Abstract
Aberrant epidermal growth factor receptor (EGFR) signaling is a major cause of tumor progression and metastasis; the underlying mechanisms, however, are not well understood. In particular, it remains elusive whether deregulated EGFR pathway is involved in epithelial-mesenchymal transition (EMT), an early event that occurs during metastasis of cancers of an epithelial origin. Here, we show that EGF induces EGFR-expressing cancer cells to undergo a transition from the epithelial to the spindle-like mesenchymal morphology. EGF reduced E-cadherin expression and increased that of mesenchymal proteins. In search of a downstream mediator that may account for EGF-induced EMT, we focused on transcription repressors of E-cadherin, TWIST, SLUG, and Snail and found that cancer cells express high levels of TWIST and that EGF enhances its expression. EGF significantly increases TWIST transcripts and protein in EGFR-expressing lines. Forced expression of EGFR reactivates TWIST expression in EGFR-null cells. TWIST expression is suppressed by EGFR and Janus-activated kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) inhibitors, but not significantly by those targeting phosphoinositide-3 kinase and MEK/ERK. Furthermore, constitutively active STAT3 significantly activates the TWIST promoter, whereas the JAK/STAT3 inhibitor and dominant-negative STAT3 suppressed TWIST promoter. Deletion/mutation studies further show that a 26-bp promoter region contains putative STAT3 elements required for the EGF-responsiveness of the TWIST promoter. Chromatin immunoprecipitation assays further show that EGF induces binding of nuclear STAT3 to the TWIST promoter. Immunohistochemical analysis of 130 primary breast carcinomas indicates positive correlations between non-nuclear EGFR and TWIST and between phosphorylated STAT3 and TWIST. Together, we report here that EGF/EGFR signaling pathways induce cancer cell EMT via STAT3-mediated TWIST gene expression.
Introduction
Accumulating evidences suggest a role of epidermal growth factor receptor (EGFR) in tumor metastasis (1–5). It has been reported that EGFR activates its downstream modules protein kinase C-δ, extracellular signal-regulated kinase (ERK), and phospholipase C-γ and facilitates migration of EGFR-overexpressing cancer cells (6, 7). It has also been shown that EGFR increases production of matrix metalloproteinase-9, possibly via phosphoinositide-3 kinase (PI3K), leading to cell migration (8, 9). However, the involvement of EGFR in epithelial-mesenchymal transition (EMT; an early step during tumor metastasis) remains elusive. Nevertheless, chronic EGF treatment has been shown to lead to down-regulation of E-cadherin expression and loss of cell-cell adherence junction (10). Down-regulation of E-cadherin is considered as a critical step and an indicator for EMT, and the down-regulation of which can be achieved by transcriptional suppression mediated by transcription factors TWIST, SLUG, and Snail (11–14).
TWIST is a basic helix-loop-helix (bHLH) transcription factor that has been known to be essential for proper gastrulation mesoderm formation, and neural crest migration (15). Intriguingly, increased TWIST expression is detected in metastatic breast cancer cells and is required for EMT and breast cancer metastasis (11). Role of TWIST in EMT has also been reported in other cancer types, including those of prostate (16) and uterus (17). High TWIST expression further correlates with tumor invasion and metastasis in breast carcinomas (11), esophageal squamous cell carcinomas (18), and gliomas (19). A recent study reported that STAT3 knockdown of mouse 4T1 mammary tumor cells led to altered expression of several genes, including those of activated src, phosphorylated Akt, c-Myc, and Twist, but not p53 nor total src (20). How mouse STAT3 knockdown led to reduced phosphorylation of src and Akt remains unknown. Whereas c-Myc is a known STAT3 target gene, mouse Twist has yet been shown to be a direct transcriptional target of STAT3. STAT-binding sites are not found in the mouse twist gene promoter, suggesting its expression reduction by STAT3 small interfering RNA was likely due to indirect effects. As for the human TWIST gene, its transcriptional regulation is yet investigated.
Given the association between EGFR overexpression and high metastatic potential, we speculate that the EGF/EGFR pathway promotes EMT via activation of EMT mediators, such as TWIST, SLUG, and Snail. Our initial examination of a breast cancer cell line, MDA-MB-468, indicated that TWIST is expressed at a higher level compared with SLUG and Snail, which prompted us to further investigate the regulatory role of the EGF/EGFR pathway in TWIST expression and in TWIST-mediated EMT. Here, we report that EGF treatment and EGFR expression are important for TWIST expression. However, unexpectedly, we also found that the underlying mechanisms involve transcriptional regulation of TWIST by STAT3, an oncoprotein constitutively activated in many human cancers and implicated in tumor progression (21–23). The information derived from these studies provides critical insights into the regulation of TWIST gene expression and into the biology of tumors with deregulated EGFR and STAT3 pathways and is important in the management of metastatic tumors.
Experimental Procedures
Cell lines and cell culture
A431 human epidermoid carcinoma cells, MDA-MB-468 human breast carcinoma cells, and EGFR-null Chinese hamster ovary (CHO) cells and Madin-Darby canine kidney (MDCK) epithelial cells were obtained from American Type Culture Collection. CHO-NEO, CHO-EGFR, and CHO-EGFR-NLS stable cells were derived from the parental CHO cells as previously described (24). HER5 cells are stable EGFR transfectants of Swiss 3T3 cells, and NR-6 cell line is an EGFR-null Swiss 3T3 variant. All cells were maintained in DMEM supplemented with 10% FCS, except that CHO-NEO, CHO-EGFR, and CHO-EGFR-NLS stable lines were supplemented with 1 mg/mL G418 additionally. PANC28 human pancreatic cancer cell line was a kind gift from Dr. Paul Chiao at the University of Texas M. D. Anderson Cancer Center.
Chemicals, plasmids, and mammalian transfection
PD158780, AG490, AG1478, and the PI3K inhibitor LY294002 were obtained from Calbiochem Corp. The specific MEK/ERK inhibitor U0126 was purchased from Promega. Iressa was purchase from AstraZeneca Pharmaceuticals. Purified EGF, transforming growth factor-α (TGF-α), HB-EGF, and all other chemicals were from Sigma-Aldrich. Transfection was carried out by a cationic liposome, SN, as previously described (25). EGFR expression vector, pcDNA3.1-EGFR, was a kind gift from Dr. David James (University of California, San Francisco, San Francisco, CA). Dominant-negative STAT3-expression vector (pCAGGS-STAT3F), which carries the mutation Y705F and constitutively active STAT3-expression plasmid (pRC-STAT3C-flag), was kindly provided by Dr. Keping Xie (M. D. Anderson Cancer Center). Control small interfering RNA and STAT3 small interfering RNA were obtained from Millipore/Pharmacon.
Reverse transcription-PCR
In these studies, cancer cells of exponential growth were starved for serum for 24 h and stimulated with EGF (100 ng/mL) for 0, 1, and 2 h. Cells were harvested, and total RNA was extracted using the SV total RNA isolation system kit (Promega). Extracted total RNA was then subjected to reverse transcription using reverse transcriptase (Invitrogen) to generated first-strand cDNA using the oligo dT primer. PCR was then carried out for 30 cycles, and each cycle was with 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. Primers with sequences 5′-TTAGCACCCCTGGCCAAGG-3′ and 5′-CTTACTCCTTGGAGGCCATG-3′ were used for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). For TWIST, primers 5′-GGAGTCCGCAGTCTTACGAG-3′ and 5′-TCTGGAGGACCTGG TAGAGG-3′ were used in PCR. For SLUG, primers 5′-TGATGAAGAGGAAAGACTACAG-3′ and 5′-GCTCACATATTCCTTGTCACAG-3 were used in the PCR reactions. Primers 5′-CGAAAGG CCTTCAACTGCAAAT-3′ and 5′-ACTGGTACTTCTTGACATCTG-3′ were used to generate Snail PCR products. For real-time PCR, SYBR-Green Master PCR mix was used in the iCycler system (Bio-Rad) in triplicates as previously described (24). All quantification was normalized to an endogenous control GAPDH. The PCR program was 95°C for 4 min, followed by 40 cycles of 95°C for 15 s and 60°C for 45 s.
Western blot analyses
Western blot analyses were done as previously described (24, 26). The antibodies used for these studies included rabbit polyclonal EGFR, TWIST, and STAT3 antibodies and goat polyclonal Snail antibody, which were purchased from Santa Cruz Biotech. Rabbit polyclonal phosphorylated STAT3 (p-STAT3; Y-705), phosphorylated EGFR (Y-1045), and Akt/phosphorylated Akt antibodies were from Cell Signaling. Additional antibodies used for Western blot were mouse monoclonal β-actin and α-tubulin (Sigma), E-cadherin (BD Pharmigen), ERK/phosphorylated ERK (Upstate), fibronectin (NeoMarkers), and vimentin (NeoMarkers) antibodies.
Construction of the human TWIST promoter-driven luciferase reporter constructs
The human genomic TWIST plasmid was a kind gift from Dr. Vincent J. Cristofalo (Allegheny University of the Health Science, Philadelphia, PA). The full-length human TWIST promoter was amplified from the genomic TWIST plasmid to contain the promoter region up to nt −824 using forward primer 5′-CGTGCTAGCATTGGACTGGGTTTCC-3′ and reverse primer 5′-AGTAAGCTTGGAGTTGGGCGAGAG-3′. The PCR product was digested with NheI and HindIII and subsequently cloned to the pGL-3 basic luciferase reporter vector (Promega), and positive clone was designated phTWIST-824. The reverse primer was used to generate other TWIST promoter segments. For phTWIST-604, phTWIST-120, and phTWIST-94 constructs, forward primers 5′-CGTGCTAGCGTTTGGCCTTT GGAAC-3′, 5′-CGTGCTAGCAAACTTTCCTATAAAAC-3′, and 5′-CGTGCTAGCTCCCTCCTCCTCACGT-3′ were used, respectively. To generate the phTWIST-120/MA mutant, forward primer 5′-AAACTTTCCTATAAAACTTCGGCCGGTCCCTCCTCC-3′ and reverse primer 5′-GGAGGGACCGGCCGAAGCCCCATAGGAAAGTTT-3′ were used. To generate the phTWIST-120/MB mutant, we used forward primer 5′-CGCGTGCTAGCAAACGGGCCTATAAAACTTCGAAAAG-3′ and reverse primer 5′-CTTTTCGAAGTTTTATAGGCCCGTTTGCTAGCACGCG-3′. Site-directed mutagenesis was done as previously described (24). All constructs were subjected to DNA sequencing to confirm no PCR-generated artifacts.
Mammalian transfection and luciferase assay
Cells were transfected using the cationic liposome method using SN as described previously (25) or Fugene HD (Roche). pRL-TK (Promega) was cotransfected as a control for transfection efficiency. After transfection and experiment treatments, cells were lysed and luciferase activity was measured using Dual Luciferase kit (Promega) according to manufacturer’s instructions (27, 28).
Biotinylated oligonucleotide precipitation assays/oligo pull-down assay
These studies were done as described previously (24, 29). The nucleotide sequences of biotinylated oligonucleotides corresponding to TWIST-120, TWIST-120/MA, and TWIST-120/MB are 5′-AAACTTTCCTATAAAACTTCGAAAAGTCCCTCCTCC-3′, 5′-AAACTTTCCTATAAAACTTCGGCCGGTCCCTCCTCC-3′, and 5′-AAACGGGCCTATAAAACTTCGGCCGGTCCCTCCTCC-3′, respectively. A functional STAT3-binding site [acute-phase response element (APRE)], known to bind to STAT3/APR factor (30, 31), was used as the positive control for STAT3 binding and was designated as STAT3-BS. The sequence for STAT3-BS is 5′-GATCCTTCTGGGAATTCCTAGATC-3′. After annealing complementary strands (1 μg), binding reactions were carried out at 4°C for 16 h to include 6.5 μg nuclear extracts isolated from the control and EGF-stimulated MDA-MB-468 cells. Protein-bound probe was precipitated using ImmunoPure streptavidin-agarose beads (Pierce), washed, and subjected to detection of p-STAT3 and EGFR via Western blot analyses as described earlier.
Chromatin immunoprecipitation assay
This procedure was done as described previously (24, 32). In brief, MDA-MB-468 cells were serum-starved overnight and treated without and with EGF (100 ng/mL) for 30 min. The cells were cross-linked with 1% formaldehyde at room temperature for 10 min, and the reaction was stopped by the addition of glycine to a final concentration of 0.125 mol/L. After washing twice with ice-cold PBS, the cells were scraped off the plate, centrifuged, and resuspended in 400 μL of cell lysis buffer [5 mmol/L Tris-HCl (pH 8.0), 85 mmol/L KCl, 0.5% NP40, 1 μg/mL aprotinin, and 1 mmol/L phenylmethylsulfonyl fluoride (PMSF)]. After incubation on ice for 10 min, the cells were dounced to isolate nuclei, and the nuclei were collected, resuspended in 200 μL nuclei lysis buffer 50 mmol/L Tris-HCl (pH 8.1), 10 mmol/L EDTA, 1% SDS, 1 μg/mL aprotinin, and 1 mmol/L (PMSF), and sonicated 4 pulses for 10 s each. After centrifugation, the supernatant was diluted 1:10 with immunoprecipitation dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mmol/L EDTA, 16.7 mmol/L Tris-HCl (pH 8.0), 167 mmol/L NaCl, 1 μg/mL aprotinin, and 1 mmol/L PMSF]. The cell lysates were precleared with normal IgG and protein A-agarose beads and subjected to immunoprecipitation with mouse monoclonal anti-EGFR antibody (Ab-13; NeoMarkers), polyclonal anti-STAT3 antibody (C-20; Santa Cruz Biotechnology), or normal IgG at 4°C overnight. Immunoprecipitated complexes were collected by adding salmon sperm DNA/protein A-agarose (Upstate) for 15 min at 4°C. Immunoprecipitates were washed twice with dialysis buffer [2 mmol/L EDTA, 50 mmol/L Tris-HCl (pH 8.0)]and four times with immunoprecipitation wash buffer [0.5 mol/L LiCl, 1% NP40, 1% sodium deoxycholate, 100 mmol/L Tris-HCl (pH 8.0)]; the DNA/protein complex was eluted twice using elution buffer (50 mmol/L NaHCO3, 1% SDS) and treated with RNase A for 30 min at 37°C. The protein/DNA cross-links were reversed by heating at 65°C for 16 h, and then the DNA were extracted using QIAquick PCR purification kit (Qiagen). For PCR, 3 μL from a 40-μL DNA preparation was used for amplifications. Specific sequences of the human TWIST promoter in the immunoprecipitates were detected by PCR with primers forward 5′- AGTCTCCTCCGACCGCTTCCTG -3′ and reverse 5′- CTCCGTGCAGGCGGAAAGTTTGG -3′. The resulting PCR product spans the −364 to −33 region that includes the putative STAT3-binding sites. The c-fos promoter has been previously shown to bind to nuclear EGFR and STAT3 and thus also was amplified as positive controls (24).
Immunohistochemical analyses, histologic scoring, and statistical analyses
The cohort of primary breast carcinoma specimens, previously stained for EGFR and iNOS, was consisted of 130 cases (24, 26). Immunohistochemical staining was done as previously described (24, 26). In this study, rabbit polyclonal TWIST and p-STAT3 (Y705) antibodies from Santa Cruz Biotech and Cell Signaling, respectively, were used. The immunoreactivity for TWIST was semiquantitatively scored using a well-established system in which H score was generated by incorporating both the percentage of positive tumor cells and the intensity of staining (33). All slides were independently viewed and scored by two pathologists (W.X. and Y.W.). Slides in which there was a scoring discrepancy of >10% were reevaluated and reconciled on a two-headed microscope. Statistica 6.0 (Statsoft) and regression analysis were used to analyze the correlation.
Results
EGF exposure induces transition of the epithelial to the mesenchymal-like phenotype in cultured breast cancer cells
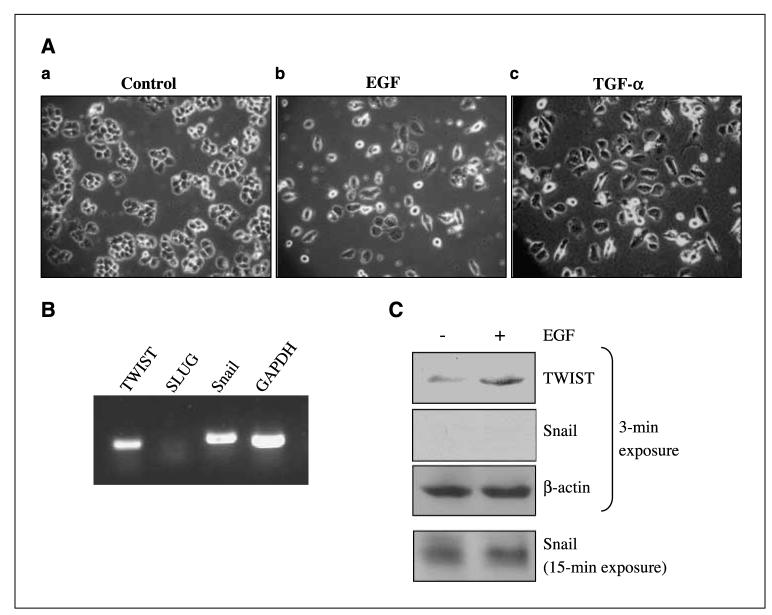
Cancer cells of epithelial origin undergo EMT as an early event that leads to local invasion and metastasis to distant sites. Despite accumulating studies showing a positive link between high EGFR expression and cancer invasion/metastasis, the specific involvement of EGFR in EMT remains elusive. We, therefore, aimed to examine whether EGF stimulation induces EMT in cancers of the breast and pancreas, two epithelial cancers that can invade and metastasize. This is also important as the survival rate for metastatic breast cancer was reported to be only 9.1% to 14.7% between 1991 and 1995 (34). As indicated in Fig. 1A, the human breast carcinoma MDA-MB-468 cells maintained in a minimal amount (0.5%) of FCS (a) retained the epithelial morphology, whereas those supplemented with EGF (b) and TGF-α (c) displayed mesenchymal-like morphology after 5-day treatments. Approximately, 500 cells were examined for each treatment, and three independent experiments were carried out. Similar observations were found in human pancreatic cancer cells PANC28 and A431 human epidermoid carcinoma cells (data not shown). Both cell lines express EGFR (Fig. 2). We next examined whether E-cadherin repressors, TWIST, SLUG, and Snail may play a role in EGF-induced EMT in MDA-MB468 cells. Using reverse transcription-PCR (RT-PCR), we found that MDA-MB-468 cells express detectable levels of TWIST and Snail transcripts, but not that of SLUG (Fig. 1B). Western blot analyses further showed that these cells contained high levels of TWIST protein, but not Snail, and that EGF enhanced TWIST protein expression (Fig. 1C). In fact, Snail expression was very low, and the signals became visible only after prolonged autoradiography. Together, these results suggest that EGFR ligands EGF and TGF-α induce EMT-like morphologic changes in human cancer cells, and among the three transcriptional repressors for E-cadherin, TWIST is the only one that expresses at a detectable level and also responds to EGF stimulation.
Figure 1.
EGF exposure induced transition of the epithelial to the mesenchymal-like phenotype in cultured breast cancer cells. Human breast carcinoma MDA-MB-468 cells were used in these studies. Three independent experiments were done. Cancer cells were serum-starved for 24 h before treatments. A, EGF-treated MDA-MB-468 cells displayed mesenchymal morphology. Cells were treated with 0.5% FCS only (a), 0.5% FCS with 50 ng/mL EGF (b), and 0.5% FCS with 50 ng/mL TGF-α (c). Cell morphology was examined and photographed daily using a phase-contrast microscope. At day 5, unstimulated MDA-MB-468 cells retained their epithelial phenotype (a). In contrast, cells treated for 5 d with EGF (b) and TGF-α (c) displayed detached mesenchymal-like morphology. B, transcripts of EMT mediators, TWIST and Snail, were detected. Total RNA was isolated and subjected to RT-PCR to detect TWIST, SLUG, Snail gene transcripts. Expression of GAPDH serves as a loading control. C, TWIST, but not Snail, protein was expressed at high levels. Cancer cells were starved for 24 h and treated without and with EGF for 6 h; total cell lysates were extracted and subjected to Western blot analyses for expression of TWIST and Snail. Note the exposure time to detect the expression of Snail was 5 times longer than that of TWIST.
Figure 2.
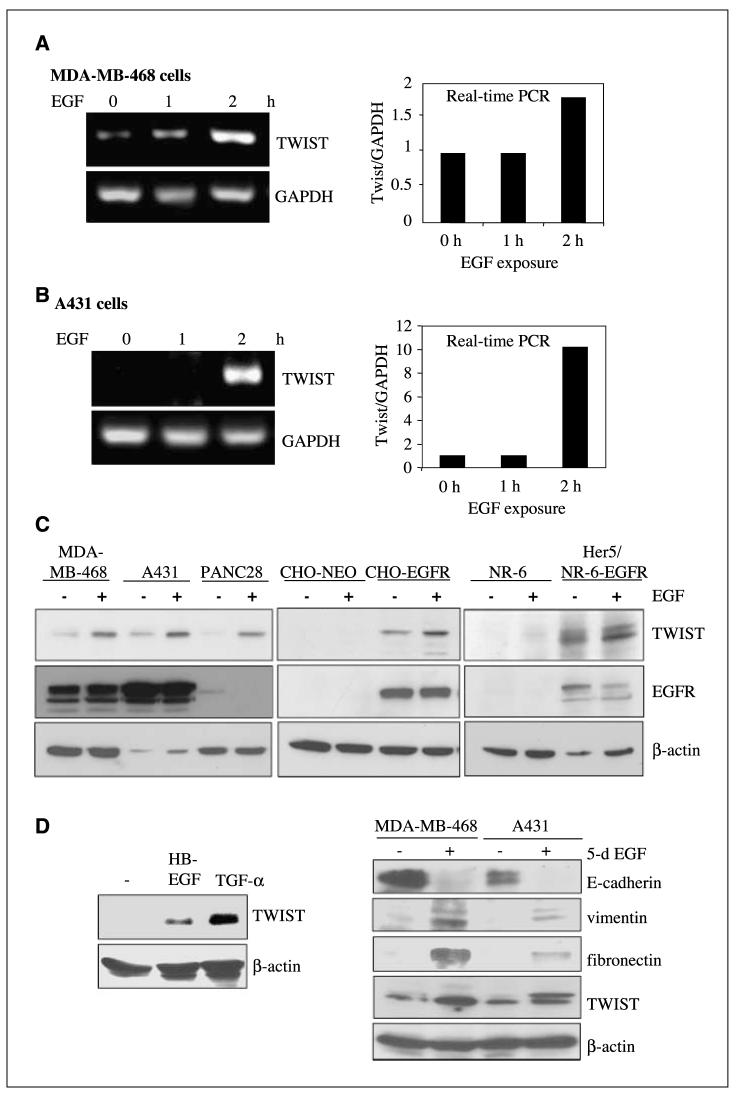
EGF stimulation induces expression of TWIST and mesenchymal markers, as well as reduction of an epithelial marker. A and B, EGF increases TWIST transcription in EGFR-expressing cancer cells. MDA-MB-468 (human breast carcinoma cells in A) and A431 (human epidermoid carcinoma cells in B) were serum-starved for 24 h, exposed to EGF (100 ng/mL) for 0, 1, and 2 h, and harvested. Total RNA was isolated and subjected to RT-PCR (left) and quantitative real-time PCR (right). Expression of GAPDH serves as a loading control. C, EGF stimulation induces expression of TWIST. All cells were serum-starved for 24 h and exposed to EGF (100 ng/mL) for 6 h, and total lysates were extracted. Expressions of TWIST, EGFR, and β-actin were examined using Western blot analyses. Left, EGF stimulation increased TWIST expression in EGFR-expressing cancer cells. A panel of EGFR-expressing cancer cell lines were used in these studies including MDA-MB-468 (human breast carcinoma), A431 (human epidermoid carcinoma), and PANC28 (human pancreatic cancer). Middle and right, forced EGFR expression in EGFR-null cells induce TWIST reexpression. CHO-NEO (middle) and NR-6 (rat fibroblasts; right) are EGFR-null, whereas CHO-EGFR and Her5 are stable lines which express EGFR. As expected, EGFR in the PANC28 cells seem to undergo down-regulation after EGF treatment, whereas those in EGFR-overexpressing MDA-MB-468 and A431 cells displayed reduced degradation (36). D, EGFR ligands HB-EGF and TGF-α activate TWIST gene expression (left). MDA-MB-468 cells were treated without or with HB-EGF (100 ng/mL) and TGF-α (100 ng/mL) for 6 h and harvested; total cell lysates were extracted and subjected to Western blot analyses. Right, Prolonged EGF treatment reduced expression of E-cadherin and increased that of vimentin, fibronectin, and TWIST. MDA-MB-468 and A431cells were treated without or with EGF (100 ng/mL) for 5 d and harvested; total cell lysates were extracted and subjected to Western blot analyses for expression of TWIST, the epithelial marker E-cadherin and mesenchymal markers, vimentin and fibronectin.
EGF induces expression of TWIST and mesenchymal markers, as well as reduction of an epithelial marker
TWIST, a bHLH transcription factor and repressor of E-cadherin expression, is frequently detected in metastatic cancer cells and is required for EMT and breast cancer metastasis (11, 16, 18). To date, human TWIST gene regulation remains largely unknown. To determine whether EGF/EGFR promotes EMT via TWIST, we first examined the effect of EGF treatment on the human TWIST gene transcription in EGFR-expressing MDA-MB-468 (Fig. 2A) and human epidermoid carcinoma A431 cells (Fig. 2B). After serum starvation for 24 h, EGF significantly increased TWIST gene transcripts at 2 h, as indicated by RT-PCR (left) and quantitative real-time PCR (right). Expression of GAPDH served as a loading control. Consistently, EGF stimulation increased the levels of TWIST protein in a panel of EGFR-expressing cancer cells, including MDA-MB-468, A431, and PANC28 (Fig. 2C, left). EGFR in the PANC28 cells seem to undergo down-regulation after EGF treatment as expected. In contrast, EGFR-overexpressing MDA-MB-468 and A431 cells contain sustained levels of EGFR. This observation is consistent with the notion that overexpression of EGFR undergoes faster recycling and impairs its down-regulation due to limiting levels of regulatory molecules that mediate rapid endocytosis and lysosomal targeting EGFR (35, 36). Interestingly, EGFR-null CHO-NEO and NR-6 cells do not contain detectable levels of TWIST, and forced EGFR expression in these cells induced TWIST expression (Fig. 2C, middle and right). CHO-EGFR and Her5 are stable lines derived from parental CHO and NR-6 cells, respectively, to express high levels of EGFR, as reported previously (37).
In addition to EGF, other EGFR ligands, HB-EGF and TGF-α, similarly activated the human TWIST gene expression in MDA-MB-468 cells (Fig. 2D, left), which is consistent with their implication in tumor invasion (38). Consistent with the phenotypic observation in Fig. 1, chronic EGF exposure reduced expression of E-cadherin, an epithelial marker, and increased levels of vimentin and fibronectin, mesenchymal markers (Fig. 2D, right). At day 5 post-EGF stimulation, both EGFR-expressing MDA-MB-468 and A431 cells expressed significantly more TWIST compared with the untreated controls. Taken together, these data indicate that EGF/TGF-α stimulation and EGFR expression are important for the human TWIST gene expression and that EGF induced gene expression changes typically found in cancer cells undergoing EMT.
EGFR-induced TWIST expression involves STAT3
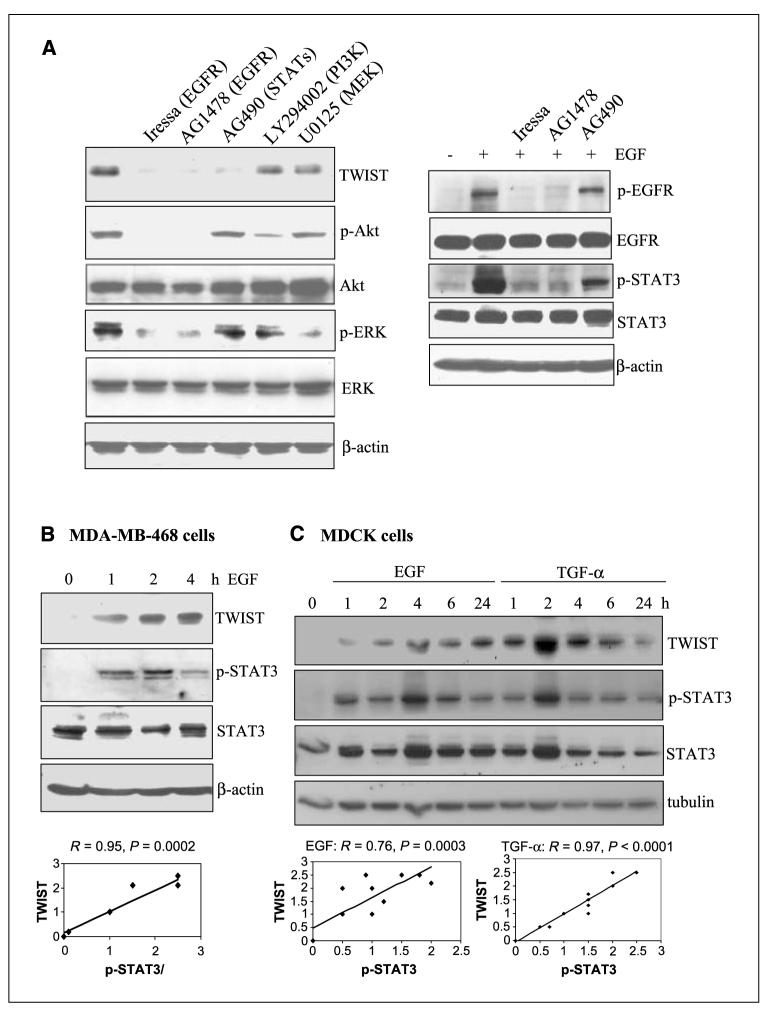
Because EGF/EGFR can activate many signaling modules to regulate gene expression, we then asked via which downstream pathways the human TWIST gene becomes activated. To address this, we pretreated MDA-MB-468 cells with inhibitors to EGFR (Iressa and AG1478), Janus-activated kinases (JAKs)/STATs (AG490), PI3K/Akt (LY294002), and MEK/ERK (U0126) and examined TWIST’s response to EGF stimulation. The results (Fig. 3A) suggest that EGF-induced TWIST expression requires EGFR and STAT3, but not those of PI3K/Akt and MEK/ERK. Effectiveness of all inhibitors is indicated by sufficient suppression of target kinases. Consistently, time course studies in Fig. 3B and C showed a concurrent TWIST expression and STAT3 activation (Y705 phosphorylation) after EGF/TGF-α treatment in MDA-MB-468 and normal epithelial MDCK cells. These experiments were done thrice; band signals were quantified by NIH ImageJ software, and the correlation (R) and P values between p-STAT3 and TWIST were determined by regression analyses. These results suggest that EGFR up-regulates the human TWIST gene expression, which seems to require STAT3 activation.
Figure 3.
EGF-induced TWIST expression involves EGFR and STAT3. A, EGFR and STATs inhibitors suppressed TWIST expression. Serum-starved MDA-MB-468 cells were pretreated for 1 h without and with various inhibitors including Iressa (EGFR; 5 μmol/L), AG1478 (EGFR; 10 μmol/L), AG490 (JAKs/STATs; 10 μmol/L), LY294002 (PI3K/Akt; 20 μmol/L), and U0125 (MEK/ERK; 10 μmol/L). Cells were then stimulated with EGF (100 ng/mL) for 5 h, harvested, and subjected to Western blot analysis. Levels of TWIST were determined. Efficiency of various inhibitors was indicated by reduced phosphorylation of EGFR (for Iressa and AG1478), STAT3 (for AG490), Akt (for LY294002), and ERK (for U0125). B, time-dependent correlation between TWIST expression and STAT3 activation in breast cancer cells. Serum-starved MDA-MB-468 cells were treated with EGF (100 ng/mL) for 0, 1, 2, and 4 h and harvested for Western blot analysis. Levels of TWIST expression, total STAT3, and p-STAT3 at Y705 (p-STAT3) were determined. These experiments were repeated thrice, and bands were quantified using the NIH ImageJ software. Regression analysis was then done to determine the correlation between p-STAT3 and TWIST. C, time-dependent correlation between TWIST expression and STAT3 activation in normal epithelial cells. MDCK epithelial cells were serum-starved and stimulated with EGFR (100 ng/mL) and TGF-α (100 ng/mL) for 0, 1, 2, 4, 6, and 24 h. Harvested cells were examined, via Western blot analysis, for expression of TWIST, p-STAT3 (Y705), STAT3, and α-tubulin. These experiments were repeated thrice, and signals were quantified using ImageJ (NIH). Regression analysis was then conducted to determine the correlation between p-STAT3 and TWIST.
EGFR and STAT3 activate the human TWIST gene promoter
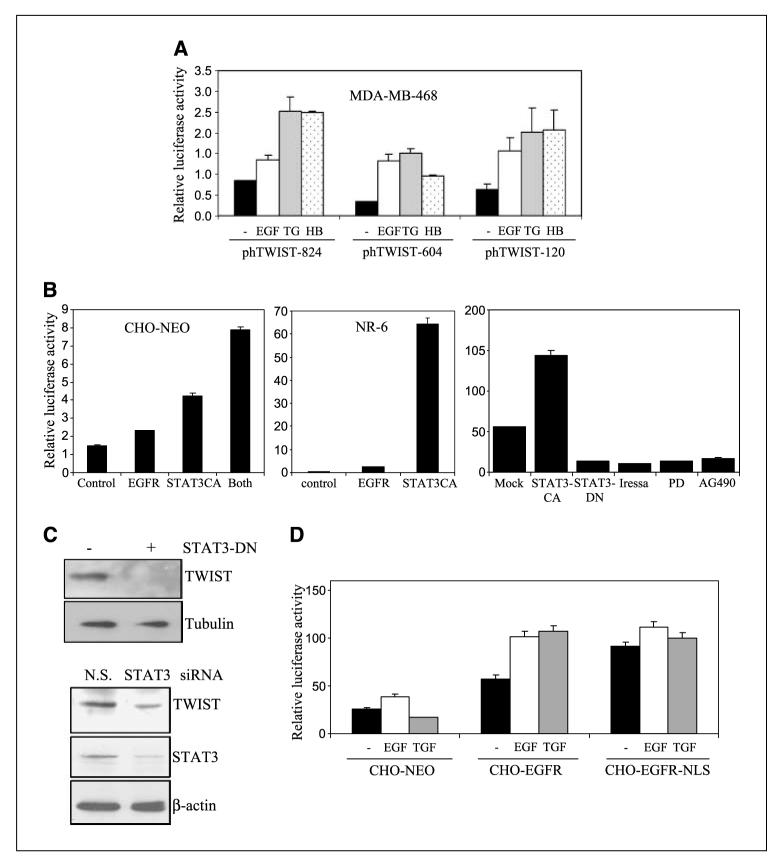
STAT3 is a DNA-binding transcription factor that elicits its physiologic function by transcriptionally regulating gene expression after activation by cytokines and growth factors signal pathways (39, 40). STAT3 becomes phosphorylated at Y705, then activated by cell-surface EGFR, and has been found to be constitutively activated in many cancers and to involve in tumorigenesis and metastasis (41–43). Given the association of STAT3 with invasive phenotype, we further aimed to examine whether STAT3 is important in activating the human TWIST gene expression at the transcriptional level. To this end, we generated the human TWIST promoter-driven luciferase constructs that contain 824, 604, and 120 bp of the promoter and designated them as phTWIST-824, phTWIST-604, and phTWIST-120, respectively. Relative luciferase activity was derived from firefly luciferase activity after normalization against the activity of the transfection efficiency control Recilla luciferase. As indicated in Fig. 4A, all three EGFR ligands (EGF, TGF-α, and HB-EGF) significantly activated the human TWIST promoter. Interestingly, the phTWIST-120 construct showed similar extents of EGF responsiveness to the phTWIST-604 plasmid, indicating that the critical DNA elements are within the −120 bp region of the promoter. Using EGFR-null CHO and NR-6 cells, we found that reexpression of EGFR and expression of constitutive STAT3 (STAT3CA) activated the TWIST-120 promoter (Fig. 4B). Coexpression of EGFR and STAT3CA achieved the highest level of induction compared with single expression (Fig. 4B, left). In CHO-EGFR cells, STAT3CA significantly induced TWIST-120 promoter activation, whereas STAT3-DN reduced the promoter activity (Fig. 4B). Consistently, EGFR kinase inhibitors (Iressa and PD158780) and JAKs inhibitor (AG490) reduced the activity of the TWIST gene promoter. Furthermore, forced expression of dominant-negative STAT3 reduced TWIST expression (Fig. 4C, left). Consistently, STAT3 expression knockdown by STAT3 small interfering RNA, but not by the control small interfering RNA, reduced TWIST expression (Fig. 4E, right). Together, these data indicate an important role of STAT3 in EGFR-mediated TWIST gene expression.
Figure 4.
EGFR and STAT3 activate the human TWIST gene promoter. The human TWIST promoter-driven luciferase constructs were engineered to contain 824, 604, and 120 bp of the promoter and designated as phTWIST-824, phTWIST-604, and phTWIST-120, respectively. All transfection and determination of luciferase activity were carried out as previously described (24). Relative luciferase activity was derived from firefly luciferase activity after normalization against the activity of the transfection efficiency control, Recilla luciferase. All data represent the mean and SD from at least three independent experiments. A, EGFR activation by various ligands induces the TWIST gene promoter. MDA-MB-468 cells in six-well culture plates were transfected with phTWIST-824, phTWIST-604, and phTWIST-120. Recilla luciferase construct was cotransfected as transfection controls. After 24 h, transfected cells were serum-starved for 20 h and stimulated with 100 ng/mL EGF, TGF-α, and HB-EGF for 4 h. Harvested cells were lysed and subjected to luciferase assay. B, expression of EGFR and constitutive STAT3 (STAT3CA) activate the TWIST promoter. EGFR-null CHO-NEO cells (left) were cotransfected with phTWIST-120 and expression plasmids, pEGFR, pSTAT3CA, or combination. EGFR-negative NR-6 cells (middle), rat fibroblasts, were cotransfected with pEGFR and pSTAT3CA. After 48 h, transfected cells were lysed and luciferase activities determined. Right, CHO-EGFR cells were cotransfected with phTWIST-120 and indicated plasmids (pSTAT3CA and pSTAT3-DN). At 48 h, serum-starved cells were stimulated with EGF (100 ng/mL) for 4 h. Additionally, aliquots of CHO-EGFR cells were transfected with phTWIST-120, serum-starved, and pretreated with EGFR inhibitors (Iressa, 5 μmol/L; PD158780/PD, 10 μmol/L) and Jak/STAT3 inhibitor (AG490, 10 μmol/L) before 4 h of EGF stimulation. Control phTWIST-120 transfected cells were treated with EGF (Mock) for 4 h before analysis for luciferase activity. C, forced expression of dominant-negative STAT3 (STAT3-DN) and STAT3 small interfering RNA reduced TWIST expression. Left, MDA-MB-468 cells were transfected with control and STAT3-DN vectors and, 48 h later, harvested and subjected to Western blot analysis for TWIST and α-tubulin expression. Right, Forced expression of STAT3 small interfering RNA reduced TWIST expression. MDA-MB-468 cells were transfected with control small interfering RNA [nonspecific (N.S.), small interfering RNA, and STAT3 small interfering RNA]. After 48 h, transfected cells were harvested and subjected to Western blot analysis for STAT3, TWIST, and β-actin expression. D, involvement of nuclear EGFR in EGF-responsiveness of the human TWIST gene promoter. CHO-NEO, CHO-EGFR, CHO-EGFR-NLS cells were transfected with phTWIST-120, serum-starved at 24 h posttransfection, and treated with 100 ng/mL EGF and TGF-α for 4 h. Harvested cells were lysed and subjected to luciferase assay.
As we recently found that nuclear EGFR contains transcriptional activity and activates expression of cyclin D1, iNOS, and B-Myb (24, 28, 32), we then examine the involvement of nuclear EGFR in EGF-induced TWIST gene activation. Using isogenic lines CHO-NEO, CHO-EGFR, CHO-EGFR-NLS (CHO with the nuclear entry—defective EGFR), we found that EGFR-NLS mutant displayed a constitutive TWIST promoter activity but failed to respond to EGF (Fig. 4D). Although the EGF responsiveness of EGFR-NLS/CHO seems to favor that nuclear localization of EGFR might be involved in the STAT-mediated TWIST up-regulation. This notion is not supported by the fact that EGFR-NLS can constitutively activate TWIST promoter (Fig. 4D). Thus, further investigation is required for a role of direct nuclear EGFR in the TWIST up-regulation.
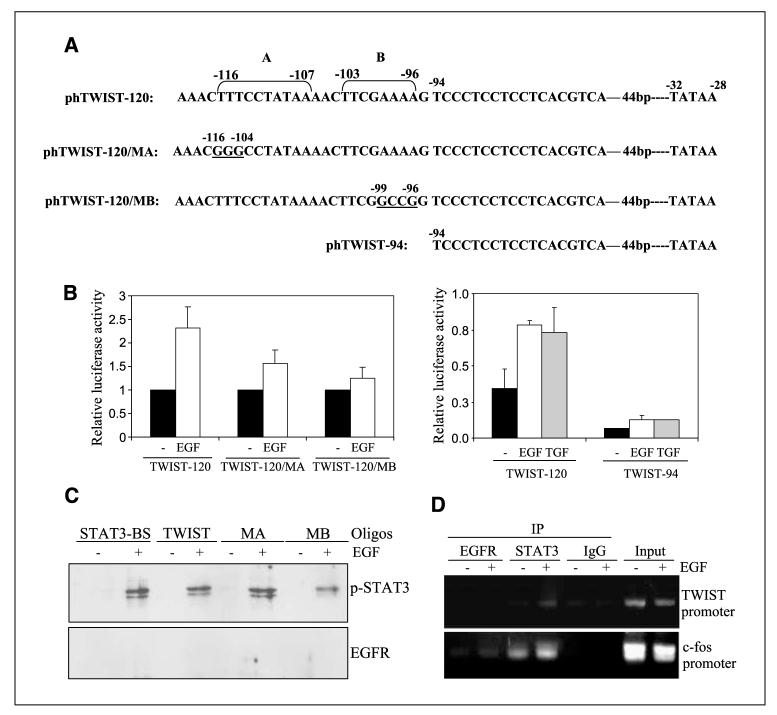
Identification of STAT3-targeted region within the human TWIST promoter
We found, thus far, that STAT3 is involved in EGFR-induced TWIST gene activation and that a 120-promoter region may be important in this involvement (Fig. 4). Next, we asked whether the TWIST gene promoter contains STAT3-binding sites. As illustrated in Fig. 5A, the proximal region (−120 bp) of the human TWIST promoter contains two putative STAT3-binding sites. A TATAA box is located at −32 to −28 bp, relative to the transcription start site. To analyze the functionality of the putative STAT3-binding sites A (−116 to −107) and B (−103 to −96), we did site-directed mutagenesis to generate the phTWIST-120/MA mutant with nucleotide substitutions (underlined) at −116 to −104 bp region and the phTWIST-120/MB mutant with nucleotide changes (underlined) at nt −99 to −96. Additionally, the pTWIST-94 was generated to remove the putative STAT3-binding sites and thus only contains the minimal promoter up to −94 bp. Using these reporters, we found that mutation at either putative STAT3-binding site, A or B, reduced but did not abolish the ability of the human TWIST promoter to respond to EGFR ligands in MDA-MB-468 cells (Fig. 5B, left). Deletion of the STAT3-binding sites reduced its basal level of promoter activity and rendered EGF-irresponsiveness of the TWIST promoter, as indicated by the failure of the phTWIST-94 construct to respond to EGF/TGF-α (Fig. 5B, right).
Figure 5.
Identification of STAT3-targeted region within the human TWIST promoter. A, schematic illustration of the proximal region of the human TWIST promoter. The human TWIST proximal promoter contains two putative STAT3-binding elements. A TATAA box is located at nt −32 to −28, relative to the transcription start site. Site-directed mutagenesis was done to generate the phTWIST-120/MA mutant that contains multiple nucleotide substitutions (underlined) at nt −116 to −107 region and the phTWIST-120/MB mutant with nucleotide changes (underlined) at nt −99 to −96. The pTWIST-94 was additionally generated to remove the putative STAT3-binding sites and thus contains the minimal promoter up to −94 bp. B, mutation at the putative STAT3-binding site II significantly reduced the ability of the human TWIST promoter to respond to EGFR ligands. MDA-MB-468 cells were transfected with phTWIST-120, phTWIST-94, phTWIST-120/MA, and phTWIST-120/MB as previously described. After 48 h, serum-starved transfected cells were stimulated with EGF (100 ng/mL) for 4 h before determination of luciferase activities. All date represent the mean and SD from three independent experiments. C, biotinylated oligonucleotides precipitation assay. These studies were done to determine the degree of the binding of STAT3 to the TWIST promoter fragments. MDA-MB-468 cells untreated and treated with EGF (100 ng/mL) for 1 h were harvested, and nuclear lysates were extracted. Nuclear extracts were then subject to binding affinity evaluation to a number of biotinylated oligonucleotides, namely, STAT3-BS/APRE (30, 31), TWIST-120/TWIST, TWIST-120/MA, and TWIST-120/MB. Biotinylated oligos were then precipitated by avidin beads, washed and subjected to Western blot analysis for p-STAT3 and EGFR. D, nuclear STAT3, but not nuclear EGFR, binds to the human TWIST gene promoter. A431 cells were serum-starved and stimulated without and with EGF (100 ng/mL) for 30 min and subjected to the in vivo binding assay ChIP, as we previously described. Briefly, EGFR monoclonal antibody (Neomarkers, Ab13) and STAT3 polyclonal antibody (Santa Cruz, C-20) were used in immunoprecipitation. IgG was used as negative control for immunoprecipitation, whereas input chromatins were used as positive controls for PCR and for equal loading. The c-fos promoter was also amplified as a positive control for both EGFR and STAT3 binding.
We further examined the ability of p-STAT3 to bind to the putative STAT3-binding site within the human TWIST promoter using the biotinylated oligonucletide precipitation/oligo pull-down assay. As indicated in Fig. 5C, EGF activated the binding of p-STAT3 to the consensus STAT3-binding site (STAT3-BS/APRE; refs. 30, 31), the putative STAT3-binding site in the TWIST promoter, the TWIST-120/MA and TWIST-120/MB oligonucleotides. Consistent with low EGF responsiveness of the TWIST-120/MB promoter (Fig. 5B, left), the TWIST-120/MB fragment was found to contain lowest binding affinity to p-STAT3 (Fig. 5C). We did not detect binding of nuclear EGFR to any biotinylated oligonucleotides that we tested. Furthermore, we examined whether nuclear STAT3 binds to the TWIST gene promoter using the in vivo protein-DNA binding chromatin immunoprecipitation (ChIP) assay, and the results are shown in Fig. 5D. Upon EGF stimulation, nuclear STAT3 binds to the human TWIST gene promoter. In contrast, nuclear EGFR did not associate with the TWIST promoter at a detectable level, consistent with the lack of EGFR binding observed in Fig. 5C. As expected, both nuclear EGFR and STAT3 bind to the c-fos promoter in an EGF-dependent fashion, as we previously reported (24). The IgG was used in immunoprecipitation as negative controls and did not yield any band signals, indicating the assay specificity. Input chromatins were also used in these assays to indicate that equal amounts of cell lysates were used from the −EGF and +EGF treatment groups. In summary, we identified and functionally characterized the STAT3-targeted region within the human TWIST promoter.
Positive correlations between EGFR/p-STAT3 and TWIST in a cohort of primary breast carcinomas
We revealed using cultured cells that EGFR activates the human TWIST gene expression via activation of STAT3. Next, we aimed to examine whether such regulation also exists in the primary tumor specimens from cancer patients. To this end, we analyzed TWIST expression via immunochemical staining analysis in a cohort of primary breast carcinomas that have been previously immunostained for EGFR and p-STAT3 (24, 26). Two pathologists independently viewed and scored all slides. All statistical analyses were done using Statistica 6.0 software. Regression analysis indicated a positive correlation between non-nuclear EGFR and TWIST (P = 0.01). Levels of nuclear EGFR do not significantly correlate with those of TWIST (P = 0.6), which is in agreement with the findings in Fig. 5C and D. To further investigate whether p-STAT3 correlates with TWIST expression, we immunostained the same cohort of tumors for p-STAT3 and analyzed the correlation between p-STAT3 and TWIST. We found that levels of TWIST correlates significantly with those of p-STAT3 (R = 0.28, P = 0.0013; Fig. 6C, left). In agreement with the role of EGFR as an upstream activator of STAT3, we found a significant, positive correlation between non-nuclear EGFR and p-STAT3 (R = 0.35, P = 0.00003; Fig. 6C, right). Furthermore, Fig. 6D shows two representative tumors in which the upper case contains high EGFR/p-STAT3/ TWIST and the lower tumor contains low EGFR/p-STAT3/TWIST. Together, we reported here that levels of EGFR and p-STAT3 correlate positively with those of the TWIST gene in breast carcinomas.
Figure 6.
Positive correlations between non-nuclear EGFR/p-STAT3 and TWIST in a cohort of primary breast carcinomas. The cohort of primary breast carcinoma specimens, previously stained for EGFR (26), was analyzed for p-STAT3 (Y705) and TWIST expression via immunochemical staining analysis. All slides were independently viewed and scored by two pathologists. When the scoring discrepancy is >10%, slides were reevaluated and reconciled by two pathologists on a two-headed microscope. All statistical analyses were done using Statistica 6.0 software. A, positive correlation between non-nuclear EGFR and TWIST. Levels of TWIST were correlated with those of non-nuclear EGFR (P = 0.01). Regression analysis was done in these analyses. B, lack of correlation between levels of nuclear EGFR and TWIST (P = 0.6). Regression analysis was similarly done as in A. C, expression of p-STAT3 correlates with those of TWIST and non-nuclear EGFR. Regression analysis was done to determine R and P values. D, representative tumors immunostained for EGFR, p-STAT3, and TWIST. Top, tumor was stained strongly for EGFR (left), p-STAT3 (middle), and TWIST (right). Bottom, tumor with negative/low expression for all three proteins.
Discussion
Metastasis is a major obstacle for cancer therapy and is a primary cause of mortality in many cancers, including that of the breast. Understanding the biology of cancer cells with high metastatic potential is, therefore, important in identifying tumors that are likely to undergo metastasis and in improving current anticancer therapy. It has been shown that cancers with the deregulated EGFR pathway possess a high likelihood for local invasion and subsequent metastasis. The specific involvement of EGFR in EMT, an event that takes place during the early stage of tumor invasion, intravasation, and subsequent metastasis to the distant organ sites, remains elusive. The current study was thus undertaken to examine the role of aberrant EGFR pathway in EMT. Here, we report that cancer cells with high EGFR expression/activity undergo EGF/TGF-α—induced EMT, and this phenotypic transition involves EGFR-mediated activation of STAT3 and subsequent STAT3-activated TWIST gene expression.
TWIST is a bHLH transcription factor that has been known as an essential player for proper gastrulation mesoderm formation and neural crest migration (15). More recently, TWIST has been found to express at high levels in a number of human tumors, including those of the breast, prostate, esophagus, lung, uterus, skin, liver, and brain (16–19, 44–46). Importantly, increased TWIST expression is associated with breast cancer metastasis to the lung and increased EMT and intravasation (11). Correlative studies further indicate an association of high TWIST expression with invasion and therapeutic response in other cancer types (17–19, 44, 47–49). Despite frequent reports of TWIST overexpression in human cancers, transcriptional regulation of the human TWIST genes is largely unknown. Moreover, regulation of the mouse Twist genes have been investigated to involve tumor necrosis factor-α/nuclear factor κB (NF-κB; ref. 50) and Wnt1/TCF/β-catenin pathways (51). However, the NF-κB and TCF/β-catenin response elements found in the mouse Twist gene promoters are not present in that of the human TWIST gene. A recent study reported that STAT3 knockdown of mouse 4T1 mammary tumor cells led to altered expression of several genes, including Twist (20). Twist has yet been shown to be a direct transcriptional target of STAT3. We did an extensive search, using TF Search and TESS transcription factor search sites, for the STAT-binding sites within the mouse twist gene promoter and did not find a putative site, suggesting its expression reduction by STAT3 small interfering RNA was likely due to indirect effects. The current study showed that EGF activated STAT3 sites in the human TWIST promoter and regulates its transcription. Because mouse TWIST promoter does not contain STAT consensus site, this raises an interesting question that the two species may use STAT to regulate TWIST expression through different molecular mechanisms.
The current report strongly supports the notion that EGFR and STAT3 oncoproteins interplay and regulate expression of a series of genes that are involved in aggressive cancer biology. Such regulation, however, seems to be highly complex and not yet fully understood. In the case of TWIST, this study indicates that STAT3 plays a direct transcriptional role by binding to the TWIST promoter and that EGFR seems to be an upstream regulator that phosphorylates and activates STAT3. Although the responsiveness of EGF of EGFR-NLS/CHO seems to favor that nuclear localization of EGFR might be involved in the STAT3-mediated TWIST up-regulation. This notion is not supported by the fact that EGFR-NLS can constitutively activate the TWIST promoter. Furthermore, nuclear EGFR does not seem to interact directly with the TWIST promoter, as indicated by the ChIP and oligo pull-down assays. IHC studies showed no significant correlation between nuclear EGFR and TWIST. Together, these evidences suggest that a role of direct nuclear EGFR in the TWIST up-regulation is unlikely. For iNOS, nuclear EGFR and STAT3 behave as transcriptional coregulators as the EGFR/STAT3 complex associated with the iNOS promoter (24). In the case of cyclin D1, nuclear EGFR seems to bind to the promoter independent of STAT3 (24). On the other hand, STAT3 can be activated by other upstream regulators, independent of EGF/EGFR, and regulates expression of Myc (52) and p21WAF1/CIP1 (53). Further investigations are indeed needed to further our understanding of EGFR-mediated and STAT-mediated gene regulation and its effect in the biology of cancer cells.
We report here that the human TWIST gene is directly up-regulated by the oncoprotein STAT3 in cancer cells with high levels of EGFR. This correlation was also found in primary breast carcinomas. Given TWIST’s function in promoting EMT, EGFR cooperates with STAT3 to induce TWIST expression leading to EMT. In this context, we observed that chronic EGF/TGF-α exposure promotes EMT in breast and pancreatic cancer cells with high EGFR levels. This finding is supported by the observation that prolonged EGF stimulation leads to loss of E-cadherin and increased invasion in A431 human epidermoid carcinoma cells (10). Consistent with our findings, tumors with high EGFR and constitutively activated STAT3 contain high potentials to undergo metastasis (1–5). Stat3 controls cell movement in Zebrafish gastrulation via increasing Zinc transporter LIV1 expression (54, 55). In line with our observations, STAT3 knockdown in mouse breast cancer cell line leads to reduced expression of Twist (20). Together, our results and those from other studies highlight an important role of EGFR and STAT3 in promoting cancer EMT via TWIST. Our findings provide important insights into the understanding of the malignant biology of tumors with deregulated EGFR and STAT3 pathways and establish new rationales for using anti-EGFR and anti-STAT3 strategies to target aggressive invasive tumors.
Acknowledgments
Grant support: NIH grants RO1 109311, P20 CA101936 (MDACC Pancreatic Cancer Specialized Programs of Research Excellence), P50 CA83639 (MDACC Ovarian Cancer Specialized Programs of Research Excellence), and P50 CA116199 (MDACC Breast Cancer Specialized Programs of Research Excellence), Department of Defense Center of Excellence W81WXH-06-2-0033, National Breast Cancer Foundation, Inc. (M.-C. Hung), Cancer Center support grant CA16672, NIH grant 1K01-CA118423-01 (H.-W. Lo), American Cancer Society grant PF TBE-109873, Wendy Will Case Cancer Fund, Elsa U. Pardee Foundation grant (H.-W. Lo), Department of Defense grant W81XWH-07-0390 (H.-W. Lo), and NIH Duke Comprehensive Cancer Center Core grant 5P30 CA14236.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Verbeek BS, Adriaansen-Slot SS, Vroom TM, Beckers T, Rijksen G. Overexpression of EGFR and c-erbB2 causes enhanced cell migration in human breast cancer cells and NIH3T3 fibroblasts. FEBS Lett. 1998;425:145–50. doi: 10.1016/s0014-5793(98)00224-5. [DOI] [PubMed] [Google Scholar]
- 2.Bruns CJ, Solorzano CC, Harbison MT, et al. Blockade of the epidermal growth factor receptor signaling by a novel tyrosine kinase inhibitor leads to apoptosis of endothelial cells and therapy of human pancreatic carcinoma. Cancer Res. 2000;60:2926–35. [PubMed] [Google Scholar]
- 3.Matsuo M, Sakurai H, Saiki I. ZD1839, a selective epidermal growth factor receptor tyrosine kinase inhibitor, shows antimetastatic activity using a hepatocellular carcinoma model. Mol Cancer Ther. 2003;2:557–61. [PubMed] [Google Scholar]
- 4.Weber KL, Doucet M, Price JE, et al. Blockade of epidermal growth factor receptor signaling leads to inhibition of renal cell carcinoma growth in the bone of nude mice. Cancer Res. 2003;63:2940–7. [PubMed] [Google Scholar]
- 5.Shintani S, Li C, Mihara M, Nakashiro K, Hamakawa H. Gefitinib (‘Iressa’), an epidermal growth factor receptor tyrosine kinase inhibitor, mediates the inhibition of lymph node metastasis in oral cancer cells. Cancer Lett. 2003;201:149–55. doi: 10.1016/s0304-3835(03)00464-6. [DOI] [PubMed] [Google Scholar]
- 6.Kruger JS, Reddy KB. Distinct mechanisms mediate the initial and sustained phases of cell migration in epidermal growth factor receptor-overexpressing cells. Mol Cancer Res. 2003;1:801–9. [PubMed] [Google Scholar]
- 7.Thomas SM, Coppelli FM, Wells A, et al. Epidermal growth factor receptor-stimulated activation of phospholipase Cγ-1 promotes invasion of head and neck squamous cell carcinoma. Cancer Res. 2003;63:5629–35. [PubMed] [Google Scholar]
- 8.Ellerbroek SM, Halbleib JM, Benavidez M, et al. Phosphatidylinositol 3-kinase activity in epidermal growth factor-stimulated matrix metalloproteinase-9 production and cell surface association. Cancer Res. 2001;61:1855–61. [PubMed] [Google Scholar]
- 9.Kondapaka SB, Fridman R, Reddy KB. Epidermal growth factor and amphiregulin up-regulate matrix metalloproteinase-9 (MMP-9) in human breast cancer cells. Int J Cancer. 1997;70:722–6. doi: 10.1002/(sici)1097-0215(19970317)70:6<722::aid-ijc15>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 10.Lu Z, Ghosh S, Wang Z, Hunter T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of β-catenin, and enhanced tumor cell invasion. Cancer Cell. 2003;4:499–515. doi: 10.1016/s1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Vernon AE, LaBonne C. Tumor metastasis: a new twist on epithelial-mesenchymal transitions. Curr Biol. 2004;14:R719–21. doi: 10.1016/j.cub.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 13.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–8. [PubMed] [Google Scholar]
- 14.Cano A, Perez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–99. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 16.Kwok WK, Ling MT, Lee TW, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–62. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 17.Kyo S, Sakaguchi J, Ohno S, et al. High Twist expression is involved in infiltrative endometrial cancer and affects patient survival. Hum Pathol. 2006;37:431–8. doi: 10.1016/j.humpath.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 18.Yuen HF, Chan YP, Wong ML, et al. Up-regulation of TWIST in Oesophageal Squamous Cell Carcinoma is associated with neoplastic transformation and distant metastasis. J Clin Pathol. 2007;60:510–4. doi: 10.1136/jcp.2006.039099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elias MC, Tozer KR, Silber JR, et al. TWIST is expressed in human gliomas and promotes invasion. Neoplasia. 2005;7:824–37. doi: 10.1593/neo.04352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling X, Arlinghaus RB. Knockdown of STAT3 expression by RNA interference inhibits the induction of breast tumors in immunocompetent mice. Cancer Res. 2005;65:2532–6. doi: 10.1158/0008-5472.CAN-04-2425. [DOI] [PubMed] [Google Scholar]
- 21.Niu G, Wright KL, Huang M, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–8. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 22.Wei D, Le X, Zheng L, et al. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–29. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- 23.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 24.Lo H-W, Hsu S-C, Ali-Seyed M, et al. Nuclear Interaction of EGFR and STAT3 in the Activation of iNOS/NO Pathway. Cancer Cell. 2005;7:575–89. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Zou Y, Peng H, Zhou B, et al. Systemic Tumor Suppression by the Proapoptotic Gene bik. Cancer Res. 2002;62:8–12. [PubMed] [Google Scholar]
- 26.Lo H-W, Xia W, Wei Y, et al. Novel Prognostic Value of Nuclear EGF Receptor in Breast Cancer. Cancer Res. 2005;65:338–48. [PubMed] [Google Scholar]
- 27.Lee CM, Lo HW, Shao RP, et al. Selective activation of ceruloplasmin promoter in ovarian tumors: potential use for gene therapy. Cancer Res. 2004;64:1788–93. doi: 10.1158/0008-5472.can-03-2551. [DOI] [PubMed] [Google Scholar]
- 28.Hanada N, Lo HW, Day CP, et al. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–7. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 29.Hata A, Seoane J, Lagna G, et al. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell. 2000;100:229–40. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 30.Wegenka UM, Buschmann J, Lutticken C, Heinrich PC, Horn F. Acute-phase response factor, a nuclear factor binding to acute-phase response elements, is rapidly activated by interleukin-6 at the posttranslational level. Mol Cell Biol. 1993;13:276–88. doi: 10.1128/mcb.13.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D, Sun M, Samols D, Kushner I. STAT3 participates in transcriptional activation of the C-reactive protein gene by interleukin-6. J Biol Chem. 1996;271:9503–9. doi: 10.1074/jbc.271.16.9503. [DOI] [PubMed] [Google Scholar]
- 32.Lin SY, Makino K, Xia W, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–8. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 33.Camp RL, Rimm EB, Rimm DL. Met expression is associated with poor outcome in patients with axillary lymph node negative breast carcinoma. Cancer. 1999;86:2259–65. doi: 10.1002/(sici)1097-0142(19991201)86:11<2259::aid-cncr13>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Tai P, Yu E, Vinh-Hung V, Cserni G, Vlastos G. Survival of patients with metastatic breast cancer: twenty-year data from two SEER registries. BMC Cancer. 2004;4:60. doi: 10.1186/1471-2407-4-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gill GN, Kawamoto T, Cochet C, et al. Monoclonal anti-epidermal growth factor receptor antibodies which are inhibitors of epidermal growth factor binding and antagonists of epidermal growth factor binding and antagonists of epidermal growth factor-stimulated tyrosine protein kinase activity. J Biol Chem. 1984;259:7755–60. [PubMed] [Google Scholar]
- 36.Wiley HS. Anomalous binding of epidermal growth factor to A431 cells is due to the effect of high receptor densities and a saturable endocytic system. J Cell Biol. 1988;107:801–10. doi: 10.1083/jcb.107.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiyokawa N, Lee EK, Karunagaran D, Lin S-Y, Hung M-C. Mitosis-specific Negative Regulation of Epidermal Growth Factor Receptor, Triggered by a Decrease in Ligand Binding and Dimerization, Can Be Overcome by Overexpression of Receptor. J Biol Chem. 1997;272:18656–65. doi: 10.1074/jbc.272.30.18656. [DOI] [PubMed] [Google Scholar]
- 38.Jayne DG, Perry SL, Morrison E, Farmery SM, Guillou PJ. Activated mesothelial cells produce heparin-binding growth factors: implications for tumour metastases. Br J Cancer. 2000;82:1233–8. doi: 10.1054/bjoc.1999.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darnell JE, Jr., Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–21. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 40.Fu XY. From PTK-STAT signaling to caspase expression and apoptosis induction. Cell Death Differ. 1999;6:1201–8. doi: 10.1038/sj.cdd.4400613. [DOI] [PubMed] [Google Scholar]
- 41.Park OK, Schaefer TS, Nathans D. In vitro activation of Stat3 by epidermal growth factor receptor kinase. Proc Natl Acad Sci USA. 1996;93:13704–8. doi: 10.1073/pnas.93.24.13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 43.Garcia R, Bowman TL, Niu G, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 44.Thomson S, Buck E, Petti F, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–62. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 45.Lee TK, Poon RTP, Yuen AP, et al. Twist Over-expression Correlates with Hepatocellular Carcinoma Metastasis through Induction of Epithelial-Mesenchymal Transition. Clin Cancer Res. 2006;12:5369–76. doi: 10.1158/1078-0432.CCR-05-2722. [DOI] [PubMed] [Google Scholar]
- 46.Hoek K, Rimm DL, Williams KR, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–82. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 47.Yauch RL, Januario T, Eberhard DA, et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res. 2005;11:8686–98. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Ling MT, Guan XY, et al. Identification of a novel function of TWIST, a bHLH protein, in the development of acquired taxol resistance in human cancer cells. Oncogene. 2004;23:474–82. doi: 10.1038/sj.onc.1207128. [DOI] [PubMed] [Google Scholar]
- 49.Diaz N, Minton S, Cox C, et al. Activation of stat3 in primary tumors from high-risk breast cancer patients is associated with elevated levels of activated SRC and survivin expression. Clin Cancer Res. 2006;12:20–8. doi: 10.1158/1078-0432.CCR-04-1749. [DOI] [PubMed] [Google Scholar]
- 50.Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-κB activity. Cell. 2003;112:169–80. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 51.Howe LR, Watanabe O, Leonard J, Brown AM. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 2003;63:1906–13. [PubMed] [Google Scholar]
- 52.Bowman T, Broome MA, Sinibaldi D, et al. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci USA. 2001;98:7319–24. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinibaldi D, Wharton W, Turkson J, et al. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: role of activated STAT3 signaling. Oncogene. 2000;19:5419–27. doi: 10.1038/sj.onc.1203947. [DOI] [PubMed] [Google Scholar]
- 54.Yamashita S, Miyagi C, Carmany-Rampey A, et al. Stat3 Controls Cell Movements during Zebrafish Gastrulation. Dev Cell. 2002;2:363–75. doi: 10.1016/s1534-5807(02)00126-0. [DOI] [PubMed] [Google Scholar]
- 55.Yamashita S, Miyagi C, Fukada T, et al. Zinc transporter LIVI controls epithelial-mesenchymal transition in zebrafish gastrula organizer. Nature. 2004;429:298–302. doi: 10.1038/nature02545. [DOI] [PubMed] [Google Scholar]