars refer to the sd of the y value.
(A) Average number of gonidia per organism calculated from 300 measurements each.
(B) Cell volume of somatic cells (parent somatic cells from the anterior end of the spheroid shortly before hatching of juveniles). In each strain, cell volumes were calculated from 100 measurements of cell diameter. The absolute cell volumes were as follows: TRef1, 568 ± 137 μm3; T175-6-1, 655 ± 147 μm3; T175-3-2, 717 ± 141 μm3; T175-4-6, 553 ± 130 μm3. The inset shows a representative micrograph of somatic cells used for the analysis (from TRef1).
(C) Cell volumes of mature gonidia. As explained in the text, the diameters of 100 four-cell embryos were used to calculate the volume of the gonidia from which those embryos were derived. The absolute cell volumes were as follows: TRef1, 68,284 ± 15,500 μm3; T175-6-1, 115,009 ± 30,477 μm3; T175-3-2, 84,395 ± 25,824 μm3; T175-4-6, 72,754 ± 23,427 μm3. The inset shows a representative micrograph of a four-cell embryo used for the analysis (from TRef1).
(D) OD of a cleared lysate of 100 organisms in a total volume of 0.5 mL. The OD was determined at wavelengths of 440 and 670 nm, so it reflects primarily the amount of chlorophyll.
(E) Fifty organisms were grown for four generations under standard conditions. Then the organisms were quantitatively collected and dried, and the dry weight was determined.
(F) Diameter of sperm packets. In each strain, 100 measurements were performed. The inset shows a representative micrograph of a sperm packet used for the analysis (from TRef1).
In addition to the increased number of gonidia, transformants also showed increased cell volume relative to the RBR1-lacking strains. Transformants T175-3-2 and T175-6-1 showed an increased volume of somatic (Figure 7B) and gonidial (Figure 7C) cells, with gonidial cells of T175-6-1 having a larger increase than those of T175-3-2. Because it is difficult in a large-scale analysis to decide whether a gonidium has reached maximum size, we used the cloverleaf-shaped four-cell stage to analyze cell volume. In V. carteri, cell cleavage begins when gonidia reach their maximum cell volume and occurs in the absence of any further growth, so the four-cell stage has approximately the same maximum diameter as the mature gonidium from which it was derived. In addition, the four-cell stage is very short relative to the rest of the life cycle and is easy to identify. Cell volumes of transformants were compared with the male control strain TRef1, a male descendant of the parent 281-1 strain used in the transformation experiments. TRef1 was transformed only with the nitA selectable marker gene and, therefore, contains no female RBR1. The average volume of gonidial cells of male transformant T175-4-6 was ∼7% higher than that of TRef1 (Figure 7C). Gonidia of transformants T175-3-2 and T175-6-1 had cell volumes ∼24 and ∼68% higher than TRef1 (Figure 7C). A t test confirmed that the differences in cell volume between the control TRef1 and transformants were statistically significant (P < 0.05), except for somatic cells of T175-4-6.
We also evaluated whether the expression of RBR1 in males influences the duration of cleavage steps. Volvox embryos undergo a rapid series of cleavage divisions (11 to 12 divisions), some of which are asymmetric (Kirk, 1998; Hoops et al., 2005). After cleavage divisions, each embryo contains all of the cells that will be present in the adult, but the flagellar ends of all cells are pointed toward the interior, rather than toward the exterior, where they need to be positioned in order to function in locomotion. In the morphogenetic process of inversion, embryos turn inside out to correct this arrangement (Kirk, 1998; Hoops et al., 2005; Hallmann, 2006a). Embryogenesis is defined as the period from the first cleavage to completion of inversion and takes ∼8 h in wild-type females or males (Kirk, 1998; Hoops et al., 2005; Hallmann, 2006a). In V. carteri, viable gonidia in the stage before cleavage can be removed mechanically from the parent spheroid. Separated gonidia develop normally under appropriate conditions and allow the microscopic observation of embryogenesis starting from an isolated gonidium. Unfortunately, it is only in the first rounds of division that the actual cleavage stage can be determined unequivocally. But ∼1 h after cleavage divisions, embryogenesis of Volvox ends with a distinctive process, the inversion. Thus, we determined the length of the period between two clearly identifiable stages (i.e., the four-cell stage and the middle of inversion) in one transformant (T175-6-1) and in the control strain TRef1. The length of this period was determined in 25 embryos each. In TRef1, the length of this period was ∼6.5 h (sd = 15 min), whereas in T175-6-1, it lasted ∼9 h (sd = 15 min). Based on these results, the total length of embryogenesis was calculated to be ∼8 h in TRef1, which is consistent with known values from wild-type V. carteri (Kirk, 1998; Hoops et al., 2005; Hallmann, 2006a), and ∼11 h in T175-6-1. In wild-type algae, each cleavage division takes ∼35 min (Kirk, 1998). The prolongation of embryogenesis by ∼3 h corresponds to an extension of each cleavage division from ∼35 min in TRef1 to ∼50 min in T175-6-1.
RBR1-expressing transformants were also distinguishable from strains that lacked RBR1 by their pigment content, particularly chlorophyll. When cleared lysates of 100 organisms were analyzed spectrophotometrically at 670 and 440 nm, the pigment concentration was determined to be ∼130% higher in the RBR1-expressing strains than in the strains that lacked RBR1 (Figure 7D). There was also a measurable difference in the produced biomass when the same number of organisms of each strain was grown for four generations (Figure 7E): the dry weight of RBR1-expressing transformants was ∼40% higher than that of strains that lacked RBR1.
Finally, the influence of RBR1 expression on the sexual development of male transformants was investigated. The main characteristic of sexual males is the production of sperm. We were not able to determine the exact number or the volume of the tiny sperm cells; therefore, we determined the diameter of sperm packets, in which the sperm are arranged in a single cell layer, forming a convex, round plate. In RBR1-expressing transformants, the diameter of sperm packets was 30 to 40% larger than that in strains that lacked RBR1 (Figure 7F).
DISCUSSION
Comparison of Volvox RBR1 and Chlamydomonas MAT3
Our phylogenetic analysis of RB-related proteins assigns RBR1 of V. carteri to an algal branch between plants and animals. This is consistent with the phylogenetic position of the species Volvox (Kirk, 2003) and with a genomic analysis of its unicellular relative Chlamydomonas, which revealed the evolution of key animal and plant functions (Merchant et al., 2007).
The comparison of the RBR from C. reinhardtii, MAT3, with RBR1 from V. carteri is of particular interest, because these proteins come from closely related species with quite different cellular strategies (i.e., unicellular versus multicellular) and rather dissimilar modes of sexual reproduction (i.e., isogamy versus oogamy). The evolution of these differences probably required the species-specific adaptation of the RB pathway.
There are both parallels and differences between RBR1 from V. carteri and MAT3 from C. reinhardtii. The parallels are as follows: (1) a similar protein sequence (59% identity), in particular within the A and B domains, and the LxCxE binding site (Figure 3A; see Supplemental Figure 4 online); (2) a similar gene structure with several conserved intron positions (Figure 1A); (3) localization at the mating-type locus (Figure 1B); (4) the influence on the cell volume: in mat3 null mutants, the cell volume was reduced by 25 to 35% compared with wild-type cells (Armbrust et al., 1995; Umen and Goodenough, 2001), but when female RBR1 was expressed as an extra protein in the male proteome, the cell volume increased by up to ∼68% (Figure 7C); (5) the influence on mass doubling time/produced biomass per time unit: in mat3 null mutants, the mass doubling time was somewhat higher (6.8 h) compared with that in wild-type cells (6.5 h) (Umen and Goodenough, 2001), corresponding to a decrease in produced biomass per time unit in mat3 null mutants, but in Volvox, the expression of RBR1 in transgenic males resulted in an increase of biomass (Figures 7D and 7E); (6) the presence of several potential CDK phosphorylation sites (Figure 2A); (7) the presence of splice variants (Figure 1A). The differences are as follows: (1) in Chlamydomonas, MAT3 was identified at the mating-type locus of both mating types (plus and minus) (Ferris et al., 2002), whereas in Volvox, RBR1 was identified only at the female mating-type locus (Figures 1B and 5); (2) MAT3 expression was constitutive during the life cycle (Bisova et al., 2005), whereas expression of RBR1 varied greatly during the life cycle and showed a strong increase during and after embryogenesis (Figure 4B); (3) in Volvox, RBR1 was expressed differentially in somatic cells and gonidia (Figure 4C), but there is only one cell type in Chlamydomonas; (4) the cell doubling time increased in mat3 null mutants (9.1 h) compared with wild-type Chlamydomonas cells (4.9 h) (Fang et al., 2006), whereas in Volvox, the duration of cleavage steps increased (up to ∼9 h) when female RBR1 was expressed as an extra protein in transgenic males compared with parent males (∼6.5 h); (5) the average number of cell divisions increased in mat3 null mutants (2.6) compared with wild-type Chlamydomonas cells (1.6) (Umen and Goodenough, 2001), whereas in Volvox, the number of gonidia increased (up to ∼13) when female RBR1 was expressed as an extra protein in transgenic males compared with parent males (∼10) (Figure 7A); (6) in Volvox, expression of RBR1 increases in response to the sex-inducer glycoprotein (Figure 4B), but there is no sex-inducer glycoprotein in Chlamydomonas.
The parallels between RBR1 and MAT3 show that these proteins descended from an ancestral, green algal RB protein. However, the differences seem to be a consequence of the divergent evolution of the multicellular, oogamous Volvox and the single-celled, isogamous Chlamydomonas requiring modifications of RBRs and the RB pathways.
Regulation of RBR1 Function
The activity of the RBR1 protein appears to be subject to transcriptional, posttranscriptional, and posttranslational control (Figures 1, 2, and 4). Transcript accumulation was shown to be strongly dependent on the developmental stage and regulated in a cell type–specific manner (Figures 4B and 4C). Transcript abundance also changes in response to light/dark, heat stress, and the presence of the sex-inducer (Figures 4B, 4D, and 4E). At the posttranscriptional level, we have identified alternative splicing events resulting in four splice variants (Figures 1A and 2B). Analysis of alternative splicing events in different lineages of eukaryotes indicates that most genes in complex eukaryotes are alternatively spliced (Brett et al., 2002; Stamm et al., 2005). During the last decade, numerous molecular analyses demonstrated that alternative splicing determines the binding properties, intracellular localization, enzymatic activity, protein stability, and posttranslational modifications of a large number of proteins; the magnitude of the effects range from a complete loss of function or acquisition of a new function to very subtle modulations, which are observed in the majority of cases reported (Stamm et al., 2005). Even an alternative splice form that causes early translational termination and an inactive protein product that is unstable can act as an important form of regulation of biological activity, and this functionality can work at low or high concentrations of the splice variant (Smith and Valcárcel, 2000; Modrek and Lee, 2002). Therefore, even the smallest RBR1-derived protein product (12 kD), resulting from splice variant 3, might be part of the RBR regulatory network.
Finally, the presence of 15 potential CDK phosphorylation sites in the RBR1 protein sequence (variant 1) suggests a posttranslational control via phosphorylation by G1-CDKs, which are themselves subject to activation by mitogens (Coqueret, 2002). Phosphorylation of RBRs is known to inhibit binding to LxCxE-containing proteins like E2F (Hinds et al., 1992; Ewen et al., 1993; Lee et al., 1998; Dahiya et al., 2000; Brown and Gallie, 2002).
The sum of probable and proven mechanisms for the regulation of RBR1 suggests that RBR1 performs a key function in Volvox development.
Expression of RBR1 in Both Gonidia and Somatic Cells
An important decision facing each cell of a multicellular organism is whether to commit to another round of replication or to exit the cell cycle. The requirements for RBR1 might be different in the two different cell types of female Volvox. In gonidia, cleavage begins when these cells reach their maximum cell volume and occurs in the absence of any further growth; the increased size of transgenic male cells expressing RBR1 suggests that RBR1 is involved in the machinery for cell cleavage and size control (Figures 7B, 7C, and 7F). Furthermore, RBR1 is likely to be involved in cell differentiation, because cell size was shown to be a key determinant in the differentiation of Volvox (Kirk et al., 1993). RBR1 expression levels were shown to be significantly higher in parent somatic cells than in gonidia or cleaving embryos (Figure 4C). This high expression level in parent somatic cells may ensure a permanent, irreversible exit from the cell cycle. This function has been shown for other RBRs in terminal differentiation and in the course of senescence (Narita et al., 2003; Sage et al., 2003; Ben-Porath and Weinberg, 2005; Blais and Dynlacht, 2007). Likewise, parent somatic cells of Volvox have to maintain the terminal differentiation and they are subject to senescence (Pommerville and Kochert, 1982). The increase of RBR1 expression during embryogenesis (Figure 4B) might reflect an acceleration of the senescence process in parent somatic cells or an increased need for an RBR in somatic cells, which keeps those cells terminally differentiated while embryos are cleaving.
Is There a Functional Analog of RBR1 in Males?
The RBR1 gene was shown to be present only in females, but vegetatively growing males show the same developmental program as vegetatively growing females. Due to the expression of RBR1 in females and the general importance of RBRs and the RB pathway, we predict that males possess a functionally related analog of RBR1, probably at the male mating-type locus. This postulated male RBR1 analog must have a sequence quite different from that of the female RBR1, because PCR and DNA gel blot analyses failed to detect an analog in males, even under low-stringency conditions (Figure 5). Nevertheless, the observation that female RBR1 functions in transgenic males (Figure 7) indicates that female RBR1 is compatible with the corresponding male signaling pathway. Expression of RBR1 in transgenic males results in enlarged reproductive cells (i.e., gonidia and sperm packets in the asexual and sexual life cycles, respectively). This result seems to reflect an additive effect, where the sum of two RBRs (i.e., the male's own [postulated] RBR1 analog) is expressed together with the heterologous female RBR1; therefore, the situation is equivalent to an overexpression of RB proteins. Larger cells have also resulted from the overexpression of an RB protein in Drosophila (RBF) (Neufeld et al., 1998). In addition to enlarged cells, RBR1-expressing transformants exhibited an extension of each cleavage division from ∼35 to ∼50 min. Similarly, induction of RBF in Drosophila increased the cell doubling time from 12.0 to 18.5 h (Neufeld et al., 1998).
There remains the question about the cause of the gender-specific RB protein status. The RBR1 gene maps to the female mating-type locus; therefore, localization of the (postulated) functionally related analog of RBR1 at the male mating-type locus seems to be reasonable. Since recombination is known to be suppressed in the vicinity of the mating-type locus of C. reinhardtii (Gillham, 1969; Ferris and Goodenough, 1994; Ferris, 1995), the same may be true for Volvox. The localization of RBRs at the mating-type loci corresponds to a connection between each RBR and gender identity. Thus, this situation allows RBRs to evolve separately in each gender. Mating type–specific alternatives carried by a species are not classical alleles, even if they would occupy allelic positions on homologous chromosomes; therefore, such alternatives with different amino acid sequences are called idiomorphs (Metzenberg and Glass, 1990). A sexual conflict (different evolutionary interests of the two sexes) occurs when variation of one idiomorph affects a trait that is present in both males and females. If there are different trait optima for the two sexes, which is very likely at least under certain conditions (e.g., sexual development), selection for sexual dimorphism is the consequence (Stockley, 1997; Parker and Partridge, 1998). Such sexual conflicts probably have been the driving force for the evolution of an isogamous Chlamydomonas-like ancestor to the oogamous Volvox. Therefore, the divergent evolution of RBRs could be not only a consequence of the maintenance of two mating types and the development of oogamy but also one of the causes. The gender-specific evolution of RBRs (and the whole mating-type loci) seems to be driven by sexual development, because during vegetative development the two sexes are indistinguishable from each other.
The evolution of gender-specific RBRs is also consistent with an increased evolutionary flexibility previously observed in sex-related proteins compared with other proteins that are not related to sex (Ferris et al., 1997). In this regard, we determined the degree of similarity between the MAT3/RBR1 putative orthologs and a couple of other Chlamydomonas–Volvox putative orthologs both related and unrelated to sex. Using the JGI Volvox genome portal 1.0, a BLAST analysis with the sex-related C. reinhardtii proteins MAT3, FUS1, MID, and SAD1 either produced hits with low similarity or failed to detect any significant putative orthologs in V. carteri (see Supplemental Table 3 online); by contrast, sex-unrelated proteins that had previously been used as controls (Ferris et al., 1997) and some additional control proteins that are unrelated to sex revealed putative orthologs with high sequence similarity (see Supplemental Table 3 online). Even if this is only a small fraction of all relevant proteins, the analysis confirms that proteins involved in sexual reproduction evolve more rapidly than other proteins.
RBRs and the Different Cleavage Programs
How does the expression of RBR1 in transgenic males influence the different cleavage programs of asexual and sexual development? The first five divisions of both male and female asexual wild-type embryos are all symmetric. In the sixth cleavage cycle, ∼16 cells of the anterior hemisphere divide asymmetrically to form sister cells of unequal size. The larger cells are gonidial initials that will give rise to one gonidium each. Every smaller sister cell and each of the symmetrically dividing cells in the posterior hemisphere is a somatic initial that will give rise to a clone of somatic cells (Starr, 1969, 1970). The gonidial initials divide asymmetrically two or three more times, generating an additional somatic initial at each division. Then the gonidial initials withdraw from the division cycle, whereas the somatic initials continue cleaving symmetrically until they have completed a total of 11 or 12 divisions. Finally, asexual male and female juveniles have ∼2000 to 4000 biflagellate somatic cells and ∼16 gonidia (Figures 4A1 and 4A3). Asexual, RBR1-expressing male transformants show an increased volume of mature gonidia (Figure 7C), which correlates with the production of an increased number of juvenile gonidia during cleavage. Kirk et al. (1993) showed an unambiguous relationship between cell size and cell fate in V. carteri: cells with a diameter of >∼8 μm at the end of cleavage always develop as gonidia, no matter where or how these cells have been produced in the embryo. Therefore, the additive effect of two RBRs in RBR1-expressing males seems to cause a few more cells to reach the critical diameter of ∼8 μm and thus to become gonidia.
Gonidia of both sexes respond to the same sex-inducer glycoprotein with different cleavage programs and substantial differences in cellular differentiation. Sexually induced female embryos fail to divide asymmetrically at the sixth division cycle, but then they do so at the seventh cycle (Starr, 1969). Thus, sexual females with ∼32 large eggs and ∼2000 to 4000 small somatic cells emerge (Figure 4A2). Sexually induced male embryos divide symmetrically six to eight times before all cells in the embryo divide asymmetrically one time. Therefore, somatic cells and germ cells of induced males (androgonidia) arise in a 1:1 ratio (Starr, 1969, 1970; Kirk, 1998). Cleavage pauses after asymmetrical division, but later, androgonidia undergo a second round of cleavage and differentiation, resulting in each androgonidium forming a packet of 64 or 128 flagellated sperm (Figure 4A4) (Starr, 1969, 1970; Hallmann et al., 1998; Kirk, 1998). In sexual development of RBR1-expressing male transformants, the diameter of sperm packets was 30 to 40% larger than that in strains that lacked RBR1 (Figure 7F). This corresponds to a doubling of cross-sectional area and suggests that the developing sperm packet in transformed males may undergo one extra round of cell division, to reach the normal cell size of sperm and thereby produce twice as many sperm.
The observation that female RBR1 functions in transgenic males and results in enlarged reproductive cells (i.e., gonidia and sperm packets in the asexual and sexual life cycles) (Figure 7) suggests that a functionally related analog of RBR1 exists in males. We have also shown that RBR1 expression increases by ∼78% in females after sex induction (Figure 4B), indicating a modified action of RBR1 in sexually induced females. These results implicate RBRs in the regulation of the gender-specific cleavage programs of the embryo. After sexual induction, both the timing of cell cleavage, particularly of the asymmetrical cleavage, and the cell type–specific cell sizes are different in male and female embryos. Therefore, the reason for the divergent evolution of RB-related proteins in females and males appears to be based on sexual development, as males and females respond to the same sex-inducer with different cleavage programs and substantial differences in cellular differentiation. The gender-specific cleavage programs in sexually induced algae probably coevolved with gender-specific RBRs, which adapted their function in a gender-specific way. Such an adaptation could be possible (1) by a gender-specific modification of RBR's interaction characteristics with hitherto existing protein binding partners, (2) through a gender-specific coevolution of RBRs and their interaction partners, or (3) by a gender-specific acquisition of new binding partners. To date, over 100 proteins have been reported to interact with RB family proteins (Morris and Dyson, 2001), and all of these interactions are potential targets when RBR functions are modulated.
Clearly, more work is necessary to give a full picture of the gender-specific RB pathway of Volvox. The next steps will be to identify and characterize the functionally related analog of RBR1 in males. This work has laid the groundwork for these future studies by establishing the gender-specific presence of RBR1 and thereby suggesting a novel function for an RB family protein.
METHODS
Strains and Culture Conditions
Female wild-type strains of Volvox carteri were Eve10 (Adams et al., 1990) and Isanuma strain Volvox 6; the latter was obtained from I. Nishii (RIKEN). Male wild-type strains of V. carteri were 69-1b, Adam (Starr, 1969), and Poona. Eve10 and Adam are descendants of HK10 (female) and 69-1b (Starr, 1969, 1970), which originated from Japan. The Poona male originated from India (Adams et al., 1990). The male V. carteri strain 281-1 with wild-type morphology was used as the DNA recipient. Strain 281-1 inherited an allele from strain HB11A, which carries a nonreversible loss-of-function mutation of nitA, the structural gene encoding nitrate reductase (Gruber et al., 1992); strain HB11A is a previously described female strain of V. carteri (Adams et al., 1990). Cultures were grown in Volvox medium (Provasoli and Pintner, 1959) at 28°C in an 8-h-dark/16-h-light (10,000 lux) cycle (Starr and Jaenicke, 1974). Prior to transformation, strain 281-1 was grown in medium supplemented with 1 mM NH4Cl; afterward, it was grown in selective medium lacking NH4Cl and containing only nitrate as a nitrogen source.
Primer Design
For all PCRs, oligonucleotide primers were designed using the primer analysis software Oligo 6 (Molecular Biology Insights), DNASIS (version 7.00; Hitachi Software Engineering), or Primer Express (Applied Biosystems).
Isolation of Genomic DNA
Genomic DNA was prepared from Volvox spheroids with a DNeasy Plant Mini kit (Qiagen). Purity check and quantification were done by agarose gel electrophoresis and UV spectrophotometry using an Ultrospec 2100 Pro UV/visible spectrophotometer (GE Healthcare).
Isolation of Total RNA
For preparation of total RNA, 1 g of frozen Volvox spheroids, 10 mL of the phenol-based TRI reagent (Sigma-Aldrich), and 3 mL of trichloromethane were used. The RNA was precipitated from the aqueous phase with isopropanol. Precipitated RNA was washed twice with 75% ethanol, air-dried, and dissolved in diethyl pyrocarbonate–treated distilled water. The total RNA was checked for purity and quantitated by agarose-formaldehyde gel electrophoresis and by measurement of absorption at 260 and 280 nm using an Ultrospec 2100 Pro UV/visible spectrophotometer (GE Healthcare).
Total RNA for RT-PCR was prepared as follows: 50 Volvox spheroids were transferred into 10 μL of RNase-free lysis buffer (50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM EGTA, and 2% SDS) (Hallmann and Sumper, 1994). After 10 min at 30°C, the remaining cell debris was pelleted at 15,000g for 3 min. The RNA in the supernatant was precipitated with 3 volumes of 100% ethanol. RNA pellets were washed with 75% ethanol, air-dried, and dissolved in 5 μL of diethyl pyrocarbonate–treated distilled water.
Standard RT-PCR
Total RNA (1 μg) was incubated with antisense primer (10 pmol) in a total volume of 14 μL at 70°C for 5 min and subsequently chilled at 4°C for 5 min. After addition of 5 μL of 5× MMLV-RT reaction buffer (250 mM Tris-HCl, pH 8.3 at 25°C, 250 mM KCl, 20 mM MgCl2, 50 mM DTT, 1.25 μL of 10 mM deoxynucleotide triphosphates, and 200 units of Moloney murine leukemia virus reverse transcriptase lacking ribonuclease H activity [Promega]), first-strand cDNA synthesis was performed at 50°C for 60 min in a total volume of 25 μL. PCR was subsequently performed using 10 μL of the reverse transcription mixture, 1 μM of sense and antisense primers, 0.2 mM deoxynucleotide triphosphates, and 2.5 units of Taq DNA polymerase in a total volume of 50 μL of 1× PCR buffer. PCR was performed in a T3 Thermocycler PCR system (Biometra) using the following conditions: 45 cycles at 95°C for 20 s, 55°C for 30 s, and 72°C for 40 s. Amplification products were cloned into pSPT18/pSPT19 vectors (Roche Applied Science) and sequenced on both strands using a GS FLX genome sequencer (Roche Applied Science) and Sanger technology.
Cloning of the RBR1 cDNA
The cDNA of RBR1 was obtained in five overlapping parts. RT-PCRs were performed using total RNA as a template. Four oligonucleotide primer pairs were used (for primer sequences, see Supplemental Table 2 online): ON01 (antisense) and ON02 (sense); ON03 (antisense) and ON04 (sense); ON05 (antisense) and ON06 (sense); and ON07 (antisense) and ON08 (sense). These pairs produced cDNA fragments of 670, 1865, 1043, and 758 bp in length, respectively. The fifth cDNA fragment was identified in an EST library (clone CBOX16325). Clone CBOX16325 was obtained from the DOE JGI. The corresponding cDNA fragment is 784 bp in length and contains the poly(A)-tailed 3′ end of the RBR1 cDNA. cDNAs were sequenced on both strands using a GS FLX genome sequencer (Roche Applied Science) and Sanger technology.
Cloning of the RBR1 Gene
The RBR1 gene was cloned by PCR using genomic DNA of V. carteri wild-type females as a template. Two oligonucleotide primer pairs, ON09 (sense) and ON10 (antisense) and ON06 (sense) and ON11 (antisense) (for primer sequences, see Supplemental Table 2 online), resulted in two genomic fragments of 4290 and 4081 bp, respectively, which were joined together via a HpaI restriction site and cloned into a pGEM-T Easy vector (Promega). The resulting plasmid, pGEM-E-Mat3-L50-7, harbors the complete RBR1 gene. The RBR1 gene was sequenced on both strands using a GS FLX genome sequencer (Roche Applied Science) and Sanger technology.
Stable Nuclear Transformation of V. carteri Males by Particle Gun Bombardment
Stable nuclear transformation of V. carteri males was performed using a particle gun as described (Schiedlmeier et al., 1994), but with several modifications according to Hallmann and Wodniok (2006). The modified V. carteri nitA gene carried on plasmid pVcNR15 (Gruber et al., 1996) was used as the selectable marker gene to rescue the nitA mutation of the male strain 281-1. Plasmid pVcNR15 was cotransformed with the plasmid pGEM-E-Mat3-L50-7 containing the RBR1 gene. Transformation of males was performed using a Biolistic PDS-1000/He particle gun (Bio-Rad), 600-μg gold microprojectiles (1 μm in diameter; Bio-Rad) coated with 1 μg of plasmid DNA of each plasmid per shot, 900-p.s.i. rupture disks, and a target distance of 16.5 cm. Screening for putative transformants was performed as described (Schiedlmeier et al., 1994). Putative transformants were tested for chlorate sensitivity by exposure to 8 mM potassium chlorate. Genomic DNA was isolated from putative transformants as described (Hallmann and Wodniok, 2006). Cotransformation and integration of the RBR1 gene was tested by genomic PCR as described (Nematollahi et al., 2006) using genomic DNA as a template and the RBR1-specific primers ON12 (sense) and ON01 (antisense) (for primer sequences, see Supplemental Table 2 online) for amplification of a 274-bp fragment.
Quantitative Real-Time RT-PCR
Total RNA (1 μg) was treated with 5 units of DNase I (Promega) in DNase I buffer (20 mM Tris, pH 8.4, 2 mM MgCl2, and 50 mM KCl) in a total volume of 10 μL at 37°C for 10 min. Addition of 1 μL of 25 mM EDTA and incubation at 65°C for 10 min stopped the reaction. For real-time RNA quantification, the QuantiTect SYBR Green RT-PCR kit (Qiagen) was used. For single-tube RT-PCR, 2× QuantiTect SYBR Green RT-PCR Master Mix was used together with the QuantiTect RT Mix. The QuantiTect SYBR Green RT-PCR Master Mix includes HotStarTaq DNA polymerase, QuantiTect SYBR Green RT-PCR buffer, the fluorescent dye SYBR Green I, and the passive reference dye ROX. The QuantiTect RT Mix contains Omniscript and Sensiscript reverse transcriptases. Reactions contained 300 ng of DNase I–treated template RNA, 0.8 μM of each primer, 12.5 μL of 2× QuantiTect SYBR Green RT-PCR Master Mix, and 0.25 μL of QuantiTect RT Mix in a total volume of 25 μL. Reverse transcription at 50°C for 30 min was followed by an incubation at 95°C for 15 min and 40 PCR amplification cycles (95°C for 20 s, 58°C for 30 s, and 72°C for 40 s) using the DNA Engine Opticon continuous fluorescence detection system (MJ Research). Results were analyzed using OpticonMonitor software (version 1.06; MJ Research). All real-time RT-PCR experiments were performed in triplicate together with controls lacking reverse transcriptase or template, and PCR results were considered valid only when controls lacking reverse transcriptase or template were free of any DNA product. The Volvox actin gene was used as a reference, because it is known to be expressed evenly throughout the life cycle (Cresnar et al., 1990) and has been used as a reference gene in several previous studies (Amon et al., 1998; Hallmann et al., 2001; Hallmann, 2006b; Nematollahi et al., 2006). All quantitative data were collected within the exponential phase of PCR amplification. To detect and exclude nonspecific amplicons, the melting curves of all PCR products were analyzed, all final products were visualized by agarose gel electrophoresis to ensure amplification of a single product of the correct size, and products were fully sequenced. Results from real-time RT-PCR experiments that met these quality control requirements were represented as cycle threshold (Ct) values (see below).
Specific primers for the four splice variants of RBR1 were as follows: ON13 (sense) and ON14 (antisense) for variant 1 (expected cDNA length, 132 bp); ON13 (sense) and ON15 (antisense) for variant 2 (expected, 129 bp); ON16 (sense) and ON17 (antisense) for variant 3 (expected, 131 bp); and ON18 (sense) and ON19 (antisense) for variant 4 (expected, 234 bp) (for primer sequences, see Supplemental Table 2 online). Actin-specific primers were ON20 and ON21 (expected, 104 bp).
Analysis of Gene Expression Using the 2−ΔΔCt Method
The expression level of a given splice variant of the RBR1 gene was analyzed using real-time RT-PCR and the 2−ΔΔCt method (Bustin, 2000; Pfaffl, 2001). The Volvox actin gene, which is known to be expressed at a constant level in both cell types throughout the life cycle (Cresnar et al., 1990), was used as an internal control in all real-time RT-PCR experiments. In order to apply the 2−ΔΔCt method (Bustin, 2000; Pfaffl, 2001), the results of real-time RT-PCRs were represented as Ct values. The Ct value was defined as the cycle at which a sample crosses a threshold that is significantly above the background fluorescence and within the exponential phase of the amplification. As there is an inverse correlation between Ct values and the amount of target mRNA, higher amounts of target mRNA have lower Ct values and lower amounts correspond to higher Ct values. The average from three Ct measurements was calculated for both the given splice variant of the RBR1 gene with a given combination of other parameters, such as strain, sex, developmental stage, cell type, and sex induction status, and for the actin gene with the same combination of parameters. ΔCt was determined as the average of the triplicate Ct values for the given splice variant of the RBR1 gene with the given combination of other parameters minus the average of the triplicate Ct values for the actin gene with the same combination of parameters. ΔΔCt represented the relative difference between samples in which one parameter was modified, more precisely ΔΔCt = ΔCt (condition 2) − ΔCt (condition 1). The x-fold higher expression of a given splice variant of the RBR1 gene in condition 2 in relation to condition 1 was calculated as 2−ΔΔCt. If the expression of a given target gene was lower in condition 2 compared with condition 1, the expression was calculated by 1/2−ΔΔCt.
DNA Gel Blot Analysis
After restriction enzyme digestion (AccI, HincII, HincII/HindIII, and KpnI), genomic DNA fragments were separated on 1% agarose gels, vacuum-transferred to nylon membranes (Hybond-N; Amersham Biosciences), and fixed to the membrane by baking for 30 min at 120°C using standard protocols (Sambrook et al., 1989). A 754-bp fragment (from nucleotide positions 1760 to 2513) of the RBR1 gene was amplified by PCR (Expand High Fidelity Plus PCR system; Roche Applied Science; primers ON35 and ON01) and simultaneously labeled using a digoxigenin DNA labeling mix (Roche Applied Science) (probe A). Likewise, an 802-bp fragment (from nucleotide positions 5807 to 6608) of the RBR1 gene was amplified by PCR (primers ON13 and ON05; probe B). For standard DNA gel blots, prehybridization, hybridization, and washing steps were performed in standard solutions (Roche Applied Science); the prehybridization and hybridization temperature was 52°C and the washing procedure was as described (Sambrook et al., 1989). For low-stringency hybridization conditions, prehybridization and hybridization were at 42°C and washing was in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 42°C. Detection of hybridizing bands was done using an anti-digoxigenin–alkaline phosphatase conjugate (1:7500 dilution) and the chemiluminescent substrate CDP Star in accordance with the instructions of the supplier of the chemiluminescence reagent (Roche Applied Science). Chemiluminescence-sensitive films (Retina XBA; Fotochemische Werke) were subsequently exposed to the membranes for 2 to 15 min.
Separation of Gonidia, Embryos, and Parent Somatic Cells
Ten-liter cultures of synchronously grown V. carteri spheroids were harvested by filtration on a 100-μm-mesh nylon screen. The spheroids were broken up using a 50-mL Dounce homogenizer with a tight-fitting pestle (B. Braun). Separation of gonidia, embryos, and parent somatic cells was as described before (Nematollahi et al., 2006).
Induction of Sexual Development
A population of vegetatively grown males of V. carteri was induced to produce and release the sex-inducer using heat shock (Kirk and Kirk, 1986). Seventy milliliters of the fluid of this culture was sterile-filtered and added to a 10-liter culture of vegetatively grown males or females at the developmental stage shortly before hatching of juveniles to induce sexual development.
Database Sequence Search
BLASTn and tBLASTx algorithms (Altschul et al., 1990) were used to search for RBR1-related sequences in genomic and EST databases of V. carteri (JGI Volvox genome portal 1.0; http://genome.jgi-psf.org/Volca1/Volca1.home.html) and Chlamydomonas reinhardtii (JGI Chlamydomonas genome portal 3.0; http://genome.jgi-psf.org/Chlre3/Chlre3.home.html) maintained by the DOE JGI (http://www.jgi.doe.gov/) and the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). Searches were performed with the latest database versions in October 2007.
Phylogenetic Analysis
Alignment of protein sequences was done using the MUSCLE (for Multiple Sequence Comparison by Log-Expectation) program (Edgar, 2004). Minor manual optimization of alignments, trimming, and management of multiply aligned data were done with BioEdit version 7.0.9 (Hall, 1999). Alignments were illustrated using GeneDoc 2.6 (Nicholas et al., 1997). The unrooted tree was calculated using PHYLIP (PHYLogeny Inference Package) (Felsenstein, 1989). In these calculations, 10,000 bootstrap resamplings of multiply aligned sequences were generated with Seqboot, distance matrices (Dayhoff's point accepted mutation matrix) were computed with Protdist, trees were constructed using the neighbor-joining method (Saitou and Nei, 1987) as implemented in Neighbor, and finally, a consensus tree was built using Consense. Phylogenetic trees were drawn with TreeView (Page, 1996).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: EU366288 (RBR1 mRNA; Volvox carteri), EF123087 (RBR1 genomic DNA; V. carteri), FD911121 (RBR1 EST clone CBOX16325), M33963 (actin; V. carteri), AY781411 (Ci MAT3; Chlamydomonas incerta), AF375824 (Cr MAT3; Chlamydomonas reinhardtii), AY675102 (Ot RBR; Ostreococcus tauri), AJ428952 (Pp RBR; Physcomitrella patens), NM_204419 (Gg RB1; Gallus gallus), NM_009029 (Mm RB1; Mus musculus), NM_000321 (Hs RB1, RB; Homo sapiens), S67171 (Hs RB2, p130; H. sapiens), L14812 (Hs RBL1, p107; H. sapiens), X96975 (Dm RBF; Drosophila melanogaster), AF133675 (Pt RBR; Populus tremula × Populus tremuloides), AF230739 (Ee RBR; Euphorbia esula), AF245395 or AT3G12280 (At RBR; Arabidopsis thaliana), AY699399 (Nb RBR; Nicotiana benthamiana), AB015221 (Nt RBR; Nicotiana tabacum), AB205136 (Sb RBR; Scutellaria baicalensis), AB012024 (Ps RBR; Pisum sativum), AJ011681 (Chr RBR; Chenopodium rubrum), X98923 (Zm RBR1; Zea mays), AJ279062 (Zm RBR2; Z. mays), AF007795 (Zm RBR2b; Z. mays), DQ124423 (Zm RBR3; Z. mays), and AY117036 (Cn RBR; Cocos nucifera).
The following protein identifiers (IDs) from this article are as given in the V. carteri sequence databases maintained by the DOE (JGI V. carteri genome portal 1.0; http://genome.jgi-psf.org/Volca1/Volca1.home.html): ID 76291 (spermine synthase), ID 127490 (protein of unknown function), ID 82799 (predicted Na+-dependent cotransporter), ID 82801 (AMP-activated protein kinase), ID 106391 (N-myristoyl transferase), ID 106397 (splicing factor 3a, subunit 2), ID 106399 (vacuolar ATP synthase, subunit C), ID 64869 (cytoplasmic dynein 1b heavy chain), ID 118743 (Hyp-rich glycoprotein), ID 127148 (retinoblastoma-related protein RBR1), ID 82819 (predicted sugar kinase), ID 127314 (putative transcription factor with RWP-RK motif), and ID 94393 (putative forkhead transcription factor).
The following protein IDs from this article are as given in the C. reinhardtii sequence databases maintained by the DOE (JGI C. reinhardtii genome portal 3.0; http://genome.jgi-psf.org/Chlre3/Chlre3.home.html): ID 206416 (spermine synthase), ID 182394 (protein of unknown function), ID 112377 (predicted Na+-dependent cotransporter), ID 112765 (AMP-activated protein kinase), ID 97790 (N-myristoyl transferase), ID 196072 (splicing factor 3a, subunit 2), ID 187322 (vacuolar ATP synthase, subunit C), ID 24009 (cytoplasmic dynein 1b heavy chain), ID 195937 (Hyp-rich glycoprotein), ID 187248 (retinoblastoma-related protein MAT3), and ID 126745 (predicted sugar kinase).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Physical Map of the Volvox RBR1 cDNA (Splice Variant 1) and Overlapping Fragments Used to Assemble the Complete Sequence.
Supplemental Figure 2. Trimmed Alignment of 24 RB-Related Proteins from Green Algae, Moss, and Higher Plants and Animals.
Supplemental Figure 3. Nucleotide Sequence Alignment of Volvox RBR1 and Chlamydomonas MAT3 Coding Sequences.
Supplemental Figure 4. Amino Acid Sequence Alignment of Volvox RBR1 and Chlamydomonas MAT3.
Supplemental Figure 5. Expression Level of RBR1 Splice Variants in Female Volvox Relative to the Background Noise Obtained in Real-Time PCR Experiments with Male Volvox.
Supplemental Figure 6. Localization of PCR Primers and DNA Gel Blot Probes within the RBR1 Gene Used to Confirm the Absence of the RBR1 Gene in Males.
Supplemental Figure 7. Presence of RBR1 in Male Transformants: PCR Results with RBR1-Specific Primers.
Supplemental Table 1. Sequence Comparison of Putative Orthologs Identified Both at the Mating-Type Locus of C. reinhardtii and on a Small Genomic Fragment of the Female V. carteri Genome (Scaffold 43) That Contains the RBR1 Gene.
Supplemental Table 2. Oligonucleotide Primers.
Supplemental Table 3. Comparison of a Subset of Sex-Related and Sex-Unrelated Proteins of C. reinhardtii with the Corresponding Putative Orthologs of V. carteri.
Supplemental Data Set 1. Trimmed Sequences of 24 RB-Related Proteins from Green Algae, Moss, and Higher Plants and Animals Corresponding to the Alignment in Supplemental Figure 2 Online.
Supplementary Material
Acknowledgments
We thank the JGI for providing Volvox and Chlamydomonas sequence information on its websites. We also thank K. Puls for technical assistance.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Armin Hallmann (armin.hallmann@gmx.de).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams, C.R., Stamer, K.A., Miller, J.K., McNally, J.G., Kirk, M.M., and Kirk, D.L. (1990). Patterns of organellar and nuclear inheritance among progeny of two geographically isolated strains of Volvox carteri. Curr. Genet. 18 141–153. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Amon, P., Haas, E., and Sumper, M. (1998). The sex-inducing pheromone and wounding trigger the same set of genes in the multicellular green alga Volvox. Plant Cell 10 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust, E.V., Ibrahim, A., and Goodenough, U.W. (1995). A mating type-linked mutation that disrupts the uniparental inheritance of chloroplast DNA also disrupts cell-size control in Chlamydomonas. Mol. Biol. Cell 6 1807–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek, J., Bartkova, J., and Lukas, J. (1996). The retinoblastoma protein pathway and the restriction point. Curr. Opin. Cell Biol. 8 805–814. [DOI] [PubMed] [Google Scholar]
- Ben-Porath, I., and Weinberg, R.A. (2005). The signals and pathways activating cellular senescence. Int. J. Biochem. Cell Biol. 37 961–976. [DOI] [PubMed] [Google Scholar]
- Bisova, K., Krylov, D.M., and Umen, J.G. (2005). Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol. 137 475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais, A., and Dynlacht, B.D. (2007). E2F-associated chromatin modifiers and cell cycle control. Curr. Opin. Cell Biol. 19 658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett, D., Pospisil, H., Valcarcel, J., Reich, J., and Bork, P. (2002). Alternative splicing and genome complexity. Nat. Genet. 30 29–30. [DOI] [PubMed] [Google Scholar]
- Brown, V.D., and Gallie, B.L. (2002). The B-domain lysine patch of pRB is required for binding to large T antigen and release of E2F by phosphorylation. Mol. Cell. Biol. 22 1390–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin, S.A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25 169–193. [DOI] [PubMed] [Google Scholar]
- Callahan, A.M., and Huskey, R.J. (1980). Genetic control of sexual development in Volvox. Dev. Biol. 80 419–435. [DOI] [PubMed] [Google Scholar]
- Chellappan, S.P., Hiebert, S., Mudryj, M., Horowitz, J.M., and Nevins, J.R. (1991). The E2F transcription factor is a cellular target for the RB protein. Cell 65 1053–1061. [DOI] [PubMed] [Google Scholar]
- Classon, M., and Harlow, E. (2002). The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2 910–917. [DOI] [PubMed] [Google Scholar]
- Claudio, P.P., Tonini, T., and Giordano, A. (2002). The retinoblastoma family: Twins or distant cousins? Genome Biol. 3 reviews3012. [DOI] [PMC free article] [PubMed]
- Coqueret, O. (2002). Linking cyclins to transcriptional control. Gene 299 35–55. [DOI] [PubMed] [Google Scholar]
- Cresnar, B., Mages, W., Müller, K., Salbaum, J.M., and Schmitt, R. (1990). Structure and expression of a single actin gene in Volvox carteri. Curr. Genet. 18 337–346. [DOI] [PubMed] [Google Scholar]
- Dahiya, A., Gavin, M.R., Luo, R.X., and Dean, D.C. (2000). Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 20 6799–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager, S.M., and Murray, J.A. (1999). Retinoblastoma proteins in plants. Plant Mol. Biol. 41 295–299. [DOI] [PubMed] [Google Scholar]
- Dick, F.A. (2007). Structure-function analysis of the retinoblastoma tumor suppressor protein—Is the whole a sum of its parts? Cell Div. 2 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, W., and Pogoriler, J. (2006). Retinoblastoma family genes. Oncogene 25 5190–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee, T., Feiler, H.S., and Gruissem, W. (2000). Retinoblastoma-related proteins in plants: Homologues or orthologues of their metazoan counterparts? Plant Mol. Biol. 43 635–642. [DOI] [PubMed] [Google Scholar]
- Dyson, N. (1998). The regulation of E2F by pRB-family proteins. Genes Dev. 12 2245–2262. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen, M.E., Sluss, H.K., Sherr, C.J., Matsushime, H., Kato, J., and Livingston, D.M. (1993). Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 73 487–497. [DOI] [PubMed] [Google Scholar]
- Fang, S.C., Reyes, C.D., and Umen, J.G. (2006). Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet 2 e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, S.C., and Umen, J.G. (2008). A suppressor screen in Chlamydomonas identifies novel components of the retinoblastoma tumor suppressor pathway. Genetics 178 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5 164–166. [Google Scholar]
- Ferris, P.J. (1995). Localization of the nic-7, ac-29 and thi-10 genes within the mating-type locus of Chlamydomonas reinhardtii. Genetics 141 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P.J., Armbrust, E.V., and Goodenough, U.W. (2002). Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P.J., and Goodenough, U.W. (1994). The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA sequences. Cell 76 1135–1145. [DOI] [PubMed] [Google Scholar]
- Ferris, P.J., Pavlovic, C., Fabry, S., and Goodenough, U.W. (1997). Rapid evolution of sex-related genes in Chlamydomonas. Proc. Natl. Acad. Sci. USA 94 8634–8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, J., and McElroy, D. (1994). Transgene inactivation: Plants fight back! Bio/Technology 12 883–888. [Google Scholar]
- Friend, S.H., Bernards, R., Rogelj, S., Weinberg, R.A., Rapaport, J.M., Albert, D.M., and Dryja, T.P. (1986). A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323 643–646. [DOI] [PubMed] [Google Scholar]
- Gilles, R., Gilles, C., and Jaenicke, L. (1984). Pheromone-binding and matrix-mediated events in sexual induction of Volvox carteri. Z. Naturforsch. 39c 584–592. [Google Scholar]
- Gillham, N.W. (1969). Uniparental inheritance in Chlamydomonas reinhardtii. Am. Nat. 103 355–388. [Google Scholar]
- Gillham, N.W., Boynton, J.E., Johnson, A.M., and Burkhart, B.D. (1987). Mating type linked mutations which disrupt the uniparental transmission of chloroplast genes in Chlamydomonas. Genetics 115 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, D.W., Wang, N.P., Qian, Y.W., Lee, E.Y., and Lee, W.H. (1991). The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell 67 293–302. [DOI] [PubMed] [Google Scholar]
- Gruber, H., Goetinck, S.D., Kirk, D.L., and Schmitt, R. (1992). The nitrate reductase-encoding gene of Volvox carteri: map location, sequence and induction kinetics. Gene 120 75–83. [DOI] [PubMed] [Google Scholar]
- Gruber, H., Kirzinger, S.H., and Schmitt, R. (1996). Expression of the Volvox gene encoding nitrate reductase: mutation-dependent activation of cryptic splice sites and intron-enhanced gene expression from a cDNA. Plant Mol. Biol. 31 1–12. [DOI] [PubMed] [Google Scholar]
- Gutiérrez, C. (1998). The retinoblastoma pathway in plant cell cycle and development. Curr. Opin. Plant Biol. 1 492–497. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Hallmann, A. (2006. a). Morphogenesis in the family Volvocaceae: Different tactics for turning an embryo right-side out. Protist 157 445–461. [DOI] [PubMed] [Google Scholar]
- Hallmann, A. (2006. b). The pherophorins: Common, versatile building blocks in the evolution of extracellular matrix architecture in Volvocales. Plant J. 45 292–307. [DOI] [PubMed] [Google Scholar]
- Hallmann, A., Amon, P., Godl, K., Heitzer, M., and Sumper, M. (2001). Transcriptional activation by the sexual pheromone and wounding: A new gene family from Volvox encoding modular proteins with (hydroxy)proline-rich and metalloproteinase homology domains. Plant J. 26 583–593. [DOI] [PubMed] [Google Scholar]
- Hallmann, A., Godl, K., Wenzl, S., and Sumper, M. (1998). The highly efficient sex-inducing pheromone system of Volvox. Trends Microbiol. 6 185–189. [DOI] [PubMed] [Google Scholar]
- Hallmann, A., and Sumper, M. (1994). Reporter genes and highly regulated promoters as tools for transformation experiments in Volvox carteri. Proc. Natl. Acad. Sci. USA 91 11562–11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann, A., and Wodniok, S. (2006). Swapped green algal promoters: aphVIII-based gene constructs with Chlamydomonas flanking sequences work as dominant selectable markers in Volvox and vice versa. Plant Cell Rep. 25 582–591. [DOI] [PubMed] [Google Scholar]
- Hanahan, D., and Weinberg, R.A. (2000). The hallmarks of cancer. Cell 100 57–70. [DOI] [PubMed] [Google Scholar]
- Harbour, J.W., and Dean, D.C. (2000). Rb function in cell-cycle regulation and apoptosis. Nat. Cell Biol. 2 E65–E67. [DOI] [PubMed] [Google Scholar]
- Helin, K., Wu, C.L., Fattaey, A.R., Lees, J.A., Dynlacht, B.D., Ngwu, C., and Harlow, E. (1993). Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7 1850–1861. [DOI] [PubMed] [Google Scholar]
- Hinds, P.W., Mittnacht, S., Dulic, V., Arnold, A., Reed, S.I., and Weinberg, R.A. (1992). Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell 70 993–1006. [DOI] [PubMed] [Google Scholar]
- Hobbs, S.L., Kpodar, P., and DeLong, C.M. (1990). The effect of T-DNA copy number, position and methylation on reporter gene expression in tobacco transformants. Plant Mol. Biol. 15 851–864. [DOI] [PubMed] [Google Scholar]
- Hoops, H.J., Nishii, I., and Kirk, D.L. (2005). Cytoplasmic bridges in Volvox and its relatives. In Cell-Cell Channels, F. Baluska, D. Volkmann, and P.W. Barlow, eds (Georgetown, TX: Eurekah.com), pp. 1–20.
- King, G.J. (2002). Through a genome, darkly: Comparative analysis of plant chromosomal DNA. Plant Mol. Biol. 48 5–20. [PubMed] [Google Scholar]
- Kirk, D.L. (1998). Volvox: Molecular-Genetic Origins of Multicellularity and Cellular Differentiation. (Cambridge, UK: Cambridge University Press).
- Kirk, D.L. (2000). Volvox as a model system for studying the ontogeny and phylogeny of multicellularity and cellular differentiation. J. Plant Growth Regul. 19 265–274. [Google Scholar]
- Kirk, D.L. (2003). Seeking the ultimate and proximate causes of Volvox multicellularity and cellular differentiation. Integr. Comp. Biol. 43 247–253. [DOI] [PubMed] [Google Scholar]
- Kirk, D.L., and Kirk, M.M. (1986). Heat shock elicits production of sexual inducer in Volvox. Science 231 51–54. [DOI] [PubMed] [Google Scholar]
- Kirk, M.M., Ransick, A., McRae, S.E., and Kirk, D.L. (1993). The relationship between cell size and cell fate in Volvox carteri. J. Cell Biol. 123 191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak, M., and Brehm, A. (2005). E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr. Opin. Genet. Dev. 15 520–527. [DOI] [PubMed] [Google Scholar]
- Lee, J.O., Russo, A.A., and Pavletich, N.P. (1998). Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature 391 859–865. [DOI] [PubMed] [Google Scholar]
- Lendvai, A., Pettko-Szandtner, A., Csordas-Toth, E., Miskolczi, P., Horvath, G.V., Gyorgyey, J., and Dudits, D. (2007). Dicot and monocot plants differ in retinoblastoma-related protein subfamilies. J. Exp. Bot. 58 1663–1675. [DOI] [PubMed] [Google Scholar]
- Lundberg, A.S., and Weinberg, R.A. (1999). Control of the cell cycle and apoptosis. Eur. J. Cancer 35 531–539. [PubMed] [Google Scholar]
- Mages, H.-W., Tschochner, H., and Sumper, M. (1988). The sexual inducer of Volvox carteri. Primary structure deduced from cDNA sequence. FEBS Lett. 234 407–410. [DOI] [PubMed] [Google Scholar]
- Merchant, S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg, R.L., and Glass, N.L. (1990). Mating type and mating strategies in Neurospora. Bioessays 12 53–59. [DOI] [PubMed] [Google Scholar]
- Mironov, V.V., De Veylder, L., Van Montagu, M., and Inze, D. (1999). Cyclin-dependent kinases and cell division in plants—The nexus. Plant Cell 11 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., and Lee, C. (2002). A genomic view of alternative splicing. Nat. Genet. 30 13–19. [DOI] [PubMed] [Google Scholar]
- Morris, E.J., and Dyson, N.J. (2001). Retinoblastoma protein partners. Adv. Cancer Res. 82 1–54. [DOI] [PubMed] [Google Scholar]
- Moses, A.M., Heriche, J.K., and Durbin, R. (2007). Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase. Genome Biol. 8 R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita, M., Nuñez, S., Heard, E., Lin, A.W., Hearn, S.A., Spector, D.L., Hannon, G.J., and Lowe, S.W. (2003). Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113 703–716. [DOI] [PubMed] [Google Scholar]
- Nematollahi, G., Kianianmomeni, A., and Hallmann, A. (2006). Quantitative analysis of cell-type specific gene expression in the green alga Volvox carteri. BMC Genomics 7 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld, T.P., de la Cruz, A.F., Johnston, L.A., and Edgar, B.A. (1998). Coordination of growth and cell division in the Drosophila wing. Cell 93 1183–1193. [DOI] [PubMed] [Google Scholar]
- Nicholas, K.B., Nicholas H.B., and Deerfield, D.W. (1997). GeneDoc: Analysis and visualization of genetic variation. Embnet News 4 14. [Google Scholar]
- Page, R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Parker, G.A., and Partridge, L. (1998). Sexual conflict and speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peach, C., and Velten, J. (1991). Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol. Biol. 17 49–60. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerville, J., and Kochert, G. (1982). Effects of senescence on somatic cell physiology in the green alga Volvox carteri. Exp. Cell Res. 140 39–45. [DOI] [PubMed] [Google Scholar]
- Provasoli, L., and Pintner, I.J. (1959). Artificial media for freshwater algae: Problems and suggestions. In The Ecology of Alga, C.A. Tyron and R.T. Hartman, eds (Pittsburgh, PA: Pymatuning Laboratory of Field Biology, University of Pittsburgh), pp. 84–96.
- Rausch, H., Larsen, N., and Schmitt, R. (1989). Phylogenetic relationships of the green alga Volvox carteri deduced from small-subunit ribosomal RNA comparisons. J. Mol. Evol. 29 255–265. [DOI] [PubMed] [Google Scholar]
- Robbens, S., Khadaroo, B., Camasses, A., Derelle, E., Ferraz, C., Inze, D., Van de Peer, Y., and Moreau, H. (2005). Genome-wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus tauri. Mol. Biol. Evol. 22 589–597. [DOI] [PubMed] [Google Scholar]
- Sage, J., Miller, A.L., Perez-Mancera, P.A., Wysocki, J.M., and Jacks, T. (2003). Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424 223–228. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schiedlmeier, B., Schmitt, R., Müller, W., Kirk, M.M., Gruber, H., Mages, W., and Kirk, D.L. (1994). Nuclear transformation of Volvox carteri. Proc. Natl. Acad. Sci. USA 91 5080–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr, C.J. (1996). Cancer cell cycles. Science 274 1672–1677. [DOI] [PubMed] [Google Scholar]
- Sherr, C.J. (2000). The Pezcoller Lecture: Cancer cell cycles revisited. Cancer Res. 60 3689–3695. [PubMed] [Google Scholar]
- Smith, C.W., and Valcárcel, J. (2000). Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 25 381–388. [DOI] [PubMed] [Google Scholar]
- Sogin, M.L. (1991). Early evolution and the origin of eukaryotes. Curr. Opin. Genet. Dev. 1 457–463. [DOI] [PubMed] [Google Scholar]
- Stamm, S., Ben-Ari, S., Rafalska, I., Tang, Y., Zhang, Z., Toiber, D., Thanaraj, T.A., and Soreq, H. (2005). Function of alternative splicing. Gene 344 1–20. [DOI] [PubMed] [Google Scholar]
- Starr, R.C. (1969). Structure, reproduction and differentiation in Volvox carteri f. nagariensis Iyengar, strains HK9 & 10. Arch. Protistenkd. 111 204–222. [Google Scholar]
- Starr, R.C. (1970). Control of differentiation in Volvox. Dev. Biol. 4 (suppl.): 59–100. [DOI] [PubMed] [Google Scholar]
- Starr, R.C. (1972). A working model for the control of differentiation during the development of the embryo of Volvox carteri f. nagariensis. Soc. Bot. Fr. Memoires 1972 175–182. [Google Scholar]
- Starr, R.C., and Jaenicke, L. (1974). Purification and characterization of the hormone initiating sexual morphogenesis in Volvox carteri f. nagariensis Iyengar. Proc. Natl. Acad. Sci. USA 71 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux, O., and Dyson, N.J. (2002). A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14 684–691. [DOI] [PubMed] [Google Scholar]
- Stockley, P. (1997). Sexual conflict resulting from adaptations to sperm competition. Trends Ecol. Evol. 12 154–159. [DOI] [PubMed] [Google Scholar]
- Tang, H., Bowers, J.E., Wang, X., Ming, R., Alam, M., and Paterson, A.H. (2008). Synteny and collinearity in plant genomes. Science 320 486–488. [DOI] [PubMed] [Google Scholar]
- Taya, Y. (1997). RB kinases and RB-binding proteins: New points of view. Trends Biochem. Sci. 22 14–17. [DOI] [PubMed] [Google Scholar]
- Tschochner, H., Lottspeich, F., and Sumper, M. (1987). The sexual inducer of Volvox carteri: Purification, chemical characterization and identification of its gene. EMBO J. 6 2203–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax, J.A., Woodbury, E.L., Quang, P.N., Paraz, M., Blethrow, J.D., Shah, K., Shokat, K.M., and Morgan, D.O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425 859–864. [DOI] [PubMed] [Google Scholar]
- Umen, J.G., and Goodenough, U.W. (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15 1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainright, P.O., Hinkle, G., Sogin, M.L., and Stickel, S.K. (1993). Monophyletic origins of the metazoa: An evolutionary link with fungi. Science 260 340–342. [DOI] [PubMed] [Google Scholar]
- Weinberg, R.A. (1995). The retinoblastoma protein and cell cycle control. Cell 81 323–330. [DOI] [PubMed] [Google Scholar]
- Weisshaar, B., Gilles, R., Moka, R., and Jaenicke, L. (1984). A high frequency mutation starts sexual reproduction in Volvox. Z. Naturforsch. 39c 1159–1162. [Google Scholar]
- Wikenheiser-Brokamp, K.A. (2006). Retinoblastoma family proteins: Insights gained through genetic manipulation of mice. Cell. Mol. Life Sci. 63 767–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater, M., Campilho, A., Perez-Perez, J.M., Heidstra, R., Blilou, I., Korthout, H., Chatterjee, J., Mariconti, L., Gruissem, W., and Scheres, B. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123 1337–1349. [DOI] [PubMed] [Google Scholar]
- Zheng, N., Fraenkel, E., Pabo, C.O., and Pavletich, N.P. (1999). Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.