Abstract
We determined the crystal structures of the T cell receptor (TCR)-like antibody 25-D1.16 Fab fragment bound to a complex of SIINFEKL peptide from ovalbumin and the H-2Kb molecule. Remarkably, this antibody directly “reads” the structure of the major histocompatibility complex (MHC)-bound peptide, employing the canonical diagonal binding mode utilized by most TCRs. This is in marked contrast with another TCR-like antibody, Hyb3, bound to melanoma peptide MAGE-A1 in association with HLA-A1 MHC class I. Hyb3 assumes a non-canonical orientation over its cognate peptide-MHC and appears to recognize a conformational epitope in which the MHC contribution is dominant. We conclude that TCR-like antibodies can recognize MHC-bound peptide via two different mechanisms: one is similar to that exploited by the preponderance of TCRs and the other requires a non-canonical antibody orientation over the peptide-MHC complex.
B cell receptor and its soluble analog, i.e. antibody molecules, typically recognize native antigens. However, some antibodies can recognize major histocompatibility complex (MHC)4-bound peptides and have been termed T cell receptor (TCR)-like antibodies. They have been derived either from large libraries containing diverse fragments encoding variable antibody regions or in the course of immunization of laboratory animals. The latter approach has proved to be less productive, suggesting that under natural conditions, TCR-like antibodies are rather rare. Because these antibodies offer attractive opportunities to track and measure particular peptide-MHC (pMHC) complexes on live cells in vitro and in vivo, a growing number of TCR-like antibodies are being developed. The molecular basis for their specificity is poorly understood, however.
The TCR-like antibody 25-D1.16 has been elicited in response to immunization of mice with a whole cell bearing pOV8-Kb complexes (1). This antibody has been shown to discriminate pOV8-Kb from other pMHC complexes on the cell surface and has been widely used to study various aspects of processing and presentation of MHC class I (MHC-I)-restricted epitopes to cytotoxic T lymphocytes. We determined the complete primary structure of this antibody and compared its parameters of binding to pOV8-Kb with those of a TCR recognizing the same ligand. This analysis led us to conclude that antibody 25-D1.16 indeed behaves like a TCR (2).
Here, we report the crystal structure of 25-D1.16 Fab bound to soluble pOV8-Kb protein. 25-D1.16 interacts with amino acid residues in conserved MHC positions that are believed to mediate the canonical TCR orientation of all TCR-pMHC structures studied thus far (3). Such a diagonal orientation facilitates direct contacts between both CDR3 loops of the Fab fragment and the Kb-bound peptide side chains, allowing the antibody to “read” the structure of MHC-bound peptide in the same way as a TCR. Because 25-D1.16 was raised without direct contribution of CD8, which could influence TCR binding to pMHC, but still utilizes the same conserved positions to contact MHC, we suggest that the MHC moiety itself “encodes” the canonical TCR orientation without co-receptor influence. In contrast, antibody Hyb3 assumes a non-canonical orientation relative to the MAGE-A1-HLA-A1 complex and forms few direct contacts with the peptide (4). The different binding modes of the two antibodies suggest that TCR-like antibodies are capable of recognizing MHC-bound peptides either by contacting the peptide directly, as a TCR usually does, or by recognizing a unique conformation of the MHC protein bound to a particular peptide.
EXPERIMENTAL PROCEDURES
Antibody 25-D1.16 and Fab Fragments—25-D1.16 hybridoma cells were grown in serum-free high glucose Dulbecco's modified Eagle's/Ham's F-12 medium (1:1) supplemented with l-glutamine, sodium pyruvate, β-mercaptoethanol, vitamins, essential and nonessential amino acids, l-ascorbic acid, and SPITE (Sigma) (5). Antibody 25-D1.16 was purified from the culture supernatant by affinity chromatography on protein G-agarose (2). Fab fragments of the antibody were isolated by papain digestion, followed by anion-exchange chromatography on a Mono Q column (GE Healthcare). The identity of the purified Fab fragments was established by SDS-PAGE and by enzyme-linked immunosorbent assay with soluble pOV8-Kb ligand.
Soluble pOV8-Kb Complex—Genes encoding the H-2Kb ectodomain including a synthetic C-terminal His6 tag and mouse β2-microglobulin (β2m) were expressed in Schneider (S2) cells as described previously (2). S2 cells that were stably transfected with plasmids containing H-2Kb, β2m, and neomycin resistance genes were expanded in Sf-900 II SFM serum-free medium (Invitrogen) and grown to a density of 1.4–2.0 × 107/ml. H-2Kb expression was induced by 1 μm CuSO4 for 72 h, and soluble H-2Kb molecules were isolated from the culture supernatant as described (2). Purity of the isolated H-2Kb was confirmed by SDS-PAGE. To load “empty” H-2Kb with pOV8, 400 μg of peptide dissolved in 40 μl of dimethyl sulfoxide were added to 4 mg of H2-Kb in 1 ml of phosphate-buffered saline (pH 7.4), and the reaction mixture was incubated overnight at room temperature (22–24 °C).
Preparation and Isolation of the Fab-pMHC Complex—The 25-D1.16-pOV8-Kb complex was formed at ∼2-fold molar excess of the Fab fragments over soluble pOV8-Kb. The reaction was allowed proceed for 1 h at room temperature. The reaction mixture was then subjected to gel filtration on Sephacryl S200 HR to separate the Fab-pMHC complex from unbound Fab (supplemental Fig. 1).
Crystallization—Crystals of 25-D1.16 Fab were grown by the hanging-drop method at 20 °C. 2 μl of protein solution (10 mg/ml) were mixed with 2 μl of reservoir buffer (0.2 m (NH4)2SO4 and 0.1 m NaOAc (pH 4.6) containing 25% (w/v) polyethylene glycol 4000). The drops were equilibrated over the same buffer. Crystals reached the maximum size of 250 × 125 × 125 μm within 2–3 weeks.
Crystals of purified Fab-pOV8-Kb complex were produced in hanging drops by vapor diffusion in 100 mm NH4OAc and 50 mm trisodium citrate dihydrate (pH 5.6) containing 13% (w/v) polyethylene glycol 4000 at 20 °C. We obtained few large needle-like single crystals (250 × 50 × 10 μm) suitable for x-ray crystallographic analysis, although the majority of crystals formed needle clusters.
25-D1.16 Fab Fragment—Suitable crystals of 25-D1.16 Fab fragments were transferred to a glass capillary. Diffraction data to 1.8-Å resolution were collected at the Kimmel Cancer Center x-ray facility using a Rigaku x-ray generator equipped with a Rigaku IV image plate detector. Data were processed and scaled with the program d'TREK.
25-D1.16-pOV8-Kb Complex—Crystals were briefly soaked in cryobuffer (reservoir solution plus 30% (v/v) dimethyl sulfoxide) and flash-cooled in liquid nitrogen. Data to 2.9-Å resolution were collected at beamline 4XA of the Brookhaven National Laboratory. All measurements were performed at 100 K. Data were processed with the program DENZO and scaled with SCALEPACK.
Structure Determination and Refinement—Crystals of the Fab fragment belong to space group P21 with cell dimensions a = 49.44 Å, b = 60.86 Å, c = 79.75 Å, α = γ = 90°, and β = 111.97°. The structure of the 25-D1.16 Fab fragment in the monoclinic space group was determined by molecular replacement using the program PHASER. The Fab structure (Protein Data Bank code 1OSP) was used as a search model (6). A clear solution was found for one Fab molecule/asymmetric unit. The molecular replacement model was subjected to automatic model building using ARP/wARP. This produced a complete and accurate model, which was manually readjusted using program O and refined using CNS and REFMAC to a final R value of 0.227 and Rfree of 0.263.
The ternary complex crystal belongs to the P212121 space group with cell dimensions a = 80.56 Å, b = 111.35 Å, c = 219.08 Å, and α = β = γ = 90°. The structure of the ternary Fab-pMHC complex was determined by molecular replacement using PHASER. Input models included the H-2Kb structure (Protein Data Bank code 2VAA) with the peptide removed and the refined 25-D1.16 Fab fragment (Protein Data Bank code 3CVI). Two MHC molecules and one Fab fragment were positioned in molecular replacement. A second Fab fragment was positioned by superimposing the MHC of the complete Fab-pMHC complex onto the incomplete Fab-pMHC complex that lacked the Fab fragment. Rigid body refinement using CNS satisfactorily adjusted the position of all fragments, giving a starting R value of 0.2933 and Rfree of 0.3458. Additional simulated annealing in CNS, ARP/wARP model update, manual model building using program O, and TLS refinement produced an excellent model with an overall R value of 0.223 and Rfree of 0.300 at 2.9-Å resolution.
RESULTS
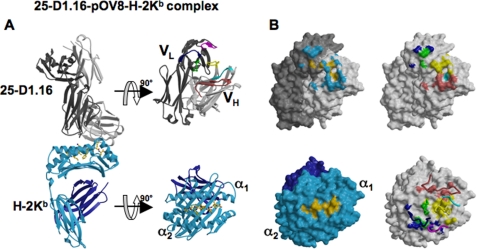
Overall Structure of the 25-D1.16-pOV8-Kb Complex—The crystal structure of the 25-D1.16-pOV8-Kb complex was determined by molecular replacement and refined to 2.9-Å resolution (Protein Data Bank code 3CVH). There are two Fab-pMHC complexes per asymmetric unit related by non-crystallographic symmetry. The modeled structures are well ordered and fit properly within calculated electron densities. Both pMHC and Fab display typical structural features of previously described immune receptors containing immunoglobulin-like domains (Fig. 1A) (for review, see Refs. 3 and 7).
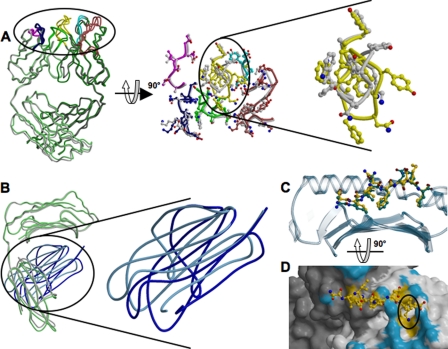
FIGURE 1.
Structure of the 25-D1.16-pOV8-Kb complex and the contact surfaces. A, the left panel shows a ribbon diagram of the 25-D1.16-pOV8-Kb complex. The antibody heavy and light chains are colored light and dark gray, respectively. The MHC heavy chain and β2m are shown in cyan and dark blue, respectively. A ball-and-stick representation of Kb-bound peptide is colored yellow. The right panel shows the individual molecules, pMHC and Fab, colored as in the left panel, but oriented to reveal their interactive surfaces. Fab CDR-H1, CDR-H2, and CDR-H3 are colored cyan, red, and yellow, whereas CDR-L1, CDR-L2, and CDR-L3 are colored blue, magenta, and green, respectively. B, the upper left panel illustrates the surface of 25-D1.16 showing contacts with the MHC moiety (cyan) and the peptide (yellow); the upper right panel shows the 25-D1.16 recognition surface with atoms from the different CDRs that directly contact the pMHC colored as in A; the lower left panel shows the contact surface of the pOV8-Kb protein (with the MHC moiety and peptide colored cyan and yellow, respectively); and the lower right panel shows areas of the pOV8-Kb contact surface that are directly contacted by 25-D1.16 CDRs (CDR color coding is as in A). The CDRs are shown as ribbons.
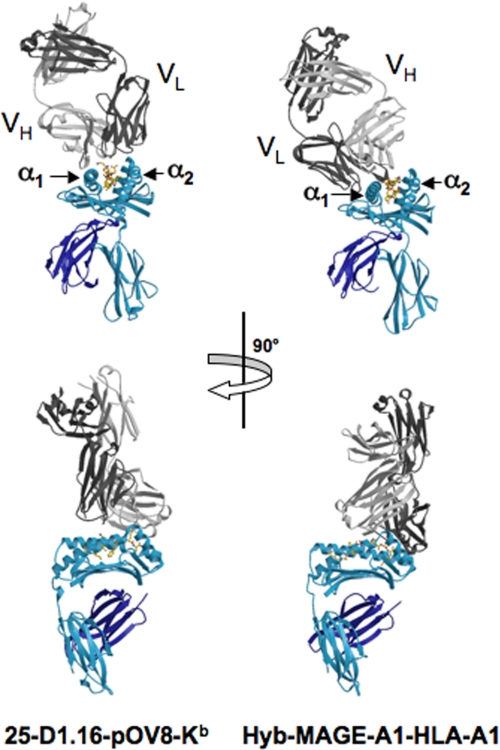
Orientation of 25-D1.16 Fab over pOV8-Kb—The contacting surfaces of both pOV8-Kb and 25-D1.16 Fab are shown in Fig. 1B. The Fab fragment is centrally positioned on the pMHC contact surface, and the long axes of symmetry of 25-D1.16 Fab and pOV8-Kb are parallel (Fig. 2). This positioning allows both CDR3 loops of Fab to contact the Kb-bound peptide, whereas the CDR1 and CDR2 loops mainly contact MHC helices α1 and α2 (Fig. 3). The variable domain of the heavy chain (VH) predominates in contacting pOV8-Kb, as it interacts with helices α1 and α2 of H-2Kb and five residues of pOV8 (P4–P8), whereas the variable light (VL) domain is less prominent, as it contacts helix α2 and merely one residue (P4) of the peptide (supplemental Tables 1 and 2).
FIGURE 2.
Positioning of 25-D1.16 and Hyb3 Fab fragments of TCR-like antibodies over cognate pMHC complexes. The left panel shows the 25-D1.16-pOV8-Kb complex, and the right panel shows the MAGE-A1-HLA-A1 complex. Fab heavy and light chains are colored light and dark gray, respectively. The MHC heavy chain and β2m are shown in cyan and dark blue, respectively. 25-D1.16 assumes an orientation that is common for TCRs, with the CDR1 and CDR2 loops contacting Kb helices and the CDR3 loops forming direct contacts with the peptide. MAGE-A1-HLA-A1-bound Hyb3 has an atypical orientation, tilting toward helix α1 without contacting helix α2 and forming few direct contacts with the peptide.
FIGURE 3.
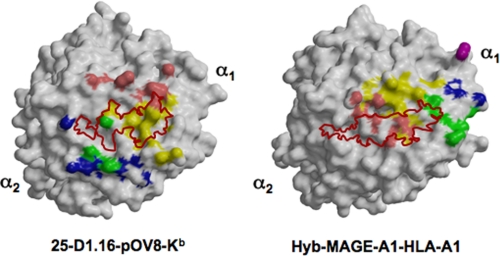
Footprints of 25-D1.16 and Hyb3 CDRs. The left panel shows the contact surfaces of pOV8-Kb, and the right panel shows MAGE-A1-HLA-A1. Direct CDR contacts (colored as described in the legend to Fig. 1A) show the inverted positions of the CDR loops relative to the corresponding pMHC molecules for the two antibodies. The corresponding peptide surfaces are outline in red.
The pMHC and Fab contact surfaces are 982 and 1032 Å2, respectively, producing a buried surface area of 2014 Å2 at the pOV8-Kb/25-D1.16 interface (Table 1). VH contributes 709 Å2 of the solvent-accessible surface area to the complex, compared with 323 Å2 contributed by VL, thus revealing the dominant role of VH in this interaction. Surfaces in the complex are well matched, as reflected in the shape complementarity (Sc) value of 0.678, and only two water molecules contribute to the complex interface. The Sc value lies within a range of values described for typical antibody-antigen complexes (3) and is very similar to the Sc value (0.7) described for the 2C TCR complex with its natural allogeneic ligand, but not syngeneic ligand (Sc = 0.41) (7). A similar Sc value (0.716) was reported previously for xenoreactive TCR AHIII 12.2 bound to cognate pMHC protein (8).
TABLE 1.
Comparison of Fab-pMHC contacts made by 25-D1.16 and Hyb3 with their cognate pMHC complexes
| Fab-pMHC contacts | 25-D1.16 | Hyb3a |
|---|---|---|
| Fab-pMHC buried area (Å2) | 2,014 | 1,902 |
| Fab total contact surface (Å2) | 1032 | 985 |
| Fab-peptide contact surface (Å2) | 257 | 168 |
| VH total contact surface (Å2) | 709 | 603 |
| VL total contact surface (Å2) | 323 | 382 |
| pMHC total contact surface (Å2) | 982 | 917 |
| Sc value | 0.678 | 0.69 |
Numbers are derived from Ref. 4.
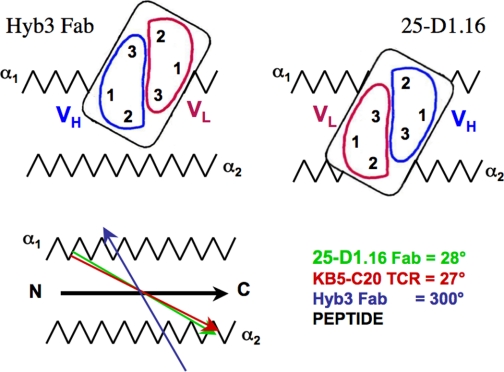
The dot product of the vector, which connects the intrachain disulfide bonds in the VH and VL domains, and the vector, which designates the peptide within the binding cleft, give an angle equal to 28° (see Fig. 5). This angle, which determines the relative orientation of Fab over pMHC, lies within the range of angles calculated for all structures of TCR-pMHC complexes, i.e. between 22° and 70° (3, 9). For TCRs recognizing different peptide-H-2Kb complexes, this angle varies between 22° and 41° (10, 11).
FIGURE 5.
Schematic representation of the positioning of 25-D1.16 and Hyb3 antibodies. 25-D1.16 and KB5-C20 TCR (11) utilize a very similar mode of recognition of MHC-bound peptide that is distinct from that used by Hyb3 antibodies (4). Red and green vectors show virtually identical orientations of 25-D1.16 and KB5-C20 TCR; the dark blue vector indicates a profoundly different orientation of Hyb3; and the black vector indicates peptide positioning in the binding groove of both H2-Kb and HLA-A1 MHC-I proteins.
Structural Changes of pOV8-Kb and 25-D1.16 Fab upon Specific Complex Formation—Major conformational differences in the structures of bound 25-D1.16 are absent (Fig. 4A); nonetheless, conformational changes in the CDR loops caused by binding of Fab to pMHC are evident (Fig. 4A and supplemental Fig. 2). The most prominent conformational differences between the free and bound Fab structures can be observed in the long CDR3 loop of VH. The structure of the CDR3 loop in intact and pMHC-bound Fab is well resolved as evident from excellent electron densities in unbiased simulated annealing omit Fo –Fc maps (supplemental Fig. 2). The backbone of loop-forming residues Pro96–Ala100B shifts by ∼3 Å, and the side chains of Tyr97, Tyr98, Asn100, and Phe100A shift by ∼9 Å, respectively. Therefore, binding of Fab to pMHC causes the exposed VH CDR3 loop to wrap around the lysine at position P7 in the peptide (Fig. 4D). CDR3 of VL did not undergo substantial conformational changes with the exception of Trp92, the side chain of which rotates by 180° and forms multiple van der Waals contacts with Thr163, Arg155, and Ala158 of helix α2. The conformational adaptations of the contacting CDR3 loops are consistent with structural changes observed at the TCR/pMHC interface (10–12) and with analyses of TCR-pMHC binding parameters (13–15). Structural plasticity of the CDR loops could allow for an adjustment of the interacting surfaces, perhaps increasing the complementarity of the pMHC/Fab interface.
FIGURE 4.
Structural changes of 25-D1.16 and pOV8-Kb upon specific complex formation. A, the left panel shows the superposition of intact (green) and pOV8-Kb-bound (gray) 25-D1.16 Fab. CDRs for both structures are colored as described in the legend to Fig. 1A. The middle panel shows a close-up view of the CDRs oriented to reveal the pMHC-binding interface. Gray CDRs correspond to the isolated Fab fragment, and the colored CDRs correspond to pOV8-Kb-bound Fab. The right panel is a close-up view of CDR-H3 and reveals the conformational change induced upon binding of Fab to pOV8-Kb. B, the right panel shows the superposition of isolated MHC (with the heavy chain colored gray and β2m colored cyan) and pOV8-Kb-bound MHC (with the heavy chain colored green and β2m colored blue). The left panel shows a close-up view of the two β2m domains, revealing distinct orientations. C, the pOV8 peptide as bound to intact Kb (with carbon atoms colored green) and in the complex with 25-D1.16 (with carbon atoms colored yellow) is shown. The polymorphic domain is shown as a blue transparent ribbon. The peptide is pulled out by the bound Fab fragment. D, shown are the molecular surface of 25-D1.16 and a ball-and-stick diagram of the MHC-bound peptide. Fab atoms are colored according to their contacts (surface contacts MHC (blue) and surface contacts the peptide (yellow)). LysP7 (highlighted by the black circle) is enveloped by CDR-H3.
25-D1.16-pOV8-Kb complex formation led to noticeable conformational differences in the peptide structure. The side chain conformation of solvent-accessible residues, i.e. P4, P6, and P7, changes, whereas the conformation of the peptide backbone remains the same (Fig. 4C).
The structures of bound 25-D1.16 and H-2Kb α1/α2 peptide-binding domains are indistinguishable. However, significant structural rearrangements are observed in the MHC moiety within the 25-D1.16-pOV8-Kb complex. The relative positioning of the non-polymorphic α3 domain and β2m domains was altered upon complex formation, whereas their structures did not change (Fig. 4B). β2m turned relative to the α3 domain by 16°, with the distal part swinging away by >10 Å from the polymorphic α1/α2 domain. Surprisingly, β2m residues Gly29–Pro33 and Phe56–Tyr63, which contact the floor of the peptide-binding groove, remained unchanged. We suggest that a number of small amino acid rearrangements affect the reorientation of β2m. Residues connecting the polymorphic and non-polymorphic domains (Leu180–Asp183), the “neck” of the MHC molecule, shifted toward β2m by 3.5 Å. The His191–Thr200 loop moved toward the C terminus by 4.9 Å, and α3 domain residues Arg234–Phe241, which directly contact β2m, shifted by 5 Å. Residues of the α3 domain that mediate contacts with the CD8 co-receptor (Gln222–Met228) (16) are dislocated by 2.5 Å toward the C-terminal end.
Similar changes can be seen in the structures of peptide-Kb proteins bound to either cognate TCR (10, 11) or the CD8αα ectodomain (17). Collectively, these data indicate that H-2Kb possesses substantial intramolecular flexibility and that the observed structural changes in H-2Kb could be induced upon binding to 25-D1.16 or TCR. The intramolecular flexibility of H-2Kb and other MHC-I proteins is consistent with the allosteric model of peptide interactions with the heavy chain/β2m heterodimer, according to which peptide binding induces a conformational transition within the heavy chain that affects heavy chain interactions with β2m (18, 19). Changes in the structure of engaged MHC-I molecules may facilitate formation of the trimolecular TCR-pMHC-I-CD8 complex (20–23) and enhance oligomerization of MHC-I proteins on the surface of target and antigen-presenting cells.
Peptide and MHC Recognition by 25-D1.16 Fab—25-D1.16 contacts both α helices of Kb and peptide pOV8 directly. Peptide contacts are predominantly mediated by the CDR3 loops of both heavy and light chains, whereas the CDR1 and CDR2 loops primarily interact with the MHC helices (Table 2 and supplemental Tables 1 and 2).
TABLE 2.
Recognition of the cognate peptide and MHC moiety by two TCR-like antibodies
|
CDR direct
contactsa
|
25-D1.16
|
Hyb3b
|
||||
|---|---|---|---|---|---|---|
| Peptide | Helixαi of H2-Kb | Helixα2 of H2-Kb | Peptide | Helixα1 of HLA-A1 | Helixα2 of HLA-A1 | |
| CDR-H1 | 1 | 0 | 0 | 0 | 3 | 0 |
| CDR-H2 | 1 | 7(5) | 0 | 2(3) | 0 | 1 |
| CDR-H3 | 9(4) | 1(1) | 6(3) | 2(1) | 6(1) | 0 |
| CDR-L1 | 0 | 1(2) | 4(1) | 0 | 0 | 0 |
| CDR-L2 | 0 | 0 | 0 | 0 | 0(1) | 0 |
| CDR-L3 | 2 | 0 | 4(1) | 0 | 2 | 4 |
The number of water-bridged hydrogen bonds is indicated in parentheses.
The number of contacts was derived from the structure of Hyb3-MAGE-A1-HLA-A1 described in Ref. 4.
25-D1.16 makes a total of 17 contacts with the peptide, including six direct hydrogen bonds, six van der Waals interactions, one salt bridge, and four water-bridged hydrogen bonds (Table 2). The salt bridge is formed between AspH50 of CDR-H2 and the ε-amino group of LysP7. The ε-amino group of LysP7 also forms a hydrogen bond with AsnH33 of CDR-H1. The aliphatic component of LysP7 directly contacts TyrH98, GlyH99, and AsnH100 of CDR-H3. Thus, the hydrophobic moiety of the side chain of LysP7 is stabilized by three van der Waals contacts with CDR-H3 and assumes an extended conformation that facilitates the formation of the salt bridge. This type of bond, characterized by a very high free energy, is called a buried salt bridge (24, 25). Overall, LysP7 makes 38% of all direct contacts between pOV8 and 25-D1.16, contributing ∼130 Å2 to the peptide contact area. Two other pOV8 residues, GluP6 and AsnP4, are also directly contacted by the CDRs of 25-D1.16. GluP6 makes one hydrogen, one van der Waals, and four water-bridged contacts with CDR-H3 (TyrH98, AsnH100, and PheH100A), whereas the side chain of AsnP4 contacts CDR-L3 (SerL93 and ThrL94) and CDR-H3 (PheH100A) via two hydrogen bonds and one van der Waals interaction. Mutations of these three residues affect pOV8-Kb binding by 25-D1.16 (1, 2) and by OT-1 TCR (26, 27), providing further evidence that 25-D1.16 is a TCR-like antibody and that both the antibody and TCR utilize a common mechanism to recognize MHC-I-bound peptide.
Recognition of the H-2Kb moiety is mediated mostly by the CDR1 and CDR2 loops. There are 23 direct contacts between the CDR loops and the H-2Kb helices (Table 2): helix α1 forms seven hydrogen bonds with CDR-H2, and helix α2 makes 14 direct contacts with CDR-H3, CDR-L1, and CDR-L3 (Table 2). The 25-D1.16 contact area with the peptide amounts to 257 Å2, of which 189 Å2 are contributed by VH and 68 Å2 by VL. Overall, 26% of the antibody contact surface directly interacts with the MHC-bound peptide. In summary, the CDR1 and CDR2 loops are almost exclusively utilized to contact helices α1 and α2 of H-2Kb, whereas the CDR3 loops are dedicated to recognition of the MHC-bound pOV8 peptide. This nearly perfect specialization of the CDR loops in determining MHC restriction and peptide recognition suggests that some variations in the CDR3 regions should not affect the interaction between the CDR1 and CDR2 loops and the H-2Kb moiety. Thus, limited mutagenesis of the 25-D1.16 CDR3 genes may allow the development of TCR-like antibodies, which could recognize other H2-Kb-restricted peptides.
DISCUSSION
The crystal structure of antibody 25-D1.16 (Fab) bound to the pOV8-Kb complex described here reveals that the antibody achieves specificity for the H-2Kb-bound pOV8 peptide by adopting a diagonal orientation over pMHC that is strikingly similar to that of many TCRs (3, 9, 28, 29). The TCR-like orientation of 25-D1.16, with CDR3 and CDR1/2 regions contacting peptide and MHC helices, respectively, is an extremely improbable event, as antibodies generally recognize a wide variety of antigens, whereas TCR has specifically evolved to recognize MHC-bound peptides. It is thus highly significant that 25-D1.16 and TCR exploit the same strategy to recognize MHC-bound peptide, suggesting that TCR and antibody have a common ancestor.
25-D1.16 has been elicited during normal immune response in vivo (1) without direct contribution from the co-receptor CD8, which is typically present on CD8+ T cells, but not on B cells. Meanwhile, it is thought that CD8/MHC-I interactions, which occur prior to pMHC recognition by TCR, could influence TCR association with MHC and, consequently, TCR orientation over MHC (22, 30). That 25-D1.16 assumes the same orientation over the pMHC ligand as TCR supports the notion that TCR has an intrinsic propensity for MHC (31) and that MHC, not CD8, determines selection of TCRs that bind with canonical orientation.
25-D1.16 is encoded by germ line genes without somatic mutations in the variable domains (2), thus resembling TCR genes, as these are naturally not subject to somatic mutations. Because 25-D1.16 belongs to IgG1, it is likely that the 25-D1.16 hybridoma was generated shortly after class switching, a very rare event. This provides an explanation as to why TCR-like antibodies are difficult to elicit. Further accumulation of somatic mutations could lead to selection of higher affinity antibodies. The MHC moiety could then become the dominant contributor to binding energy, and peptide specificity would be lost (32). It is likely that only the canonical orientation of TCR over pMHC agrees with a sizable contribution of the peptide to binding energy, thus ensuring recognition of MHC-bound peptide (33). Such an orientation ensures that the energy contribution from MHC is sufficient to account for MHC restriction, but minimizes TCR interactions with MHC to preclude autoimmunity.
The canonical diagonal TCR orientation appears to be essential for TCR and TCR-like antibodies to discriminate between different MHC-bound peptides, as it focuses CDR3 loops on the center of MHC-bound peptide. Although the central positioning of CDR3 loops can be achieved at many different TCR orientations, the latter is very similar in most known TCR-pMHC complexes (3). It has been suggested that the canonical diagonal orientation is determined by a small number of polymorphic residues at conserved positions on MHC helices. These residues are thought to form energetically dominant TCR-MHC bonds, suggesting that TCR has an inherent predisposition to interact with MHC helices (3, 7, 12, 29, 31, 34, 35). Three conserved MHC residues, two on helix α1 and one on helix α2, are thought to contribute the minimal set of interactions that determine MHC restriction and the conserved TCR orientation over MHC-I (12). The same residues (Arg62, Gln65, and Arg155 in H2-Kb) directly contact the CDRs of 25-D1.16 (supplemental Table 1), and these interactions are presumably responsible for the canonical TCR-like orientation of 25-D1.16 over pMHC. Such an orientation also allows comparable contributions of MHC helices α1 and α2 to the interactions with the CDR1 and CDR2 loops (Fig. 3 and Table 2). In marked contrast, the majority of the interactions between Hyb3 and MAGE-A1-HLA-A1 are mediated by contacts between HLA-A1 helix α1 and the VH and VL domains (Fig. 3 and Table 2). The asymmetric positioning of Hyb3 over MAGE-A1-HLA-A1 is a distinct feature of this complex that results in the tilting of Hyb3 toward helix α1 (Fig. 2) and the non-canonical reversed orientation of the Fab molecule relative to TCRs as bound to pMHC (4). None of the HLA-A1 residues that are thought to determine the canonical TCR orientation form contacts with Hyb3 CDRs (4). As a consequence, the vector connecting intrachain disulfide bridges within VH and VL domains of Hyb3 points in the opposite direction compared with the corresponding vectors of 25-D1.16 and canonical TCR. The angle of Hyb3 over MAGE-A1-HLA-A1 protein is 300° compared with 28° for 25-D1.16 relative to pOV8-Kb (Fig. 5). Thus, Hyb3 assumes diagonal but reversed orientation over cognate pMHC (Fig. 5), a feature that has not been observed thus far for any TCR-pMHC complex. The orientation of Hyb3 over MAGE-A1-HLA-A1 is reminiscent of the unusual orientation of a TCR specific for myelin basic protein peptide in association with human DRB1 MHC-II protein, the only one example of a TCR with non-canonical orientation over cognate pMHC observed thus far (36). In another example, the semi-invariant TCR from natural killer T cells docks in parallel to the binding groove of the MHC-I-like CD1d molecule presenting α-galactosylceramide, a non-peptide strong agonist (37). Notably, none of the three positions in CD1d, which are homologous to those in MHC-I postulated to mediate MHC restriction and conserved TCR orientation (12), are involved in the TCR/CD1d interactions (37).
The contact and buried surface areas for both Fab-pMHC complexes are very similar, as are the Sc values (Table 1). Accordingly, 25-D1.16 and Hyb3 Fab fragments bind their respective ligands in a peptide-dependent manner with almost identical affinities (2 × 10–8 and 1.4 × 10–8 m, respectively) (2, 4). However, although 26% of the total contact area occurs between 25-D1.16 and pOV8, only 17% of the total contact area (168 Å2) is buried between Hyb3 and MAGE-A1 peptide (Table 1). There are four direct contacts between Hyb3 CDRs and the MAGE-A1 peptide, one of which represents a hydrogen bond between the side chain of HisP7 and GlyH55 in CDR-H2. The other three contacts involve the peptide backbone (Table 2 and supplemental Tables 1 and 2). On the other hand, there are 12 direct contacts between 25-D1.16 and pOV8 (Table 2). Apparently, Hyb3 contacts with MAGE-A1 peptides are limited, suggesting that direct Fab/peptide interactions play a minor role in determining specificity of Hyb3. Thus, TCR-like antibodies are able to discriminate between various MHC-bound peptides at least via two different mechanisms, either by contacting the peptide directly or by recognizing a unique conformational epitope induced when a particular peptide binds to the MHC moiety. The latter can be associated with a non-canonical orientation of Fab over the pMHC ligand.
Supplementary Material
Acknowledgments
We thank Angel Porgador and Ronald Germain for providing the 25-D1.16 hybridoma. We also thank Dr. Helen Pace for excellent technical assistance during the initial phase of this project. We are grateful to Wayne Hendrickson, Ronald Germain, and David Kranz for critical reading of the manuscript and meaningful comments.
This work was supported, in whole or in part, by National Institutes of Health Research Grant AI058755 (to Y. S.). Beamline X4A at the National Synchrotron Light Source, a Department of Energy facility, was supported by the New York Structural Biology Center. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Tables 1 and 2. The atomic coordinates and structure factors (codes 3CVI and 3CVH) have been deposited in the Protein Data Bank, Research Collaboratory for Stuctural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Footnotes
The abbreviations used are: MHC, major histocompatibility complex; TCR, T cell receptor; pMHC, peptide-MHC; MHC-I, MHC class I; CDR, complementarity-determining region; β2m, β2-microglobulin; VH, variable domain of heavy chain; VL, variable domain of light chain; Sc, shape complementarity.
References
- 1.Porgador, A., Yewdell, J. W., Deng, Y., Bennink, J. R., and Germain, R. N. (1997) Immunity 6 715–726 [DOI] [PubMed] [Google Scholar]
- 2.Mareeva, T., Lebedeva, T., Anikeeva, N., Manser, T., and Sykulev, Y. (2004) J. Biol. Chem. 279 44243–44249 [DOI] [PubMed] [Google Scholar]
- 3.Rudolph, M. G., Stanfield, R. L., and Wilson, I. A. (2006) Annu. Rev. Immunol. 24 419–466 [DOI] [PubMed] [Google Scholar]
- 4.Hulsmeyer, M., Chames, P., Hillig, R. C., Stanfield, R. L., Held, G., Coulie, P. G., Alings, C., Wille, G., Saenger, W., Uchanska-Ziegler, B., Hoogenboom, H. R., and Ziegler, A. (2005) J. Biol. Chem. 280 2972–2980 [DOI] [PubMed] [Google Scholar]
- 5.Markvicheva, E. A., Kuptsova, S. V., Mareeva, T. Y., Vikhrov, A. A., Dugina, T. N., Strukova, S. M., Belokon, Y. N., Kochetkov, K. A., Baranova, E. N., Zubov, V. P., Poncelet, D., and Rumsh, L. D. (2000) Appl. Biochem. Biotechnol. 88 145–157 [DOI] [PubMed] [Google Scholar]
- 6.Li, H., Dunn, J. J., Luft, B. J., and Lawson, C. L. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3584–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colf, L. A., Bankovich, A. J., Hanick, N. A., Bowerman, N. A., Jones, L. L., Kranz, D. M., and Garcia, K. C. (2007) Cell 129 135–146 [DOI] [PubMed] [Google Scholar]
- 8.Buslepp, J., Kerry, S. E., Loftus, D., Frelinger, J. A., Appella, E., and Collins, E. J. (2003) J. Immunol. 170 373–383 [DOI] [PubMed] [Google Scholar]
- 9.Garcia, K. C., and Adams, E. J. (2005) Cell 122 333–336 [DOI] [PubMed] [Google Scholar]
- 10.Garcia, K., Degano, M., Pease, L., Huang, M., Peterson, P., Teyton, L., and Wilson, I. (1998) Science 279 1166–1172 [DOI] [PubMed] [Google Scholar]
- 11.Reiser, J. B., Gregoire, C., Darnault, C., Mosser, T., Guimezanes, A., Schmitt-Verhulst, A. M., Fontecilla-Camps, J. C., Mazza, G., Malissen, B., and Housset, D. (2002) Immunity 16 345–354 [DOI] [PubMed] [Google Scholar]
- 12.Tynan, F. E., Burrows, S. R., Buckle, A. M., Clements, C. S., Borg, N. A., Miles, J. J., Beddoe, T., Whisstock, J. C., Wilce, M. C., Silins, S. L., Burrows, J. M., Kjer-Nielsen, L., Kostenko, L., Purcell, A. W., McCluskey, J., and Rossjohn, J. (2005) Nat. Immunol. 6 1114–1122 [DOI] [PubMed] [Google Scholar]
- 13.Willcox, B., Gao, G., Wyer, J., Ladbury, J., Bell, J., Jakobsen, B., and van der Merwe, P. (1999) Immunity 10 357–365 [DOI] [PubMed] [Google Scholar]
- 14.Boniface, J. J., Reich, Z., Lyons, D. S., and Davis, M. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11446–11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anikeeva, N., Lebedeva, T., Krogsgaard, M., Tetin, S. Y., Martinez-Hackert, E., Kalams, S. A., Davis, M. M., and Sykulev, Y. (2003) Biochemistry 42 4709–4716 [DOI] [PubMed] [Google Scholar]
- 16.Gao, G. F., Willcox, B. E., Wyer, J. R., Boulter, J. M., O'Callaghan, C. A., Maenaka, K., Stuart, D. I., Jones, E. Y., van der Merwe, P. A., Bell, J. I., and Jakobsen, B. K. (2000) J. Biol. Chem. 275 15232–15238 [DOI] [PubMed] [Google Scholar]
- 17.Kern, P. S., Teng, M. K., Smolyar, A., Liu, J. H., Liu, J., Hussey, R. E., Spoerl, R., Chang, H. C., Reinherz, E. L., and Wang, J. H. (1998) Immunity 9 519–530 [DOI] [PubMed] [Google Scholar]
- 18.Gakamsky, D. M., Bjorkman, P. J., and Pecht, I. (1996) Biochemistry 35 14841–14848 [DOI] [PubMed] [Google Scholar]
- 19.Gakamsky, D. M., Boyd, L. F., Margulies, D. H., Davis, D. M., Strominger, J. L., and Pecht, I. (1999) Biochemistry 38 12165–12173 [DOI] [PubMed] [Google Scholar]
- 20.Luescher, I. F., Vivier, E., Layer, A., Mahiou, J., Godeau, F., Malissen, B., and Romero, P. (1995) Nature 373 353–356 [DOI] [PubMed] [Google Scholar]
- 21.Cho, B. K., Lian, K. C., Lee, P., Brunmark, A., McKinley, C., Chen, J., Kranz, D. M., and Eisen, H. N. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 1723–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anikeeva, N., Lebedeva, T., Clapp, A. R., Goldman, E. R., Dustin, M. L., Mattoussi, H., and Sykulev, Y. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16846–16851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia, C. K., Scott, C. A., Brunmark, A., Carbone, F. R., Peterson, P. A., Wilson, I. A., and Teyton, L. (1996) Nature 384 577–581 [DOI] [PubMed] [Google Scholar]
- 24.Huber, R., Kukla, D., Bode, W., Schwager, P., Bartels, K., Deisenhofer, J., and Steigemann, W. (1974) J. Mol. Biol. 89 73–101 [DOI] [PubMed] [Google Scholar]
- 25.Marquart, M., Waletr, J., Deisenhofer, J., Bode, W., and Huber, R. (1983) Acta Crystallogr. Sect. B 39 480–490 [Google Scholar]
- 26.Jameson, S. C., Carbone, F. R., and Bevan, M. J. (1993) J. Exp. Med. 177 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alam, S., Davies, G., Lin, C., Zal, T., Nasholds, W., Jameson, S., Hogquist, K., Gascoigne, N., and Travers, P. (1999) Immunity 10 227–237 [DOI] [PubMed] [Google Scholar]
- 28.Housset, D., and Malissen, B. (2003) Trends Immunol. 24 429–437 [DOI] [PubMed] [Google Scholar]
- 29.Marrack, P., Scott-Browne, J. P., Dai, S., Gapin, L., and Kappler, J. W. (2008) Annu. Rev. Immunol. 26 171–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yachi, P. P., Lotz, C., Ampudia, J., and Gascoigne, N. R. (2007) J. Exp. Med. 204 2747–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huseby, E. S., White, J., Crawford, F., Vass, T., Becker, D., Pinilla, C., Marrack, P., and Kappler, J. W. (2005) Cell 122 247–260 [DOI] [PubMed] [Google Scholar]
- 32.Reay, P. A., Matsui, K., Haase, K., Wulfing, C., Chien, Y. H., and Davis, M. M. (2000) J. Immunol. 164 5626–5634 [DOI] [PubMed] [Google Scholar]
- 33.Eisen, H. N., Sykulev, Y., and Tsomides, T. J. (1997) in Antigen-Binding Molecules: Antibodies and T-cell Receptors (Haber, E., ed) pp. 1–56, Academic Press, San Diego, CA
- 34.Feng, D., Bond, C. J., Ely, L. K., Maynard, J., and Garcia, K. C. (2007) Nat. Immunol. 8 975–983 [DOI] [PubMed] [Google Scholar]
- 35.Mazza, C., Auphan-Anezin, N., Gregoire, C., Guimezanes, A., Kellenberger, C., Roussel, A., Kearney, A., van der Merwe, P. A., Schmitt-Verhulst, A. M., and Malissen, B. (2007) EMBO J. 26 1972–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hahn, M., Nicholson, M. J., Pyrdol, J., and Wucherpfennig, K. W. (2005) Nat. Immunol. 6 490–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borg, N. A., Wun, K. S., Kjer-Nielsen, L., Wilce, M. C., Pellicci, D. G., Koh, R., Besra, G. S., Bharadwaj, M., Godfrey, D. I., McCluskey, J., and Rossjohn, J. (2007) Nature 448 44–49 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.