Abstract
In most cells, 100-1000 Okazaki fragments are produced for each replicative DNA polymerase present in the cell. For fast-growing cells, this necessitates rapid recycling of DNA polymerase on the lagging strand. Bacteria produce long Okazaki fragments (1-2 kb) and utilize a highly processive DNA polymerase III (pol III), which is held to DNA by a circular sliding clamp. In contrast, Okazaki fragments in eukaryotes are quite short, 100-250 bp, and thus the eukaryotic lagging strand polymerase does not require a high degree of processivity. The lagging strand polymerase in eukaryotes, polymerase δ (pol δ), functions with the proliferating cell nuclear antigen (PCNA) sliding clamp. In this report, Saccharomyces cerevisiae pol δ is examined on model substrates to gain insight into the mechanism of lagging strand replication in eukaryotes. Surprisingly, we find pol δ is highly processive with PCNA, over at least 5 kb, on Replication Protein A (RPA)-coated primed single strand DNA. The high processivity of pol δ observed in this report contrasts with its role in synthesis of short lagging strand fragments, which require it to rapidly dissociate from DNA at the end of each Okazaki fragment. We find that this dilemma is solved by a “collision release” process in which pol δ ejects from PCNA upon extending a DNA template to completion and running into the downstream duplex. The released pol δ transfers to a new primed site, provided the new site contains a PCNA clamp. Additional results indicate that the collision release mechanism is intrinsic to the pol3/pol31 subunits of the pol δ heterotrimer.
Chromosome replication in eukaryotes utilizes both DNA polymerase (pol)2 ε and DNA pol δ, which are thought to function on the leading and lagging strands of the replication fork, respectively (1, 2). During replication fork movement, discontinuous lagging strand fragments are initiated by RNA primers generated by the primase function of DNA polymerase α/primase (pol α/primase), and the primers are extended by DNA pol α to a total length of 30-35 nucleotides. Unlike pol α, pol δ and pol ε both utilize PCNA, a ring-shaped sliding clamp that increases the processivity of these DNA polymerases, and the PCNA clamp is assembled onto DNA by the 5-subunit RFC clamp loader (3-7). In these respects the eukaryotic replication apparatus is similar to the bacterial replisome, which contains sliding clamps and a clamp loader, except the bacterial replisome utilizes two identical copies of a DNA polymerase (pol III) for the leading and lagging strands (8). Eukaryotes also contain a heterohexameric MCM 2-7 complex, which is thought to be the replicative helicase, analogous to the Escherichia coli DnaB homohexamer (9).
Despite the similarities between bacterial and eukaryotic replisomes, they appear to diverge significantly in many other respects. For example, the bacterial clamp loader is tightly associated with the leading and lagging stand polymerases, whereas the RFC clamp loader lacks stabile interactions with either pol δ or pol ε. Furthermore, the eukaryotic replication fork utilizes numerous factors that have no apparent homolog in bacteria, including the GINS complex and Cdc45 protein, among others (10). Also, RPA, the eukaryotic single strand DNA-binding protein, is a heterotrimer whereas the bacterial SSB is a homotetramer.
We have a long term interest in understanding the detailed mechanism by which DNA polymerases act at a replication fork. In this report we focus on Saccharomyces cerevisiae pol δ and its expected actions on the lagging strand during chromosome replication. Most cells produce 100-1000 Okazaki fragments for each replicative DNA polymerase, and this necessitates efficient recycling of the scarce DNA polymerase. It is particularly fascinating when an enzyme with high processivity functions on the lagging strand, because it must dramatically loosen its tight grip to DNA at the end of each lagging strand fragment to dissociate from DNA and recycle to new primed sites. A clear example of this is the bacterial pol III replicase, which functions with the β sliding clamp to achieve processivity of over 50 kb, yet Okazaki fragments are only 1-2 kb. Recycling of the tightly bound pol III on the lagging strand is solved by a “collision release” mechanism that disengages pol III from the β clamp upon completing a DNA template (11). The released pol III then rapidly associates with a new β clamp assembled at an upstream RNA primed site. Collision release involves the τ subunit, which facilitates separation of pol III from the clamp upon completing a DNA segment (12). A second type of polymerase recycling mechanism, referred to as “premature release” (also called “signaling release”) occurs when the lagging strand polymerase releases from the clamp prior to finishing an Okazaki fragment. Premature release is the dominant, if not sole, mechanism of polymerase recycling in the T4 phage replication system (13). Premature release may be required in the T4 phage system due to production of very short 5-nucleotide RNA primers that would dissociate if they were not immediately extended by polymerase. Primers in E. coli are much longer (∼12 nucleotides) and thus remain on DNA. However, premature release can be observed in the E. coli system when the Okazaki fragment cycle is perturbed or the lagging strand polymerase is stalled (14, 15). Premature release is thought to be signaled by events related to priming or by assembly of clamps on new RNA primers.
The current report uses pure recombinant S. cerevisiae replication proteins to study pol δ-PCNA processivity, and to gain insight into the mechanism of polymerase recycling on the lagging strand in eukaryotes. We expected pol δ to lack a high degree of processivity, partly based on previous reports of human pol δ (16, 17), but also based on the small size of Okazaki fragments in eukaryotes (18, 19). Instead, we find that yeast pol δ is remarkably processive with PCNA and extends a single primer around an entire 5.4-kb primed ϕX174 ssDNA genome. Unlike bacterial pol III, the high processivity of pol δ with PCNA does not require specific contact to the RPA ssDNA-binding protein. Instead, pol δ is just as rapid and processive with PCNA when E. coli SSB is used in place of RPA. This result may explain the ability of pol δ to replicate both leading and lagging strands of the SV40 viral genome (20-22). The high processivity of pol δ-PCNA also suggests that pol δ may substitute for pol ε in cells that lack the N-terminal polymerase domain of pol ε, yet remain viable (23, 24).
The high processivity of yeast pol δ with PCNA raises the question of how pol δ rapidly recycles to new primed sites upon completion of each short Okazaki fragment. We examine this issue here and find that the highly processive pol δ-PCNA contains an intrinsic capacity for collision release. Study of the collision release mechanism shows that pol δ does not recognize the 5′ RNA terminus to induce collision release; 5′ DNA is just as efficient. Collision release also does not require RPA, nor does it require the ssDNA binding region of the RFC1 subunit of RFC, a putative functional analogue of the E. coli τ subunit involved in collision release. We also ask if premature release of pol δ can be signaled by stalling pol δ-PCNA in the presence of a primed challenge DNA but find that the stalled pol δ retains its tight grip to PCNA. In addition, the current report demonstrates that the pol32 subunit of pol δ is not required for processivity or collision release. Overall, the study indicates that the unexpected high processivity of pol δ and the collision release mechanism are intrinsic properties of pol δ; additional factors are not required, and the responsibility for these actions is relegated to one or both of the pol3 and pol31 subunits of pol δ.
EXPERIMENTAL PROCEDURES
DNA Substrates—M13mp18 single strand circular DNA was prepared as described (25). ϕX174 virion single strand circular DNA was purchased from New England Biolabs (Ipswich, MA). Unless otherwise noted, sequences of oligonucleotide primers used to initiate synthesis on circular ssDNA were: primer 1B (ϕX174) 5′-ACCAACATAAACGTTATTGCCCGGCGTACG-3′, primer 3B (M13mp18) 5′-AGTTAAAGGCCGCTTTTGCGGGATCGTCACCC-3′, and the RNA/DNA primer (ϕX174) 5′-rCrArArGrCrArGrUrArGdTdAdAdTdTdCdCdTdGdCdTdTdTdAdTdCdAdAdG-3′.
Genes encoding the three subunits of S. cerevisiae pol δ (pol3, pol31, and pol32) were cloned into separate compatible E. coli expression plasmids under control of the bacteriophage T7 promoter. The POL32 gene was cloned in-frame with an N-terminal GST affinity tag followed by codons for the amino acids LEVLFQGPH and then by the coding sequence of POL32. The italicized residues are recognized by the PreScission Protease (GE Healthcare, Piscataway, NJ), which cleaves between the adjacent Q and G, leaving three extra amino acid residues at the N terminus of the pol32 protein after proteolytic removal of the GST moiety. The pol3 protein used in these studies has the sequence Glu-Leu at positions 78 and 79, respectively, which differs from the sequence Asp78-Val79 reported in the Saccharomyces Genome Data Base (www.yeastgenome.org), as previously described (26).
Proteins—To express and purify pol δ, the pol3, pol31, and GST-pol32 plasmids were co-transformed into electrocompetent E. coli strain BL21(DE3) (Lucigen, Middleton, WI) by electroporation, and co-transformants were selected at 37 °C on LB agar plates containing ampicillin, kanamycin, and streptomycin. After overnight growth of individual transformants in liquid medium, cultures were diluted into 12 liters of fresh selective medium and grown at 37 °C to an A600 of ∼0.7, then placed on ice for 30 min, induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and grown overnight at 15 °C. Induced cultures were harvested by centrifugation, resuspended in 3 ml of Tris-sucrose (50 mm Tris, pH 7.5, 10% sucrose), and stored at -80 °C. Frozen cell pellets were thawed, adjusted to 0.5 m NaCl, 2 mm phenylmethylsulfonyl fluoride, and 1× GBB (GST binding buffer: 50 mm Tris, pH 7.5, 1 mm EDTA, 10% glycerol). Cells were lysed in a cold French Press. After lysis, spermidine was added to 30 mm, and the lysate was clarified by centrifugation at 12,500 rpm for 1 h at 4 °C in a Sorvall SLA-1500 rotor. The clarified supernatant (100-150 ml) was applied onto a 7-ml column of glutathione-Sepharose 4B (GE Healthcare) equilibrated with 1× GBB plus 0.5 m NaCl. This and all subsequent steps were performed at 4 °C. The column was washed with 5-7 column volumes of 1× GBB buffer containing 0.5 m NaCl, then equilibrated with 3 column volumes of elution buffer (50 mm Tris, pH 7.5, 1 mm EDTA, 10% glycerol, and 5 mm DTT) containing 0.3 m NaCl. Protein was eluted with 2 column volumes of elution buffer containing 40 mm reduced l-glutathione, and fractions of 0.3 ml were collected and analyzed by SDS-PAGE. Peak fractions (∼4 ml) were pooled, mixed with 40 units of PreScission Protease to remove the GST tag from pol32, and dialyzed overnight against 4 liters of Buffer A (20 mm Tris, pH 7.5, 0.5 mm EDTA, 2 mm DTT, 10% glycerol) containing 300 mm NaCl. After dialysis and protease cleavage, pooled fractions were diluted with Buffer A to a conductivity equivalent to Buffer A containing 80 mm NaCl and applied at 0.25 ml/min to a 1-ml mono Q column equilibrated with Buffer A plus 80 mm NaCl. The mono Q column was washed with 10 column volumes of Buffer A plus 80 mm NaCl, and bound protein was eluted with a 20-column volume gradient of Buffer A plus 80 mm NaCl to Buffer A plus 200 mm NaCl. Fractions containing the two-subunit form of pol δ (pol3 and pol31) eluted early from the column and were stored individually at -80 °C. Peak fractions of stoichiometric pol δ heterotrimer were pooled, aliquoted, and stored at -80 °C (total yield ∼ 0.25 mg). Recombinant RFC and the RFC mutant containing a truncated RFC1 subunit lacking the ssDNA binding region (27, 28), PCNA (29), and E. coli SSB (30) were prepared as described.
DNA Replication Assays—DNA replication reactions were performed as follows: PCNA (240 fmol, as trimer), RFC (360 fmol, as pentamer), ϕX174 circular ssDNA (165 fmol, as circles) primed with a DNA 30-mer, either RPA (640 nm, as heterotrimer) or E. coli SSB (65 pmol, as tetramer), and the indicated amount of pol δ (as heterotrimer) were incubated 5 min at 30 °C in 135 μl of Replication Buffer (20 mm Tris-Cl (pH 7.5), 50 mm potassium glutamate, 5 mm DTT, 0.1 mm EDTA, 40 μg/ml bovine serum albumin, 8 mm MgCl2, 0.5 mm ATP, 5% glycerol, and 60 μm each of dGTP and dCTP) unless indicated otherwise in the legends. DNA synthesis was initiated by adding 15 μl of Start Buffer (60 μm dATP, 20 μm dTTP, and 15 μCi of [α-32P]dTTP in Replication Buffer) and incubated at 30 °C (all concentrations are with respect to the final 150-μl reaction). At the indicated times, 25-μl aliquots were removed and quenched by addition of an equal volume of 1% SDS/40 mm EDTA. Total incorporation of [α-32P]dTTP (as dTMP) was determined by spotting a portion of the reaction on DE81 filters. Filters were washed and dried, and total incorporation was analyzed in a liquid scintillation counter. RFII products were analyzed in a 0.8% agarose gel in 1× TBE. Gels were dried, exposed to PhosphorImager screens, and imaged using a Typhoon 9400 Laser scanner (GE Healthcare).
For challenge experiments, separate donor and challenge reactions were prepared containing (donor, challenge): primed circular ssDNA (68 fmol ϕX174, 334 fmol M13mp18), either RPA (40 pmol, 130 pmol), or E. coli SSB (30 pmol, 120 pmol), PCNA (180 fmol, 690 fmol), and RFC (190 fmol, 725 fmol) in Replication Buffer (potassium glutamate was omitted in reactions containing SSB). Donor and challenge reactions were incubated separately for 5 min at 30 °C and then mixed. pol δ (270 fmol) was added either to the donor reaction before pre-incubation or to the combined reaction after mixing as indicated. DNA synthesis was initiated upon adding Start Buffer as described above, either immediately or after co-incubation of the mixed reaction for the indicated times, and then aliquots were quenched at various times after initiation and analyzed as described above.
Analysis of Terminal Replication Products—For analysis of the terminal products of replication, the basic replication assay was modified as follows: pol δ (900 fmol), PCNA (408 fmol), RFC (416 fmol), and SSB (68 pmol) were incubated along with singly primed ϕX174 (175 fmol) and 40 units of XhoI restriction endonuclease in 125 μl of Replication Buffer for 5 min at 30 °C. The primer for these reactions has the sequence 5′-CAAGCAGTAGTAATTCCTGCTTTATCAAG-3′. DNA synthesis was initiated by adding 25 μl of Start Buffer. A parallel reaction (75 μl) using E. coli pol III* (275 pmol) and β-clamp (1.1 pmol as dimer) along with singly primed ϕX174 (87 fmol) and 20 units of XhoI was performed as a control. To confirm cutting by XhoI, 25-μl aliquots were removed at the indicated times, quenched, and analyzed as above. For analysis of the terminal products of replication, 12.5-μl aliquots were removed at the indicated times and quenched by addition of an equal amount of 2× denaturing PAGE loading buffer. Samples were loaded onto a 12% polyacrylamide/8 m urea gel. Gels were exposed and analyzed as above. For comparison, a DNA ladder was generated using the 3′-5′ exonuclease minus derivative of Klenow fragment and a 5′-end radiolabeled primer (5′-GAGCTTCTCGAGCTGCGCAAGG-3′) was annealed to ϕX174 ssDNA in the presence of chain-terminating mixtures of dNTPs/ddNTPs.
RESULTS
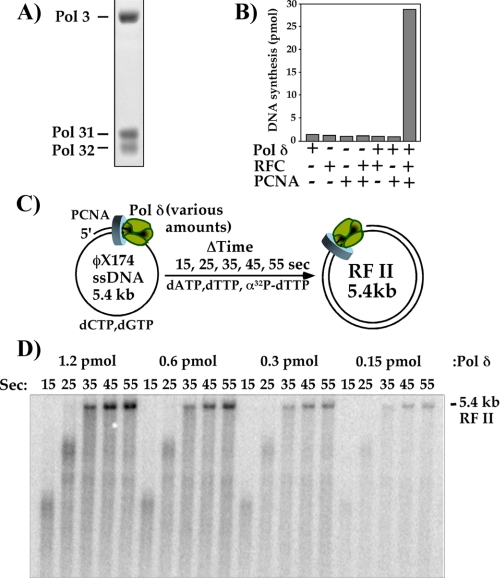
DNA Synthesis by pol δ-PCNA Is Remarkably Processive with PCNA—The studies of this report utilize a recombinant S. cerevisiae pol δ heterotrimer produced in E. coli, which consists of the pol3 polymerase and its two small subunits, pol31 and pol32. The purified recombinant pol δ contains all three subunits in approximately equal molar ratio (Fig. 1A). We have previously expressed recombinant S. cerevisiae RFC and PCNA to high levels in E. coli (28, 29). The recombinant pol δ shows robust replication activity that depends on both PCNA and RFC (Fig. 1B).
FIGURE 1.
pol δ is highly processive with the PCNA clamp. A, SDS-PAGE of recombinant pol δ heterotrimer. B, assays using primed ϕX174 ssDNA and the indicated components. C, scheme illustrating the reactions shown in D. D, effect of dilution of pol δ with respect to PCNA clamped onto a constant concentration of primed circular ϕX174 ssDNA. Reactions were performed as described under “Experimental Procedures.” Aliquots from each reaction were quenched at the indicated times and analyzed in a 0.8% native agarose gel followed by autoradiography. The position of the fully replicated RF II duplex product is indicated.
In the experiment of Fig. 1C, we examine the processivity of pol δ with PCNA during synthesis. The assay utilizes the 5.4-kb ϕX174 bacteriophage ssDNA genome primed with an oligonucleotide and coated with RPA. The RFC clamp loader was first used to assemble a PCNA clamp onto the primed site, and then a series of time-course experiments was performed using progressively lower amounts of pol δ and a constant amount of RPA, RFC, PCNA, and DNA (see scheme in Fig. 1C). To visualize DNA products, [α-32P]dTTP was added to the dNTP mixture, and time points were withdrawn and analyzed in a neutral agarose gel.
If the action of pol δ-PCNA is not processive for the full length of the 5.4-kb primed template, pol δ will come on and off DNA during extension of the single primer around the full circle. At high concentrations of pol δ, as soon as one pol δ dissociates from PCNA, another will rapidly take its place and resume synthesis of the full-length product. But as pol δ becomes limiting, a new pol δ will not take the place of a pol δ that dissociates. Furthermore, once pol δ dissociates from a partially extended substrate, it does not necessarily reassociate with the same partially replicated substrate. Instead, the dissociated pol δ will sample the multiple primed substrates, and thus will require a longer time to form full-length 5.4-kb duplex RFII product compared with reactions that contain a higher amount of pol δ. The result in Fig. 1D shows that the rate of RFII duplex product formation is the same for all pol δ concentrations, even when pol δ is limiting. Hence, pol δ is fully processive with PCNA on the 5.4-kb substrate and extends the primer full circle in a single DNA-binding event. The time course also shows that the pol δ-PCNA replicase forms 5.4-kb RFII product within 35 s (Fig. 1D), indicating a rate of incorporation of ∼150 nucleotides/s. This result implies that complete extension of a 100- to 250-bp Okazaki fragment requires only 1-2 s.
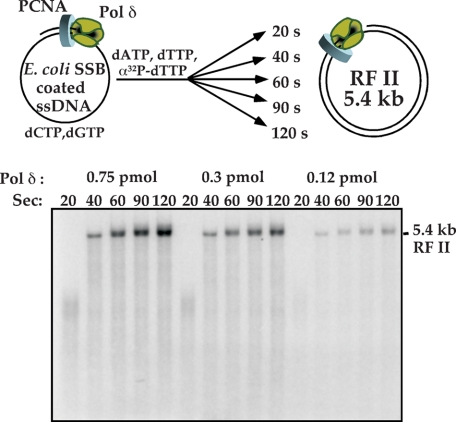
High Processivity of pol δ-PCNA Does Not Require Its Cognate ssDNA-binding Protein—Processivity of pol δ-PCNA during synthesis of an entire 5.4-kb ϕX174 ssDNA genome is unexpected given the short length of Okazaki fragments in eukaryotes. This high degree of processive synthesis is also observed with the E. coli pol III replicase, which functions with the circular β clamp, similar to pol δ with PCNA (31). E. coli pol III requires its cognate SSB for high processivity and is no longer highly processive when heterologous single-stranded DNA-binding proteins are used to coat the ssDNA, such as human RPA or phage T4 gene 32 protein (32). In the E. coli system, the pol III replicase contains the χ subunit, which binds to SSB and contributes to the processivity of E. coli pol III with its β clamp (32, 33). Eukaryotes have no apparent sequence homolog of the E. coli χ subunit (34), and thus may not exhibit a strong requirement for its cognate RPA single strand-binding protein. Although previous studies have shown that pol δ-PCNA can function on E. coli SSB-coated DNA, processivity was not measured (6).
In Fig. 2 we test whether pol δ-PCNA requires RPA for processive function by replacing RPA with SSB from E. coli using primed ϕX174 ssDNA. The results demonstrate that as pol δ is titrated down to limiting amounts in the reaction, full-length 5.4-kb RFII product is still formed at the same rate as at high pol δ concentration. Therefore, pol δ-PCNA does not require its cognate RPA single strand DNA-binding protein for rapid and processive function with PCNA. Furthermore, pol δ-PCNA displays essentially the same rate of elongation on the E. coli SSB-coated ssDNA template as on RPA-coated primed ssDNA (compare Fig. 1D with Fig. 2).
FIGURE 2.
pol δ-PCNA does not require RPA for speed and processivity. A, scheme illustrating reactions performed using E. coli SSB in place of yeast RPA. pol δ-PCNA is assembled on primed ϕX174 ssDNA coated with E. coli SSB, then dATP, dTTP, and [32P]dTTP are added to initiate synthesis. Time points are collected as indicated in the figure. B, reaction conditions are detailed under “Experimental Procedures,” and DNA products were analyzed in a 0.8% native agarose gel followed by autoradiography. The position of the fully replicated RFII duplex product is indicated.
pol δ Dissociates from DNA Specifically upon Completing DNA—The high processivity of pol δ with PCNA stands in contrast to its action on the lagging strand where pol δ must rapidly dissociate upon finishing an Okazaki fragment so it can transfer to a new primer for extension of the next Okazaki fragment. Rapid transfer to a new primed site requires that pol δ lose its tight grip on DNA upon completing a DNA template.
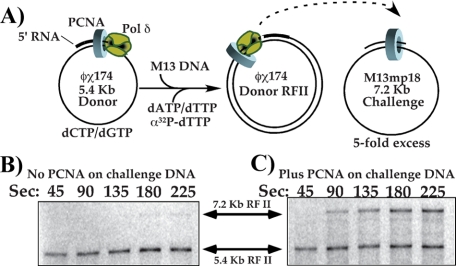
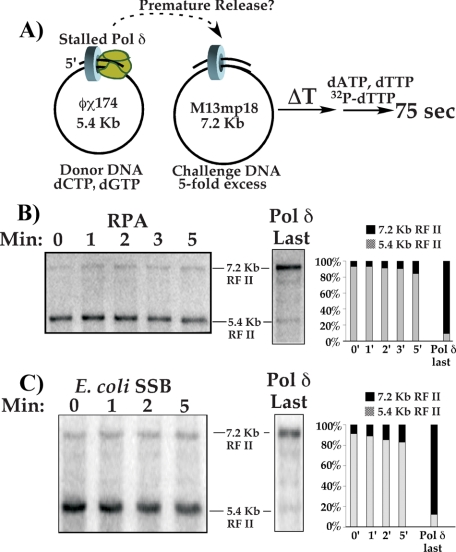
To test whether pol δ loses affinity for DNA upon completing DNA synthesis, we assembled pol δ with PCNA on a donor ϕX174-primed DNA and added it to a 5-fold excess of primed M13mpl8 challenge DNA. For this experiment, the ϕX174 template was primed using an oligonucleotide that contains 5′ ribonucleotides to mimic the structure pol δ encounters during normal replication, Okazaki fragments terminated at the 5′ terminus by RNA synthesized by pol α/primase (35). After mixing the two DNAs and initiating DNA synthesis (see scheme in Fig. 3A), time points were withdrawn and analyzed in a native agarose gel. If pol δ and its associated PCNA clamp remain tightly bound to the donor ϕX174 RFII duplex product, it will not transfer to the challenge DNA, in which case synthesis of M13mp18 7.2 kb RF II will not be observed. On the other hand, if pol δ-PCNA dissociates from ϕX174 DNA upon completing replication and the polymerase transfers to the M13mp18 challenge DNA along with its clamp, the 7.2-kb M13mp18 RFII product will become visible shortly after the 5.4-kb ϕX174 RFII product appears. Due to their different sizes, the duplex RFII products of the ϕX174 and M13mp18 are readily distinguished in a native agarose gel.
FIGURE 3.
Collision release by pol δ enables it to transfer to new primed sites endowed with a PCNA clamp. A, reaction scheme: polδ was assembled with PCNA on the RPA-coated 5.4-kb ϕX174 donor ssDNA primed with an oligonucleotide containing 5′ ribonucleotides, and then a 5-fold molar excess of M13mp18 challenge primed template was added; the challenge DNA either contained PCNA or lacked PCNA. Replication was initiated immediately after addition of the challenge DNA, and reactions were quenched at times indicated in B and C. B, challenge DNA lacks PCNA, and aliquots from the reaction were quenched at the indicated times, then products were analyzed in a 0.8% native agarose gel. C, the experiment in B was repeated, but the challenge M13mp18 primer contained a pre-loaded PCNA clamp.
The result shows that pol δ-PCNA does not transfer to the challenge DNA, because the M13mp18 RFII product is not formed (Fig. 3B). This observation indicates that pol δ and/or PCNA retains its tight grip to DNA even after it is finished replicating it. However, a very different outcome is observed when a PCNA clamp is first assembled onto the challenge M13mp18 ssDNA. In this event, M13mp18 RFII product is formed directly following the appearance of ϕX174 RFII (Fig. 3C). This finding implies that pol δ dissociates from the donor ϕX174 RFII DNA upon completing replication, but that the PCNA clamp remains tightly bound to the completed DNA. Once released from ϕX174 RFII DNA, pol δ transfers to PCNA on the challenge DNA to form M13mp18 RFII duplex products.
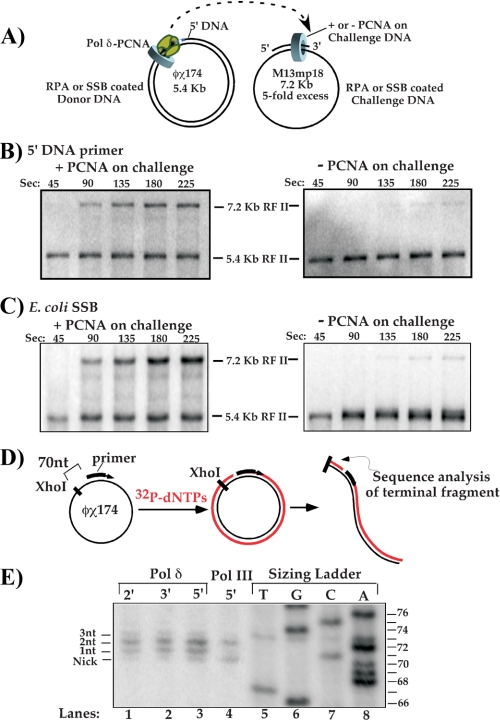
Characterization of pol δ Release from PCNA on Replicated DNA—The results of Fig. 3 indicate that the highly processive pol δ-PCNA complex breaks apart upon completing replication. This action frees pol δ to release from PCNA so it can transfer to new primed sites that contain a PCNA clamp. Characterization of these events in the E. coli system reveal that release of E. coli pol III from its β clamp only occurs upon complete replication of DNA (11). In addition, separation of pol III from the β clamp is facilitated by the C-terminal region of the τ clamp loading subunit; this region is not essential for clamp loading (36, 37) but interacts with ssDNA and pol III (12, 38). In the next few experiments we examine the degree to which pol δ-PCNA completes a primed ϕX174 ssDNA circle, and we also ask whether the ssDNA binding region of the RFC1 clamp loader subunit is involved in pol δ release from PCNA. In addition we determine if collision with a 5′ DNA, instead of 5′ RNA, is sufficient to trigger collision release of pol δ from PCNA.
First, we determine whether pol δ-PCNA needs to collide with a 5′ RNA to trigger dissociation from DNA (Fig. 4A). To test this, the donor ϕX174 ssDNA was primed using a DNA oligonucleotide instead of a primer containing 5′ ribonucleotides. In this case, when pol δ-PCNA goes full circle around the ϕX174 donor it will encounter a 5′ DNA terminus. If pol δ must encounter a 5′ RNA terminus to trigger ejection from PCNA, it will no longer dissociate from the ϕX174 RFII duplex, and the M13mp18 acceptor will not be replicated to an RFII product. The result clearly shows production of M13mp18 RFII, and therefore pol δ dissociates from the ϕX174 DNA product upon encountering 5′ DNA (Fig. 4B, left panel). To determine if PCNA still remains on the donor DNA, this experiment was repeated in the absence of a PCNA clamp on the M13mp18 challenge DNA (Fig. 4B, right panel). The result shows no M13mp18 RFII and thus confirms that transfer of pol δ to the M13mp18 DNA requires a PCNA clamp on the M13mp18 challenge DNA.
FIGURE 4.
Characterization of the collision release reaction. A, scheme of collision release assay and variables tested in the subsequent experiments in the panels below. B, test of collision release upon pol δ-PCNA encounter with a 5′ DNA terminus. In this case, the donor ϕX174 ssDNA was primed with a DNA oligonucleotide, and reactions were performed as described under “Experimental Procedures” using pol δ, PCNA, and RPA, except the RFC mutant lacking the ssDNA binding region of RFC1 was used to load PCNA onto both the donor and challenge DNAs. Left panel: PCNA was assembled on the challenge DNA. Right panel: PCNA is not present on the challenge DNA. C, reactions were performed as in B, except both donor and challenge templates are coated with E. coli SSB instead of RPA. Left panel: PCNA is assembled on the challenge DNA. Right panel: PCNA is not present on the challenge DNA. D, scheme for determining the extent of synthesis by pol δ upon converting primed ϕX174 ssDNA to duplex DNA. DNA is replicated in the presence of [α-32P]dTTP, then linearized 70 bp upstream from the 5′ terminus of the initiating primer using XhoI. The small replicated fragment (indicated) is then analyzed in a sequencing gel. E, termination sites of synthesis by pol δ-PCNA are analyzed in a sequencing gel (lanes 1-3). For comparison, a sizing ladder was produced in a separate reaction as described under “Experimental Procedures” (lanes 5-8). E. coli pol III holoenzyme is included as a control (lane 4). The contrast of the pol δ and pol III lanes in the autoradiogram was adjusted to approximate the signal from the sizing ladder.
Provided pol δ-PCNA completely replicates the donor ϕX174 DNA, there should be no ssDNA for RPA (or E. coli SSB) to bind. In this case, RPA is not expected to play a role in collision release of pol δ from PCNA upon completing DNA synthesis. This prediction is tested in the two template pol δ recycling assay by replacing RPA with E. coli SSB (Fig. 4C, left panel). The results demonstrate that use of E. coli SSB does not alter the collision release process; pol δ transfers to the acceptor M13mp18 DNA after completing the ϕX174 template. In addition, PCNA is required on the challenge DNA to observe the M13mp18 RFII product, indicating that pol δ leaves PCNA behind on the donor ϕX174 RFII product (Fig. 4C, right panel).
The E. coli replicase τ subunit is used in the collision release mechanism to recognize ssDNA and sense when replication in complete; it then enables separation of pol III from the β clamp. The ssDNA binding region of τ is an extension that protrudes from the clamp loader, and is not required for clamp loading activity. RFC also has one subunit, RFC1, that contains a 30-kDa N-terminal region that binds ssDNA, yet is not essential for clamp loading activity (39, 40). In the experiments of Fig. 4 (B and C), we used an RFC complex that contains an RFC1 subunit in which the N-terminal 30-kDa region is deleted. The fact that pol δ efficiently recycles from the ϕX174 donor to the M13mp18 acceptor in these experiments demonstrates that the RFC1 subunit N-terminal extension is not required for pol δ to dissociate from PCNA upon completing DNA.
pol δ-PCNA Duplicates Every Nucleotide of Product DNA and Does Not Release Prematurely—We next sought to determine whether pol δ-PCNA completely extends the primer around the ϕX174 ssDNA circle before dissociating from the ϕX174 duplex product. Alternatively, pol δ may undergo “premature release,” and leave a short ssDNA gap in the RFII. To examine this issue, we analyzed the terminal region of the replicated 5.4-kb ϕX174 RFII DNA in a sequencing gel. To decrease the size of the replicated DNA, enabling it to enter a sequencing gel, the RFII product was treated with XhoI, which yields a 70-base radioactive DNA fragment if pol δ were to completely replicate the ϕX174 template (see scheme in Fig. 4D). To determine the exact size of the XhoI fragment, we compared it to a reference ladder produced by a DNA sequencing reaction using a 5′ radiolabeled primer that abuts the precise position where XhoI cleaves ϕX174. Thus, the bands in the ladder have exactly the same sequence as DNA produced by XhoI cleavage of the ϕX174 duplex product. The result shows that pol δ does not release prematurely; it replicates the ϕX174 DNA to completion and sometimes proceeds 1-3 nucleotides beyond the 5′ terminus by strand displacement synthesis (Fig. 4E). The E. coli pol III replicase control also produced nicked circles and 2 nucleotide strand displacement products.
The fact that the pol δ-PCNA fully replicates ϕX174 DNA, even in the presence of challenge DNA (i.e. Figs. 3 and 4), indicates that pol δ does not prematurely release from DNA while it is moving. To determine whether premature release of pol δ can be triggered by stalling pol δ-PCNA in the presence of new primed sites containing PCNA clamps, we performed the experiment illustrated in Fig. 5A. In one reaction, limiting pol δ is assembled onto primed 5.4-kb ϕX174 ssDNA containing a PCNA clamp, but in the presence of only 2 dNTPs to mimic a block to chain elongation. The other reaction contained a 5-fold molar excess (relative to ϕX174 DNA) of 7.2-kb challenge M13mp18 primed ssDNA containing a PCNA clamp. The two reactions were then mixed and co-incubated for varying amounts of time (see scheme in Fig. 5A). If premature release of the stalled pol δ from PCNA is induced by challenge primed sites (containing PCNA), the stalled pol δ should transfer to the 5-fold excess challenge M13mp18 DNA. Timed aliquots were withdrawn from the co-incubation, and a short 75-s pulse of DNA synthesis was initiated by addition of the final two dNTPs. The 75-s pulse of DNA synthesis is sufficient time for pol δ-PCNA to replicate whichever DNA it is on, either ϕX174 or M13mp18 DNA, but not both.
FIGURE 5.
PCNA on challenge DNA does not signal premature release of stalled pol δ. A, scheme to test premature release by a stalled pol δ-PCNA. pol δ-PCNA is stalled at a primer terminus by omission of two dNTPs, then mixed with a 5-fold molar excess of a primed M13mp18 challenge template to which PCNA has been attached. The two DNAs are then co-incubated for the indicated times before initiation of replication; only sufficient time of synthesis is provided to replicate one DNA (i.e. whichever DNA substrate pol δ is attached to at initiation of synthesis). B, pol δ-PCNA is stalled on RPA-coated ssDNA. Left gel: autoradiogram of a 0.8% agarose gel which resolves the 5.4-kb ϕX174 donor RFII and 7.2-kb M13mp18 challenge RFII products. The gel to the right is a control in which PCNA clamps were loaded on 1× donor and 5× challenge templates in separate reactions and then mixed before addition of pol δ. The graph shows the relative incorporation on each substrate. C, pol δ-PCNA was stalled on E. coli SSB-coated ssDNA, and reactions were performed as described above and under “Experimental Procedures.”
The result shows that the stalled pol δ is remarkably stabile with PCNA on ϕX174 DNA, because ϕX174 RFII is the most abundant product (84-93%) at all time points (Fig. 5B, left). Thus, stalled pol δ remains stably bound to PCNA and is not induced to release prematurely by exogenous new primed sites. As a control, PCNA was loaded onto both templates before adding pol δ, then a 75-s pulse of replication was initiated with all 4 dNTPs (Fig. 5B, right). As expected, at least 5-fold greater M13mp18 RFII is produced compared with ϕX174 RF II products, consistent with the presence of excess challenge M13mp18 DNA in these reactions.
Does the stability of the stalled pol δ-PCNA complex require RPA, for example by specific interaction between pol δ and RPA, thereby preventing premature release of a stalled polymerase? In Fig. 5C we examine the stability of stalled pol δ-PCNA on primed ϕX174 ssDNA coated with E. coli SSB rather than yeast RPA. The result demonstrates that pol δ-PCNA remains on E. coli SSB-coated ϕX174 primed ssDNA throughout the entire time course. Hence, RPA does not suppress premature release of a blocked pol δ-PCNA, nor does it enhance its stability on DNA. The control reaction in which pol δ is added last to a mixture of the two DNAs gives the expected excess of M13mp18 RF II product over ϕX174 RFII (Fig. 5C, right panel).
Perhaps premature release requires other eukaryotic replisome factors, or occurs in the context of the replication fork when the leading polymerase continues moving forward, as observed in the E. coli system (14). In this case, the mechanism of premature release during eukaryotic lagging strand synthesis will require study of complete eukaryotic replisomes, which have yet to be reconstituted in vitro.
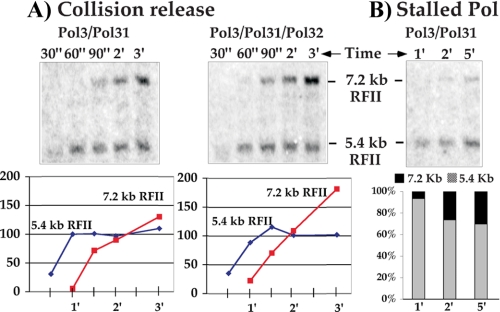
A Two-subunit Form of pol δ Lacking the pol32 Subunit Is Competent for Collision Release—The pol32 subunit of the pol δ heterotrimer contains a consensus PCNA interacting motif, suggesting that it may be important for one or more of the pol δ actions with PCNA explored in this study (41, 42). Therefore, we used a pol3-pol31 two-subunit form of pol δ (lacking the pol32 subunit) that elutes separately during purification of the three-subunit enzyme on mono Q and tested it in the two template collision release assay. In the experiment of Fig. 6A, we examine the two-subunit pol δ (pol3/pol31) for collision release from PCNA upon completing DNA. The result shows that the two-subunit pol δ has comparable efficiency to the three-subunit pol δ in completing the initial donor template (ϕX174) and transferring to PCNA on the challenge M13mp18 DNA (Fig. 6A). Furthermore, the fact that the donor ϕX174 is replicated to an RFII duplex in the presence of the challenge M13mp18 DNA indicates that the two-subunit pol δ is highly processive with PCNA and does not dissociate from the donor template until it has completed it.
FIGURE 6.
The pol32 subunit of pol δ is not required for collision release. A, the two template collision release assay of the two-subunit form of pol δ (pol3 and pol31) (left panel) is compared with that of the pol δ heterotrimer (pol3, pol31, and pol32) (right panel). Reactions were performed on DNA-primed ssDNA templates coated with E. coli SSB as detailed under “Experimental Procedures.” Graphs below each gel show relative quantitation of the indicated bands on an arbitrary scale. B, the two-subunit form of pol δ was examined for premature release when stalled at a primed site in the presence of excess challenge DNA to which PCNA is attached. Reactions were performed as in Fig. 5 using DNA-primed ssDNA templates coated with E. coli SSB. The graph below shows the relative incorporation on each substrate.
The experiment in panel B (Fig. 6) demonstrates that the stalled two-subunit pol δ-PCNA remains on the initial donor ϕX174 DNA, and rapid premature release is not induced by stalling or the presence of the 5-fold excess challenge M13mp18 DNA. Thus high processivity of pol δ-PCNA and its ability to undergo collision release are intrinsic to the pol3 and pol31 subunits and do not require the pol32 subunit.
DISCUSSION
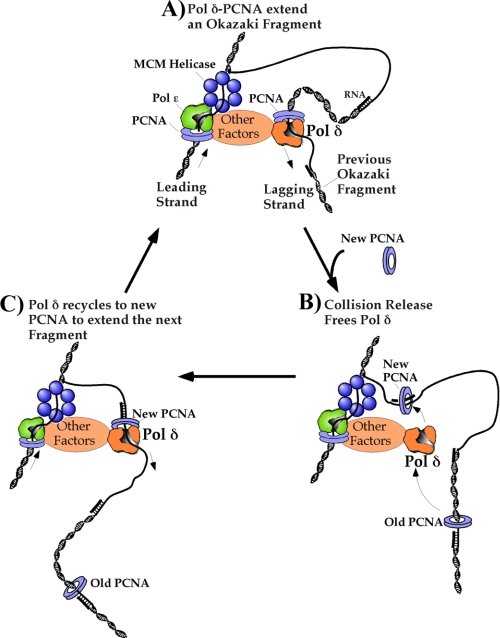
The current study utilizes pure recombinant S. cerevisiae pol δ in studies that explore its function with PCNA during lagging strand synthesis (Fig. 7). We were particularly interested in the processivity of pol δ with the PCNA clamp, because Okazaki fragments are quite small in eukaryotes and a high degree of processivity should not be required. However, we find that pol δ-PCNA is exceedingly processive, much more so than is needed during lagging strand synthesis. Therefore a mechanism must exist that signals pol δ to dissociate from DNA upon completing each short 100- to 250-bp Okazaki fragment (Fig. 7B), enabling it to recycle to new primers made by pol α/primase (Fig. 7C). Interestingly, we find that the processive pol δ-PCNA harbors an intrinsic collision release mechanism that enables it to disengage from PCNA upon completing replication. This enables pol δ to transfer to new primed sites provided they are endowed with a PCNA clamp. We presume a mechanism must also exist for premature release of a blocked enzyme, but find that a stalled pol δ-PCNA is exceedingly stabile on DNA; excess primed sites (with PCNA on them) do not elicit premature release of pol δ idling on an incomplete ssDNA template. We discuss these properties of pol δ below, as well as mechanistic details of these processes. The actions of pol δ with PCNA are compared and contrasted to those that occur in E. coli, several important features of which are different between the two organisms.
FIGURE 7.
Scheme of pol δ action on the lagging strand during replication fork movement. A, the architecture of the eukaryotic fork is still unknown, but the MCM helicase is thought to function on the leading strand with other proteins (Cdc45 and GINS). pol ε-PCNA and pol δ-PCNA are thought to be the leading and lagging strand polymerases, respectively. Okazaki fragment extension forms a DNA loop on the lagging strand, provided the lagging strand polymerase is connected to the leading strand polymerase or the helicase. B, when pol δ-PCNA finishes an Okazaki fragment and collides with the 5′ terminus of the previous Okazaki fragment, pol δ undergoes collision release and dissociates from PCNA and DNA. A fresh PCNA clamp is loaded onto the new upstream RNA/DNA primer synthesized by pol α/primase on the lagging strand. C, pol δ associates with the new upstream PCNA clamp to begin synthesis of the next Okazaki fragment, leaving the old PCNA clamp behind on the completed Okazaki fragment.
pol δ Binds Tightly to DNA and PCNA for Exceedingly High Processivity—In eukaryotes, the discontinuous lagging strand is synthesized as a series of short Okazaki fragments in the range of 100-250 bp (18, 19). Thus the lagging strand pol δ does not necessarily require a high degree of processivity to perform its function. Indeed, previous studies of pol δ from human and yeast indicate that pol δ is a very poor polymerase when acting alone, but is stimulated by PCNA which increases the processivity of pol δ by holding it to DNA during synthesis (7, 43). The analogous situation exists in bacteria, in which the pol III replicase is held to DNA by the β sliding clamp and exhibits high processivity that enables it to fully extend a primer full circle around a 7.2-kb M13mp18 ssDNA genome in one binding event (31, 44). A recent study has measured the processivity of pol δ with PCNA as ∼600 nucleotides for yeast pol δ (45). Although this degree of processivity is sufficient for synthesis of eukaryotic Okazaki fragments, it is low compared with the bacterial replicase.
The bacterial pol III-β replicase achieves enhanced stability on DNA, and therefore increased processivity, through interaction of the clamp loader χ subunit with SSB (32, 33), and the τ subunit with ssDNA (12). The E. coli pol III replicase has a half-life for binding the β clamp of under 1 min without τ, and ∼5 min with all subunits including τ (46). The current study demonstrates that yeast pol δ is exceedingly stabile with PCNA on DNA, with a half-life of >5 min. Hence, the studies of this report suggest that the heterotrimeric pol δ has a stability with PCNA comparable to that of pol III with β.
The very high processivity of yeast pol δ with PCNA shown here contrasts with the much lower processivity number observed in the earlier study of pol δ (45). We presume the reason of this difference lies in the use of 125 mm sodium acetate in the earlier study compared with 50 mm potassium glutamate used in several reactions of the current study. We find that addition of too much sodium chloride or sodium acetate reduces the processivity of pol δ (data not shown). The same is true in the bacterial pol III system (32). In vivo, pol δ-PCNA likely functions in the company of other proteins, and this may be expected to enhance the stability and processivity of pol δ-PCNA in elevated ionic strength.
Eukaryotic pol δ does not associate tightly with RFC, if at all, although preliminary experiments indicate a weak interaction may exist (34, 47). Indeed, RFC has been shown to eject from DNA after the clamp loading step (40), whereas the E. coli clamp loader remains with pol III-β on DNA after loading the clamp through contacts between the τ subunit and both pol III and ssDNA (12, 38, 48). Early studies showing that ATPγS inhibits pol δ suggested human RFC may travel with pol δ-PCNA (49). But human RFC is now known to unload PCNA (3), and this fact combined with the relatively low processivity of human pol δ-PCNA suggests that RFC-ATPγS may have unloaded PCNA during the experiments using ATPγS. Inhibition by ATPγS can therefore be explained without invoking that RFC travels with pol δ-PCNA. Furthermore, if RFC traveled with pol δ-PCNA and participated in processivity, one may expect RFC-RPA and RFC-ssDNA contacts to participate in pol δ-PCNA stability and processive action, as observed for the χ and τ subunits of the pol III replicase (8). However, we demonstrate that pol δ-PCNA stability and processivity is indifferent to replacement of RPA for E. coli SSB. In addition, we show that the actions of pol δ-PCNA using either wtRFC or RFC deleted for the N-terminal ssDNA binding region of RFC1, show no difference in the processive behavior of pol δ-PCNA (Fig. S1) or the ability of pol δ to recycle by collision release (Fig. 4C).
It is interesting to note that cells containing pol ε mutants that lack the DNA polymerase domain remain viable, suggesting that some other polymerase can substitute for pol ε in replication of the leading strand (23, 24). pol δ is suggested as the most attractive candidate for a pol ε substitute, based on in vitro studies of SV40 virus replication (50). Specifically, pol δ replicates both the leading and lagging strands of the SV40 virus genome (20). The very high processivity of pol δ-PCNA observed here fits nicely with the use of pol δ in leading strand synthesis, particularly in cells that lack pol ε polymerase activity.
Mechanism of pol δ-PCNA Collision Release upon Completing a DNA Template—The high processivity and tight grip of pol δ to PCNA on DNA is advantageous for action needed on the leading strand, where synthesis is continuous and in the same direction as replication fork movement. However, too tight of a grip to DNA is conceivably a disadvantage to lagging strand actions where synthesis is discontinuous and polymerase must dissociate from DNA after extension of each Okazaki fragment. This same problem is faced by the highly processive pol III replicase of E. coli. Upon completing a DNA fragment, the E. coli pol III replicase rapidly releases from the β clamp. This mode of polymerase recycling is referred to as “collision release,” because the replicase collides with the 5′ terminus of the downstream fragment. Studies of the collision release mechanism in E. coli show that the τ subunit of the clamp loader disengages pol III from the β clamp upon completing replication (12).
The studies of the current report demonstrate that yeast pol δ has a functional collision release mechanism, but appears to be quite different from the mechanism of the E. coli system, because pol δ does not require a factor analogous to E. coli τ, and thus the collision release mechanism appears to be intrinsic to the pol δ heterotrimer. We show here that the pol32 subunit of the pol δ heterotrimer is not required for collision release, indicating that the collision release mechanism resides in the pol3/pol31 subunits. Although participation of RFC cannot be ruled out entirely, RFC is reported to dissociate after clamp loading and does not appear to participate in elongation by the pol δ-PCNA complex (6, 40). The E. coli τ subunit separates pol III from β in collision release, and the region of τ that performs this function binds ssDNA and pol III and is not required for clamp loading (12, 36, 51). The RFC1 subunit contains an ssDNA binding region that can be deleted without effect on clamp loading activity (39, 40), and we show here that collision release of pol δ is still observed using the RFC mutant, which lacks the ssDNA binding region of the RFC1 subunit.
There also exists a second type of polymerase recycling mechanism, referred to as “premature release,” in which the polymerase is signaled to release from the clamp before the Okazaki fragment is complete (14). In fact, the T4 phage system predominately recycles by this mechanism (13). The signal for premature release is hypothesized to be the RNA primer, or the presence of a new clamp on the upstream primed site (13, 14). Premature release may be necessary in phage T4 due to the small 5-bp size of the RNA primer, which would dissociate from DNA without rapid recruitment of the polymerase, even if the downstream Okazaki fragment were not complete. Premature release could also be important if the lagging strand polymerase were slower than the leading strand polymerase, for example due to single strand DNA-binding protein that coats the lagging strand. In this case premature release would keep the replication fork moving without waiting for each Okazaki fragment to finish. Because phage contain plentiful DNA polymerase, the resulting ssDNA gaps left in the wake of the moving fork could simply be filled in by the excess viral DNA polymerase. The E. coli primase generates longer RNA primers (∼12 bp), and the pol III replicase is sufficiently rapid on SSB-coated ssDNA that premature release may not be needed. However, premature release has been observed for the E. coli replicase when the lagging pol III stalls at a block (52, 53) and also upon forcing production of an abnormally long Okazaki fragment by limiting primase (14). Hence, premature release in the E. coli system may be used to prevent fork collapse due to aberrant lagging strand synthesis.
The small size of Okazaki fragments in eukaryotes would appear to make premature release unnecessary. In addition, the current study shows that pol δ-PCNA extends a primed site on RPA-coated ssDNA at a rate of 150 nucleotides/s, similar to the fastest observed rate of fork movement in vivo (54). Although premature release may not be required for normal replication fork progression in eukaryotes, one may presume that the eukaryotic replication fork will have a mechanism for premature release based on the bacterial and phage studies. The current study examines pol δ for premature release upon stalling in a simple model system. pol δ-PCNA was stalled by limiting dNTPs, and its ability to release from PCNA was examined in the presence of challenge primed sites containing PCNA. A primed site with a clamp is conceivably a signal for premature release in the T4 phage system (13). However, the stalled pol δ does not release from PCNA, but remains tightly attached to PCNA at its original primed site (Fig. 5). Thus, premature release of pol δ from PCNA, if it occurs, must receive some other signal at a eukaryotic replication fork.
What Does pol δ Recognize to Release from PCNA upon Completing Replication?—The studies of this report indicate that the collision release mechanism of pol δ is inherent within the heterotrimer pol δ replicase. There are many possible signals that could be sensed by pol δ-PCNA specifically upon closing a ssDNA gap to a nick. A brief consideration of these suggests the following: (i) The absence of template ssDNA. In this case, pol δ will be unable to pair a bound dNTP to a template base, and a bound unpaired dNTP could signal that replication is complete. (ii) pol δ may bind template ssDNA for additional strength in binding at a primed site. Loss of ssDNA upon complete conversion to duplex DNA could lower the affinity of pol δ for PCNA and DNA. (iii) pol δ may recognize the 5′ RNA of the previously synthesized downstream Okazaki fragment that it “collides” with. 5′ RNA recognition may invoke a signal for release of pol δ from PCNA. (iv) pol δ may recognize the downstream duplex structure as a signal for release. In this case, pol δ recognition of duplex DNA, or an RNA-DNA duplex, may be sufficient to signal pol δ that replication is complete. (v) Replication fork factors that are unique to eukaryotes could perform the task of separating pol δ from PCNA upon replication. (vi) Upon finishing replication, pol δ will stall at the 3′ terminus that can no longer be extended. A stalled pol δ may signal it to release its attachment to a PCNA clamp.
The studies of the current report rule out some of the possibilities listed above, but not all of them. For example, we have tested a 5′ RNA versus 5′ DNA and find that pol δ ejects from PCNA upon colliding with either 5′ RNA or 5′ DNA (compare Figs. 3C with 4B). Hence, 5′-end recognition of an RNA terminus is not required for pol δ collision release. However, recognition of a 5′ duplex, be it RNA or DNA, may still be considered. We have also ruled out the use of replication factors that are specific to eukaryotes (e.g. GINS, Cdc45, and others), because no extra replication proteins need to be added for collision release to occur. However, it remains possible that other factors may control collision release, either preventing it, or triggering it to occur prematurely to free a lagging strand pol δ that is stalled at a template lesion. Given the demonstrated role of pol δ in processing Okazaki fragments to remove the downstream RNA primer for ligation, the presence of Fen1 and Ligase, which are also involved in this process, may keep pol δ from recycling until RNA removal is complete, although this possibility remains to be explored (55). This may be contrasted with the mechanism of collision release in the E. coli system which, for example, uses additional subunits (see reference 56 for further experiments of Pol III in collision release).
Detailed future studies will be required to understand the precise mechanism by which pol δ recognizes that DNA is complete and triggers it to disengage from PCNA specifically upon completing replication. It should also be noted that PCNA may play a role in recognition that DNA is complete, although PCNA is located on the duplex DNA behind the primed template junction bound to the active site of pol δ. Presumably, the recognition and collision release process is mediated by a conformational change in pol δ that lowers its affinity for PCNA and/or DNA. Future studies will be required to determine the detailed nature of this conformational change, and to identify which subunits participate in this process.
Supplementary Material
Acknowledgments
We are grateful to Jeff Finkelstein for making the expression clones of pol δ, other members of the laboratory for helpful advice throughout this work, and Frank Leu for artwork in Fig. 7 and for further experiments in the E. coli system (56).
This work was supported, in whole or in part, by National Institutes of Health Grant GM38839. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The online version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: pol, polymerase; PCNA, proliferating cell nuclear antigen; ssDNA, single strand DNA; GST, glutathione S-transferase; DTT, dithiothreitol; ATPγS, adenosine 5′-O-(thiotriphosphate); RFC, replication factor C; RPA, replication protein A; SSB, single strand-binding protein.
References
- 1.Pursell, Z. F., Isoz, I., Lundstrom, E. B., Johansson, E., and Kunkel, T. A. (2007) Science 317 127-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nick McElhinny, S. A., Gordenin, D. A., Stith, C. M., Burgers, P. M., and Kunkel, T. A. (2008) Mol. Cell 30 137-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao, N., Turner, J., Kelman, Z., Stukenberg, P. T., Dean, F., Shechter, D., Pan, Z. Q., Hurwitz, J., and O'Donnell, M. (1996) Genes Cells 1 101-113 [DOI] [PubMed] [Google Scholar]
- 4.Burgers, P. M., and Yoder, B. L. (1993) J. Biol. Chem. 268 19923-19926 [PubMed] [Google Scholar]
- 5.Podust, L. M., Podust, V. N., Sogo, J. M., and Hubscher, U. (1995) Mol. Cell. Biol. 15 3072-3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgers, P. M. (1991) J. Biol. Chem. 266 22698-22706 [PubMed] [Google Scholar]
- 7.Prelich, G., Tan, C. K., Kostura, M., Mathews, M. B., So, A. G., Downey, K. M., and Stillman, B. (1987) Nature 326 517-520 [DOI] [PubMed] [Google Scholar]
- 8.O'Donnell, M. (2006) J. Biol. Chem. 281 10653-10656 [DOI] [PubMed] [Google Scholar]
- 9.Labib, K., Tercero, J. A., and Diffley, J. F. (2000) Science 288 1643-1647 [DOI] [PubMed] [Google Scholar]
- 10.Bauerschmidt, C., Pollok, S., Kremmer, E., Nasheuer, H. P., and Grosse, F. (2007) Genes Cells 12 745-758 [DOI] [PubMed] [Google Scholar]
- 11.Stukenberg, P. T., Turner, J., and O'Donnell, M. (1994) Cell 78 877-887 [DOI] [PubMed] [Google Scholar]
- 12.Leu, F. P., Georgescu, R., and O'Donnell, M. (2003) Mol. Cell 11 315-327 [DOI] [PubMed] [Google Scholar]
- 13.Yang, J., Nelson, S. W., and Benkovic, S. J. (2006) Mol. Cell 21 153-164 [DOI] [PubMed] [Google Scholar]
- 14.Li, X., and Marians, K. J. (2000) J. Biol. Chem. 275 34757-34765 [DOI] [PubMed] [Google Scholar]
- 15.McInerney, P., and O'Donnell, M. (2004) J. Biol. Chem. 279 21543-21551 [DOI] [PubMed] [Google Scholar]
- 16.Masuda, Y., Suzuki, M., Piao, J., Gu, Y., Tsurimoto, T., and Kamiya, K. (2007) Nucleic Acids Res. 35 6904-6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podust, V. N., Podust, L. M., Muller, F., and Hubscher, U. (1995) Biochemistry 34 5003-5010 [DOI] [PubMed] [Google Scholar]
- 18.DePamphilis, M. L., and Wassarman, P. M. (1980) Annu. Rev. Biochem. 49 627-666 [DOI] [PubMed] [Google Scholar]
- 19.Bielinsky, A. K., and Gerbi, S. A. (1998) Science 279 95-98 [DOI] [PubMed] [Google Scholar]
- 20.Waga, S., and Stillman, B. (1994) Nature 369 207-212 [DOI] [PubMed] [Google Scholar]
- 21.Weinberg, D. H., Collins, K. L., Simancek, P., Russo, A., Wold, M. S., Virshup, D. M., and Kelly, T. J. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 8692-8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, S. H., Eki, T., and Hurwitz, J. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 7361-7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kesti, T., Flick, K., Keranen, S., Syvaoja, J. E., and Wittenberg, C. (1999) Mol. Cell 3 679-685 [DOI] [PubMed] [Google Scholar]
- 24.Dua, R., Levy, D. L., and Campbell, J. L. (1999) J. Biol. Chem. 274 22283-22288 [DOI] [PubMed] [Google Scholar]
- 25.Turner, J., and O'Donnell, M. (1995) Methods Enzymol. 262 442-449 [DOI] [PubMed] [Google Scholar]
- 26.Morrison, A., and Sugino, A. (1992) Nucleic Acids Res. 20 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, A., Yao, N. Y., Bowman, G. D., Kuriyan, J., and O'Donnell, M. (2006) J. Biol. Chem. 281 35531-35543 [DOI] [PubMed] [Google Scholar]
- 28.Finkelstein, J., Antony, E., Hingorani, M. M., and O'Donnell, M. (2003) Anal. Biochem. 319 78-87 [DOI] [PubMed] [Google Scholar]
- 29.Ayyagari, R., Impellizzeri, K. J., Yoder, B. L., Gary, S. L., and Burgers, P. M. (1995) Mol. Cell. Biol. 15 4420-4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao, N., Hurwitz, J., and O'Donnell, M. (2000) J. Biol. Chem. 275 1421-1432 [DOI] [PubMed] [Google Scholar]
- 31.Fay, P. J., Johanson, K. O., McHenry, C. S., and Bambara, R. A. (1981) J. Biol. Chem. 256 976-983 [PubMed] [Google Scholar]
- 32.Kelman, Z., Yuzhakov, A., Andjelkovic, J., and O'Donnell, M. (1998) EMBO J. 17 2436-2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glover, B. P., and McHenry, C. S. (1998) J. Biol. Chem. 273 23476-23484 [DOI] [PubMed] [Google Scholar]
- 34.Yuzhakov, A., Kelman, Z., Hurwitz, J., and O'Donnell, M. (1999) EMBO J. 18 6189-6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conaway, R. C., and Lehman, I. R. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 4585-4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maki, S., and Kornberg, A. (1988) J. Biol. Chem. 263 6555-6560 [PubMed] [Google Scholar]
- 37.Onrust, R., Finkelstein, J., Naktinis, V., Turner, J., Fang, L., and O'Donnell, M. (1995) J. Biol. Chem. 270 13348-13357 [DOI] [PubMed] [Google Scholar]
- 38.Studwell-Vaughan, P. S., and O'Donnell, M. (1991) J. Biol. Chem. 266 19833-19841 [PubMed] [Google Scholar]
- 39.Uhlmann, F., Cai, J., Gibbs, E., O'Donnell, M., and Hurwitz, J. (1997) J. Biol. Chem. 272 10058-10064 [DOI] [PubMed] [Google Scholar]
- 40.Podust, V. N., Tiwari, N., Stephan, S., and Fanning, E. (1998) J. Biol. Chem. 273 31992-31999 [DOI] [PubMed] [Google Scholar]
- 41.Gerik, K. J., Li, X., Pautz, A., and Burgers, P. M. (1998) J. Biol. Chem. 273 19747-19755 [DOI] [PubMed] [Google Scholar]
- 42.Johansson, E., Garg, P., and Burgers, P. M. (2004) J. Biol. Chem. 279 1907-1915 [DOI] [PubMed] [Google Scholar]
- 43.Bauer, G. A., and Burgers, P. M. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 7506-7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Donnell, M. E., and Kornberg, A. (1985) J. Biol. Chem. 260 12884-12889 [PubMed] [Google Scholar]
- 45.Chilkova, O., Stenlund, P., Isoz, I., Stith, C. M., Grabowski, P., Lundstrom, E. B., Burgers, P. M., and Johansson, E. (2007) Nucleic Acids Res. 35 6588-6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Donnell, M. E. (1987) J. Biol. Chem. 262 16558-16565 [PubMed] [Google Scholar]
- 47.Pan, Z. Q., Chen, M., and Hurwitz, J. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, S., Dallmann, H. G., McHenry, C. S., and Marians, K. J. (1996) J. Biol. Chem. 271 21406-21412 [DOI] [PubMed] [Google Scholar]
- 49.Tsurimoto, T., and Stillman, B. (1991) J. Biol. Chem. 266 1961-1968 [PubMed] [Google Scholar]
- 50.Stillman, B. (2008) Mol. Cell 30 259-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsuchihashi, Z., and Kornberg, A. (1989) J. Biol. Chem. 264 17790-17795 [PubMed] [Google Scholar]
- 52.Higuchi, K., Katayama, T., Iwai, S., Hidaka, M., Horiuchi, T., and Maki, H. (2003) Genes Cells 8 437-449 [DOI] [PubMed] [Google Scholar]
- 53.Pages, V., and Fuchs, R. P. (2003) Science 300 1300-1303 [DOI] [PubMed] [Google Scholar]
- 54.Raghuraman, M. K., Winzeler, E. A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D. J., Davis, R. W., Brewer, B. J., and Fangman, W. L. (2001) Science 294 115-121 [DOI] [PubMed] [Google Scholar]
- 55.Garg, P., and Burgers, P. M. (2005) Crit. Rev. Biochem. Mol. Biol. 40 115-128 [DOI] [PubMed] [Google Scholar]
- 56.Leu, F. P. (2002) Chromosomal Replication in Escherichia coli: Mechanisms of Lagging Strand DNA Synthesis Ph.D. Thesis, Cornell Medical College, New York
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.